Abstract
Breast cancer remains the leading cause of cancer mortality in females, and about 70% of the primary breast cancer patients are diagnosed ERα-positive, which is the most common type of breast cancer. MicroRNA-34a (miR-34a) has been shown to be a master regulator of tumor suppression in many types of cancers including breast cancer. However, the role of miR-34a in ERα-positive breast cancer has not been elucidated. Here, we find that in MCF-7, which is an ERα-positive breast cancer cell line, miR-34a is remarkably downregulated after E2 treatment. Overexpression of miR-34a by lentivirus suppresses cell proliferation, S phase ratio, and tumor formation in an E2-dependent manner in vitro. According to the mRNA sequence, lemur tyrosine kinase 3 (LMTK3), which is an important regulator of estrogen receptor alpha (ERα), is a predicted target of miR-34a. This is confirmed by dual luciferase reporter assay and the decrease of LMTK3 mRNA and protein levels after overexpression of miR-34a. Moreover, miR-34a overexpression decreases AKT signaling pathway and increases ERα phosphorylation status. Taken together, these results suggest that miR-34a inhibits breast cancer proliferation by targeting LMTK3 and might be used as an anti-ERα agent in breast cancer therapy.
Introduction
With an annual incidence of 1.3 million cases worldwide and 465,000 deaths, breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death in females (Stebbing et al., 2013). Approximately 70% of primary breast cancer patients were positive for estrogen receptor alpha (ERα) at diagnosis, and the presence of ERα appeared to contribute to endocrine resistance by regulating the transcriptional activity and stability of estrogen-regulated genes (Wu et al., 2004; Jiang et al., 2007). ERα seems to contribute to tumorigenesis primarily by activating MAPK (ERK1/2) and/or AKT signaling pathways (Thomas and Gustafsson, 2011). Although endocrine therapies are designed to limit estrogen biosynthesis or inhibit aromatase activity, a number of the patients with primary breast cancer and most of those presenting with recurrence acquire resistance during the course of treatment are with tumor progression and eventual death. A better understanding of acquired resistance is important to find the new treatment of endometrial carcinoma.
MicroRNAs (miRNAs) are 21 and 22-nucleotide single-stranded noncoding RNAs and regulate gene expression via base-pairing with complementary sequences within 3′-UTR (untranslated region) of target mRNAs in the post-transcriptional stages (Eulalio et al., 2008; Winter et al., 2009). Mature miRNAs, target mRNAs, and many other associated proteins compose into the RNA-induced silencing complex, which mediates gene silencing. MiRNAs have been shown to play an important role in a wide variety of biological processes such as apoptosis, development, aging, and cancer. There is also a wealth of evidence to suggest that miRNAs are involved in the initiation, development, and metastasis of cancers. Hsa-miR-34 comprises three family members: miR-34a, miR-34b, and miR-34c in which miR-34a accounts for the majority of total miR-34. The miR-34a gene is located on chromosome 1p36.22, and miR-34b and miR-34c are expressed from a polycistronic transcript encoded on chromosome11q23.1. Previous reports have already shown that miR-34a was downregulated in many types of malignancies, including cancers of the lung, breast, kidney, bladder, brain, skin, cervix, and lymph system (Bader, 2012); these studies indicated that miR-34a behaves as a tumor-suppressor by regulating many cellular processes, such as differentiation, proliferation, apoptosis, cellular transformation, carcinogenesis, and metastasis (Bader, 2012).
Recent work described that miR-34 suppresses invasion and metastasis of breast cancer by directly targeting Fra-1 (Yang et al., 2012); other reports have shown that p53/miRNA-34 axis regulates snail-dependent epithelial–mesenchymal transition in tumorigenesis (Kim et al., 2011). Ectopic miR-34 expression was shown to induce apoptosis, cell-cycle arrest, and senescence (Hermeking, 2010). Interestingly, antagonizing miR-34a increases the radiation sensitivity of breast cancer cells (O'Day and Lal, 2010). In triple-negative breast cancer, which is characterized mainly by ERα, PgR, and HER2 absence, miR-34b, but not miR-34a or miR-34c was reported to be negative correlated with disease-free survival and an overall survival (Svoboda et al., 2012). A microarray conducted in breast cancer has shown that miR-34a was significantly downregulated in DCIS and IDC compared with benign tissue (Yang et al., 2012). However, the role of miR-34a in breast cancer still needs to be further elucidated. In this study, we have found that E2 treatment can dramatically downregulate the expression level of miR-34a. And we proved that lemur tyrosine kinase 3 (LMTK3) might be a target of miR-34a and overexpression of miR-34a would inhibit cell proliferation, S phase ratio, and tumor formation in an E2-dependent manner in breast cancer cell line MCF-7. Besides this, overexpression of miR-34a also inhibits AKT signaling pathway and increases ERα phosphorylation status. Therefore, our results revealed the potential medicinal value of miR-34a in ERα-positive breast cancer.
Materials and Methods
Plasmids construction
pLL3.7-miR-34a: The fragment including the pre-miR-34a sequence (http://mirbase.org/) plus 198 bp at both 5′-and 3′-flanking regions (chr1:9211529:9212034: -1) was PCR-amplified from human genomic DNA and cloned into pLL3.7 with HpaI/Xhol.
psiCHECK™-2-LMTK3-3′UTR: The full length 3′UTR of LMTK3 (Genbank Accession: NM_001080434.1) was PCR-amplified from human genomic DNA and cloned into psiCHECK-2 dual-luciferase reporter plasmid immediately downstream of the stop codon of the Renilla luciferase gene with Xhol/NotI.
psiCHECK-2-LMTK3-3′UTR-mut: Three nucleotides of 3′UTR of LMTK3, which are the perfect binding sites with miR-34a seed sequence, was mutated at the position of 76–78, from CACTGCC to CAgacCC by site-directed PCR mutation.
psiCHECK-2-LMTK3-3′UTR-re: The reverse orientation sequence of wild-type LMTK3-3′UTR was cloned into psiCHECK-2 vector within the same site between Xhol/NotI, serving as a negative control.
pcDNA3.1(+)-LMTK3-CDS: The coding sequence of LMTK3 was PCR-amplified from human cDNA and cloned into pcDNA3.1(+) with HindIII/EcoRI.
All successful constructed plasmids were confirmed by DNA sequencing.
Cell culture, infection, and transfection
Human breast cancer MCF-7 cells and human embryonic kidney HEK-293T cells were purchased from the Chinese Academy of Science and cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Gibco BRL) in a humidified atmosphere (5% CO2; 37°C). A final concentration of 10 nM E2 (Sigma) was added into cultured cells 24 h before future analysis for E2 treatment.
To establish stable miR-34a-expressing cells, pLL3.7-miR-34a lentiviruses were generated and used to infect MCF-7 cells as described previously (Jia et al., 2011). Infection efficiency was monitored by the expression of GFP with fluorescent microscopy. The cells were sorted 72 h post infection on a FACS Canto (BD Biosciences) on the basis of GFP expression of the pLL3.7. Cells with stable expression of either pLL3.7 vector (empty vector, EV) or miR-34a were under sterile condition and as a stable model were used in following experiments.
Cell transfection was performed using Lipofectamine 2000 (Invitrogen). Cells were harvested at 48 h post transfection for protein analysis or luciferase activity assay.
RNA extraction and quantitative polymerase chain reaction
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen), and reverse transcription (RT) was performed according to the manufacturer's instructions (Invitrogen) and proceeded to real-time PCR with gene-specific primers in the presence of 2×SYBR Green Master Mix (DBI Bioscience). The relative abundance of mRNA was calculated by normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. Specific stem-loop RT primers were used for reverse transcription reaction of miR-34a. The RNU6 was an internal control for normalizing the expression of miR-34a. All primer information can be seen in Table 1. All reactions were carried out in triplicate and all experiments were performed three independent times.
Table 1.
Information on the Primers Used for Reverse Transcription, Real-Time Polymerase Chain Reaction, and Construction of Plasmids
| Primer name | Sequence (5′-3′ orientation) |
|---|---|
| MiR-34a- RT primer | 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA CAA CC-3′ |
| MiR-34a mimics | 5′- TGG CAG TGT CTT AGC TGG TTG T -3′ |
| Mimics control | 5′-AGT GTG AGT TCT ACC ATT GCC AAA-3′ |
| MiR-34a inhibitor | 5′-ACA ACC AGC TAA GAC ACT GCC A-3′ (2′Ome) |
| MiR-34a -F | 5′-CGG GCT GGC AGT GTC TT-3′ |
| MiR-34a -R | 5′-GTG CAG GGT CCG AGG T-3′ |
| MiR-223 | 5′-TGT CAG TTT CTC AAA TAA ACC CCA-3′ |
| RNU6- RT | 5′-AAA ATA TGG AAC GCT-3′ |
| RNU6-F | 5′-CTC GCT TCG GCA GCA CA-3′ |
| RNU6-R | 5′-AAC GCT TCA CGA ATT TGC GT-3′ |
| GAPDH-F | 5′-AGG GCA TCT TGG GCT ACA C-3′ |
| GAPDH-R | 5′-TGG TCC AGG GTT TCT TAC TCC-3′ |
| lmtk3-F4172 | 5′-TCG ACC AGG AGA CGC C-3′ |
| lmtk3-R4329 | 5′-CTC GAA ACT GCC TCC AAA G-3′ |
| LMTK3-3'UTR-F-Xhol | 5′-AAA ACT CGA GAT TCC CCG AAG ACC CG-3′ |
| LMTK3-3'UTR-R-Not I | 5′-AAA AGC GGC CGC TTC CTC CAA AAA GTT GTT-3′ |
| LMTK3-CDS-F-Hind III | 5′-CCC CAA GCT TAT GAG GCA AGT GCT GTG G-3′ |
| LMTK3-CDS-R-EcoR I | 5′-AAA AGA ATT CTC AAT TCT CCA CGG GG-3′ |
The underlined letters are the endonuclease sites and the bases before these sites are protection bases.
RT, reverse transcription.
In vitro cell growth, viability, and cell cycle progression assay
For proliferation assay, after 6 h transfection, 3×103 cells were seeded in 96-well plates. CCK-8 (Cell Counting Kit-8) reagent was added at the time point of 24, 48, 72, 96, and 120 h after seeding and incubated at 37°C for half to 4 h according to the color change. The absorbance at 450 nm was measured by a microplate reader.
Cell viability was assayed by trypan blue staining. Cells were incubated with a final concentration of 0.04% (w/v) trypan blue solution at room temperature for 3 min and then directly observed using a microscope (Olympus). Cell viability (%)=(viable counts/total counts)×100%. Six random filed are chosen to count the amount of cells in each group.
Cell cycle analysis was detected using propidium iodide (PI) staining and flow cytometry. After treatment, cells were trypsinized, rinsed with PBS, fixed in 70% ethanol at 4°C overnight, and treated with RNaseA (0.02 mg/mL) in the dark at room temperature for 30 min. Then, cells were resuspended in 0.05 mg/mL PI. Cell cycle analysis was measured using a Cytomic FC 500 flow cytometer (Beckman Coulter) and analyzed using Modifit LT software.
Plate colony formation assay
Single cells were seeded at 2×103 cells/well in a 35 mm-diameter culture dish with complete medium for 14 days. The colonies were fixed with ethyl alcohol, stained with 0.5% crystal violet for 20 min, and washed thrice. A cluster with more than 50 cells was defined as one colony. The number of clones in 10 random view fields was counted under a light microscope (Olympus) and the average representing the 95% confident region was calculated. All experiments were performed in triplicate at least.
In vivo tumor growth
Female BALB/c athymic nude mice (5–6 weeks old) were purchased from Sino-British Sippr/Bk Lab Animal Ltd. and miR-34a or EV cells were injected mice skin under the front legs of same mouse for comparation and assigned EV group (right) or the miR-34a group (left). All experiments were carried out according to the NIH Guide for the Care and Use of Laboratory Animals and local institutional ethical guidelines for animal experiments. In total, 5×106 miR-34a- or EV-infected MCF-7 cells suspended in 150 μL sterile PBS were subcutaneously injected into the front legs of the mice. Tumor growth was measured every 3 days for 5 weeks once it became palpable (approximately 10 days after injection). Five weeks after inoculation, all the mice were sacrificed and the tumor volume (V) was determined by measuring the length (L) and width (W) with a caliper and using the formula V=1/2 (L×W2).
Luciferase reporter assay
For luciferase reporter assay, HEK-293T cells (3×103/well) were plated in a 96-well plate (Corning) for 24 h before transfection. Cells were co-transfected with 20 nM miRNA mimics of either miR-34a, miR-223, miR-34a inhibitor (synthesized by Genepharma Co. Ltd) or control of miR-34a inhibitor and 60 ng of psiCHECK-2-vector (Promega), psiCHECK-2-LMTK3 3′-UTR-wt, psiCHECK-2-LMTK3 3′-UTR-mut, or psiCHECK-2-re. After 48 h infection, cells were lysed and luciferase activity was measured using a dual-luciferase reporter assay system (catalog no.E1960; Promega) following the manufacturer's instructions. The luciferase activity was measured by a Lumat 9507 illuminometer (Berthold). Transfection efficiency was normalized to thymidine kinase-driven Renilla luciferase activity. All experiments were performed in triplicate at least.
Western blot
Cells were collected at logarithmic growth phase or 48 h post transfection. In brief, 50 μg of total protein was separated on a 10% running gel and transferred to a polyvinylidene difluoride membrane (Millipore). After blocking with 1% bovine serum albumin, the blots were incubated with antibodies from abcom and bioworld companies including against LMTK3 (ab110516), Ki67(ab58380), GAPDH (ab9485), ERα (ab37438), p-ERα (p-S167) (ab31478) ER, AKT (AP0059), p-AKT (p-S473) (BS4006), Cyclin D1 (BS1741), GSK3β (ab18893), and p-GSK3β (p-Ser9) (ab9769). After incubation with horseradish peroxidase-conjugated secondary antibody, protein bands were visualized using the Chemilucent Plus Western Enhancing Kit (Millipore). Band intensity was quantified using the Image-Pro Plus software.
Statistical analysis
Significant differences in tumor growth were assessed by Student's t-test. All results were expressed as mean±SD unless otherwise stated. The differences among the three groups were considered statistically significant when *p<0.05, **p<0.01. All experiments were performed in triplicate at least.
Results
E2 downregulated miR-34a expression in MCF-7 cell line
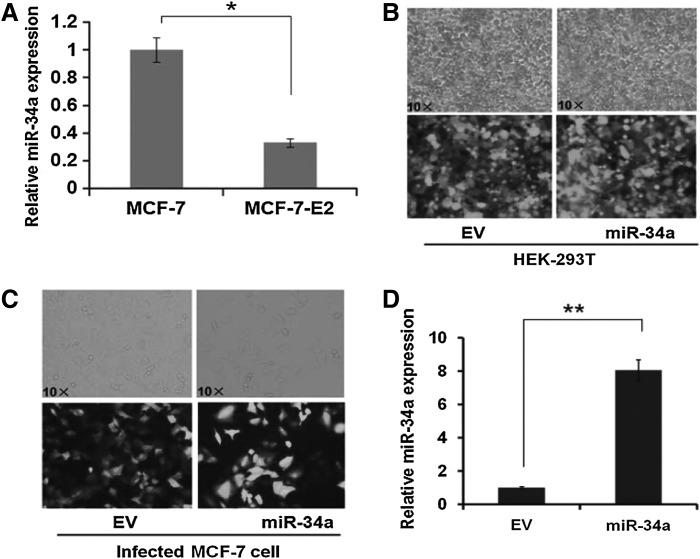
To investigate whether miR-34a regulates breast cancer in an E2-dependent manner, we treated MCF-7, which are ERα-positive human breast cancer cell line, with 10 nM E2 for 24 h. The expression level of miR-34a was analyzed by quantitative PCR with specific stem-loop RT primer. Significant decrease of miR-34a to 0.33-fold was observed compared with control group (treated without E2) (p<0.05; Fig. 1A). Therefore, to further investigate the effect of E2 on miR-34a expression and underline potential mechanism, miR-34a overexpression model was generated on MCF-7 cell based on lentivirus vector pLL3.7. As shown in Figure 1B, the lentivirus of miR-34a or EV was packaged in HEK-293T cells and the efficiency was about 90–95%. After infecting MCF-7 cells with lentivirus of either EV (Fig. 1C, left) or pLL3.7-miR-34a (right), the infected cells were screened and sorted by FACS based on the expression of green fluorescent protein (GFP), which indicated the presence of the plasmids pLL3.7-miR-34a or the EV under sterile condition. The expression of miR-34a in MCF-7 cells stably expressing miR-34a was 7.86-folds compared with EV group (p<0.01; Fig. 1D). The sorted miR-34a-infected MCF-7 cells were then used as the stable miR-34a overexpression model in the following experiments. We referred to the infected cells by either MCF-7-miR-34a or MCF-7-EV as miR-34a or EV groups, respectively, in the following experiments.
FIG. 1.
The decrease of miR-34a expression in MCF-7 cells after E2 treatment and establishment of stable miR-34a-expressing MCF-7 cells. (A) Total RNA was extracted from MCF-7 cells after E2 treatment as indicated, and quantitative PCR was performed to determine miR-34a expression. *p<0.05. (B) The lentivirus of miR-34a or empty vector (EV) was packaged in HEK-293T cells, which were less than 20 generations. The packaging efficiency was evaluated by GFP fluorescent signal. Original magnification 10×. (C) MCF-7 cells were infected by the EV or miR-34a. The GFP fluorescent signal could be stably observed 72 h after infection. Infected MCF-7 cells were sorted by FACS at sterile condition and collected for the following experiment. (D) Total RNA was extracted from MCF-7 cells of EV or miR-34a, and quantitative PCR was performed to determine miR-34a expression. **p<0.01. GFP, green fluorescent protein.
Overexpression of miR-34a suppresses MCF-7 cell growth, cell viability, and cell cycle progression in vitro and in vivo in the presence of E2
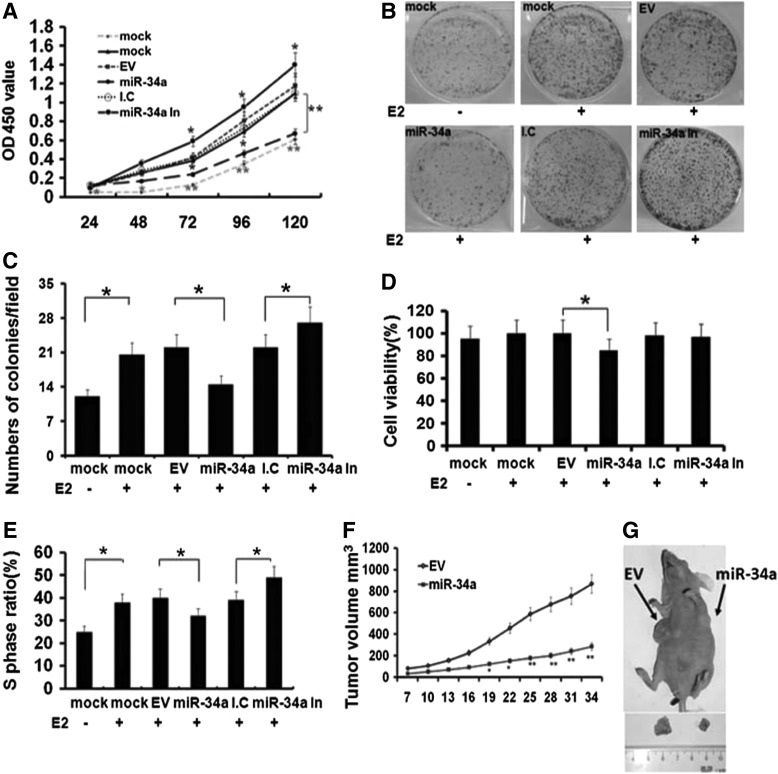
To verify the relationship between miR-34a expression and ERα-positive human breast cancer, the effect of miR-34a on cell proliferation was determined by the following experiments in the presence of E2(10 nM) for 24 h with mock MCF-7 cell without E2 treatment as a control. First, cell growth rate of 5 days was evaluated by CCK-8 assay. Cell proliferation was significantly increased with E2 treatment for 24 h compared with mock cell in the absence of E2 treatment (p<0.05; Fig. 2A). Stable expression of miR-34a mediated by pLL3.7 vector caused significant decreased proliferation, which became significant from 72 to 120 h compared with EV group in the presence of E2 for 24 h (p<0.05, p<0.01; Fig. 2A). Inhibition of miR-34a with complementary inhibitors at 50 nM significantly increased cell growth 72 h post transfection compared with inhibitor control (p<0.05; Fig. 2A). Second, cell viability and cell cycle were detected, respectively. As shown in Figure 2D. Overexpression of miR-34a significantly suppressed cell viability to 85% compared with EV group (p<0.05; Fig. 2D). DNA replication in S phase of cell cycle is indicator of cell proliferation. The ratio of cells in S phase was strikingly increased from 44% to 56% in the presence of E2 (p<0.05; Fig. 2E). But, overexpression of MiR-34a decreased the ratio of S phase from 53% to 37%. In contrast, inhibition of miR-34a remarkably increased S ratio from 42% to 53% (p<0.05; Fig. 2E). Third, colony formation was significantly increased with E2 treatment. The number of colonies decreased to 0.43-fold in miR-34a group compared with EV (p<0.05; Fig. 2B, C) and neutralization of miR-34a with its inhibitor strikingly increased the number of colonies (p<0.05; Fig. 2B). These results indicated that inhibitory effect of miR-34a on MCF-7 cell proliferation is in an E2-dependent manner, indicating that miR-34a inhibited breast cancer cell proliferation and DNA replication in vitro in the presence of E2. To further confirm the effect of miR-34a, a nude mice model was performed by subcutaneous injection of 5×106 cells of either MCF-7-miR-34a or MCF-7-EV into the mouse skin under the front right or left legs respectively, The tumor mass became palpable after 8 to 11 days inoculation in all (5/5) mice of both groups (one representative mouse in Fig. 2G). After 5 weeks inoculation, all mice were sacrificed and the tumor masses were weighted. The average tumor weight of miR-34a group was significantly less than control group. Tumor volumes in two groups was measured every 3 days after tumors were palpable and the results indicated that tumor volumes in miR-34a group only achieved 0.33-fold compared with EV control (p<0.05; Fig. 2F). Taken together, these data suggested that miR-34a suppressed cell proliferation in an E2-dependent manner both in vitro and in vivo.
FIG. 2.
MiR-34a suppresses cell proliferation in MCF-7 cells in vitro and in vivo and inhibition of miR-34a promotes cell proliferation. (A) Growth curves of MCF-7 cells were conducted by CCK-8 assay in the presence of 10 nM E2 as indicated. The OD value at 450 nm represented the viable cell numbers. Mock: parental MCF-7 cells without infection or transfection; E2-: without E2 treatment; E2+: treatment with E2; EV and MiR-34a: MCF-7 cells stably expressing pLL3.7 vector or miR-34a; I.C: miR-34a Inhibitor Control; MiR-34a In: MiR-34a Inhibitor. The transfection was performed with Lipofectamine 2000 and 50 nM of miR-34a inhibitor or inhibitor control were used for transfection. All experiments were carried out thrice independently. *p<0.05, **p<0.01. (B) Colony formation was performed with MCF-7 cells as indicated; Cells were cultured with or without E2 treatment for 2 weeks. The experiment was repeated thrice independently. (C) Quantification of colony formation assay. The colonies consisting of more than 50 cells were counted in 10 random visual fields. The experiment was repeated thrice independently. *p<0.05. (D) Cell viability was performed with Trypan blue staining. After treatment as indicated, cells were incubated with a final concentration of 0.04% (w/v) Trypan blue solution at room temperature for 3 min and then directly observed using a microscope (Olympus). Cell viability (%)=(viable counts/total counts)×100%. Each experiment was repeated thrice independently. *p<0.05. (E) Cell cycle analysis using propidium iodide staining and flow cytometry. The ratio of G1, S, and G2/M phase was analyzed. MiR-34a groups significantly reduced the ratio of S phase. Each experiment was repeated thrice independently. *p<0.05. (F) Tumor size observation in nude mice after the inoculation. 5×106 either miR-34a (left) or EV group (right) infected MCF-7 cells were injected subcutaneously in nude mice (n=5). The tumor volume was measured every 3 days with calipers after tumor appeared. The average size of the tumors was measured every 3 days and shown in the curves. The error bars show SD (standard deviation). *p<0.05, **p<0.01. (G) One representative nude mouse was shown. Arrows indicate tumors that resulted from either EV or miR-34a stably expressing MCF-7 cells.
LMTK3 is a new functional target of miR-34a in MCF-7 cells
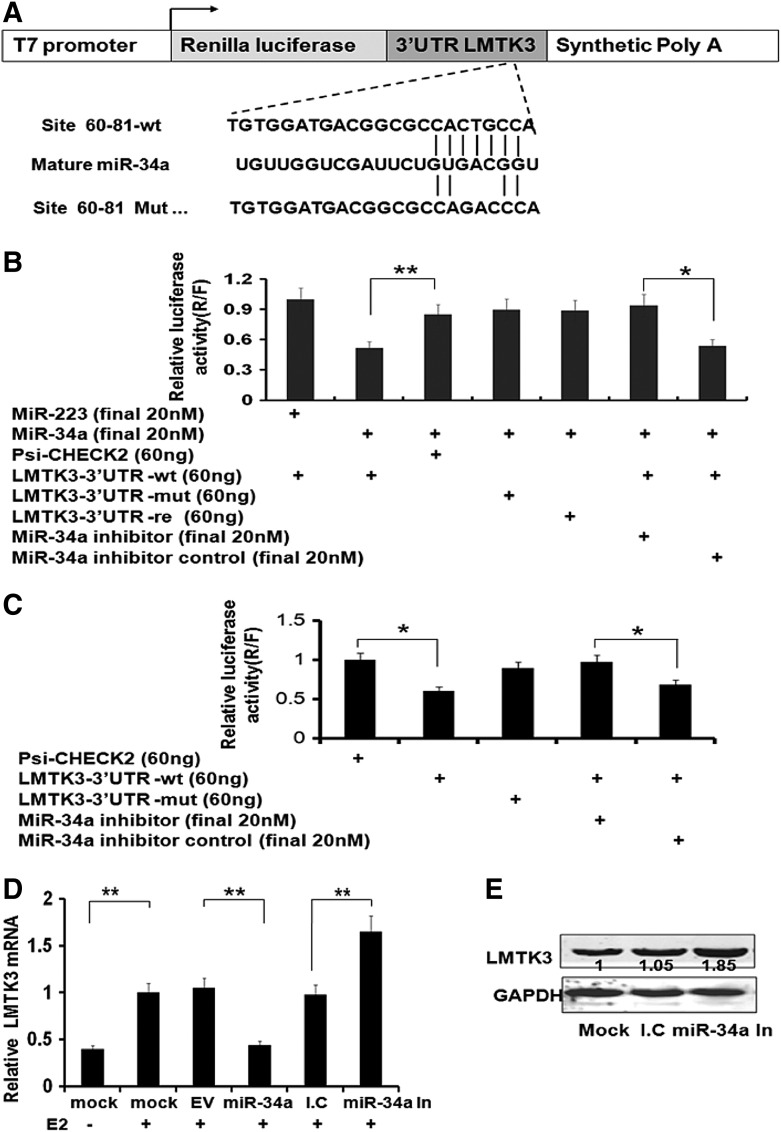
To identify potential targets of miR-34a both for experimental validation and functional studies in ERα-positive breast cancer MCF-7 cell, we performed in silico analysis of a range of miRNA target prediction databases. The target prediction of miR-34a was performed by using the following databases: TargetScan (www.targetscan.org) and MicroCosm (www.ebi.ac.uk/). From the databases analyzed, all presented LMTK3 as a converging target of miR-34a, which plays an important role in regulating ERα. To validate the interaction between miR-34a and its putative target gene LMTK3, we used psiCHECK-2 as a vector for dual luciferase reporter assay. This vector contains a Firefly luciferase (F), which serves as internal control and a Renilla luciferase (R), which serves as changing factor. The suppression of target gene by miRNA was measured by the ratio of R/F. The entire wild-type 3′UTR of LMTK3 (psiCHECK-2-LMTK3-3′UTR-wt), the mutant 3′UTR (psiCHECK-2- LMTK3 3′UTR-mut) with a 3-nucleotide mutations (Fig. 3A), and 3′UTR in reverse orientation by exchange endonucleases Xhot/NotI to NotI/Xhot at the forward and reverse primers (psiCHECK-2-LMTK3-3′UTR-re) were cloned immediately downstream of Renilla luciferase. HEK-293T cells, which exhibit low levels of miR-34a expression, were used for transient transfections with psiCHECK-2-LMTK3-3′UTR-wt, psiCHECK-2-LMTK3-3′UTR-mut, and psiCHECK-2-LMTK3-3′UTR-re. Co-transfection with miR-34a (a synthetic miR-34a mimic) resulted in a significant decrease (to 59.46%) in luciferase gene expression from the reporter vector containing the wild-type 3′UTR of LMTK3 when compared with a vector control (p<0.01; Fig. 3B), indicating that LMTK3 was the target of miR-34a (p<0.01; Fig. 3B). LMTK3-3′UTR failed to respond to miR-223, a microRNA that mainly expressed in blood system and predicted no targeting on LMTK3, as a negative control. Consistent with the data, no decreasing in luciferase activity was observed when miR-34a was co-transfected with the mutant 3′UTR of LMTK3 reporter. Besides, miR-34a specific inhibitor, which is the antisense oligonucleotides of miR-34a, can almost abolish the inhibition effect compared with inhibitor control (p<0.05; Fig. 3B).
FIG. 3.
LMTK3 is a direct target of miR-34a. (A) The highly conserved mature miR-34a sequence in mammals and the potential seed matching between miR-34a and LMTK3-3′UTR sequence are shown. The mutated bases in LMTK3-3′UTR-mut are indicated. The structure and cloning site of psiCHECK™-2 vector are also shown. (B) The transfection was done as indicated in 293T cells, followed by luciferase activity assay 48 h later. MiR-223 mimics serve as a negative control for miR-34a. Data of three independent experiments were expressed as mean±SD. *p<0.05, **p<0.01. (C) The Luciferase reporter assay was also performed in MCF-7 cells. Data of three independent experiments were expressed as mean±SD. *p<0.05. (D) Total RNA was extracted from MCF-7 cells after treatment as indicated, and quantitative PCR was performed to determine LMTK3 expression. Data of three independent experiments were expressed as mean±SD. **p<0.01. (E) MCF-7 cells were treated as indicated. Endogenous LMTK3 protein levels were determined by WB along with the GAPDH control. Relative protein levels were normalized by GAPDH and quantified by the ratio compared with controls.
These results indicated that LMTK3 might be a direct target of miR-34a and responsible for miR-34a targeting in its 3′UTR.
To verify whether miR-34a regulated LMTK3 in MCF-7 cells, which express higher miR-34a levels compared with HEK-293T cells, we performed another luciferase report experiment on MCF-7 cells. In agreement with finding observed in HEK-293T cells, the ratio of R/F in LMTK3-3′UTR-wt group was significantly decreased, compared with vector psiCHECK-2 group or LMTK3-3′UTR-mut group (p<0.05; Fig. 3C). The decrease of R/F ratio by LMTK3-3′UTR-wt could be rescued by the suppression of endogenous miR-34a with its inhibitor (final 20 nM), but not the inhibitor control (p<0.05; Fig. 3C). The results made it evident that miR-34a suppressed LMTK3 expression by directly binding to the 3′UTR region of LMTK3 and validate that LMTK3 was a direct target of miR-34a.
Target genes are usually directly regulated at transcriptional and/or post-transcriptional level. Therefore, we measured the LMTK3 mRNA and protein levels in MCF-7 cells with different treatment. LMTK3 mRNA level was upregulated to 2.63-fold when treated with E2 (p<0.05; Fig. 3D). Overexpressing of miR-34a in MCF-7 cells could abolish the increase of LMTK3 mRNA level resulted from E2 treatment (p<0.05; Fig. 3D). Inhibition of miR-34a with inhibitor enhanced expression of LMTK3 higher than inhibitor control (mRNA: 1.65-fold; protein: 1.85-fold) (p<0.05; Fig. 3D, E). These data indicated that miR-34a could directly suppress LMTK3 expression at both mRNA and protein level.
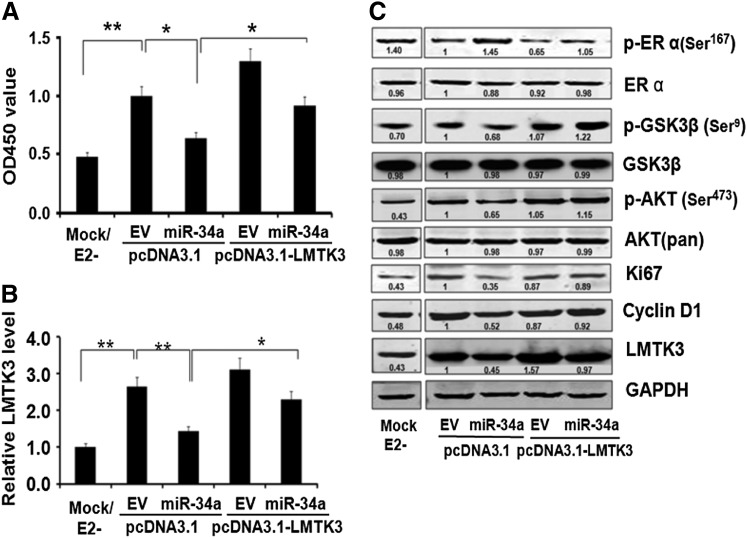
To further explore whether LMTK3 was a functional target of miR-34a, we re-introduced LMTK3 coding sequence without 3′UTR through transient transfection and measured the cell growth of 72 h by CCK-8. The cell proliferation increased after re-introducing LMTK3, and the change was similar to LMTK3 mRNA and protein levels (Fig. 4B, C). These results indicated that LMTK3 was a functional target of miR-34a, which was directly regulated by miR-34a.
FIG. 4.
Reintroducing LMTK3 rescued the inhibitory effect by miR-34a and affected AKT signal pathway and ERα phosphorylation status. (A) Cell proliferation of MCF-7 cells after transfection and E2 treatment as indicated was determined by Cell Counting Kit-8. Data of three independent experiments were expressed as mean±SD. *p<0.05, **p<0.01. (B) Total RNA was extracted from MCF-7 cells after transfection and E2 treatment as indicated, and quantitative PCR was performed to determine LMTK3 expression. Data of three independent experiments were expressed as mean±SD. *p<0.05, **p<0.01. (C) MCF-7 cells were transfected or treated as indicated. Endogenous LMTK3, the major molecules in AKT signaling pathway and ERα phosphorylation status were determined by WB along with the GAPDH control. Relative protein levels were normalized by GAPDH and quantified by the ratio compared with controls. Data of each experiment is repeated thrice independently.
The inhibitory effect of miR-34a on MCF-7 proliferation involved in AKT signaling pathway and ERα phosphorylation
To further investigate the underlying mechanisms in which miR-34a inhibited MCF-7 cell proliferation, the effects of miR-34a on expression of Ki67 were detected. As shown in Figure 4C, Ki67, a proliferation marker affected by LMTK3, was significantly downregulated to 0.35-fold compared with EV group. Similarly, Cyclin D1, a nuclear protein playing a pivotal role in cell proliferation and downstream of PI3K/AKT in miR-34a was also downregulated to 0.52-fold. LMTK3 expression was reactivated by reduced p-ERaSer167 to 0.65-fold in EVgroup and almost restored in miR-34a group. The expression of p-AKTSer473 was coincident with LMTK3 while total AKT had no change. The protein level of GSK3β, the major kinase downstream of PI3K/AKT signal pathway had no change, but its phosphrylation status (p-GSK3βSer9) was consistent with p-AKTSer473 downregulated to 0.68-fold. Interestingly, total ERα protein was slightly reduced to 0.88-fold compared with EV group, but p-ERαSer167 was significantly upregulated to 1.45-fold. Rescued p-ERaSer167 to 0.65-fold in EV group and almost restored in miR-34a group are observed by reintroduction of LMTK3 transfection. However, they are not consistent in the expression of cyclin D1, Ki67, p-AKTSer473, and p-GSK3βSer9 in the EV group with overexpression of LMTK3. These results implicated that the suppression of miR-34a on MCF-7 cell proliferation involved AKT signal pathway and ERα phosphorylation status.
Discussion
MiRNAs play a key role in the regulation of major cell functions, including development, differentiation, apoptosis, and metastasis. Based on their function, miRNAs can act as oncogenes or tumor suppressors and contribute to cancer initiation and progression (Shenouda and Alahari, 2009). The present study showed that miR-34a functions as a negative regulator or tumor suppressor in vitro and in vivo in breast cancer MCF-7 cells. The underlying mechanism was that LMTK3 was a novel functional target of miR-34a involved in cell proliferation and cell cycle progression. Several lines of evidences support this notion. First, bioinformatics from online prediction Websites, including TargetScan and MicroCosm, predicted that LMTK3, a kinase that phosphorylates ERα, is a putative target of miR-34a. Second, miR-34a repressed luficerase reporter gene containing LMTK3 3′UTR through its binding site. And mutation of binding site, reversed 3′UTR or specific inhibitor can almost abolish the inhibitory effect. Thirdly, overexpression of miR-34a downregulated the expression of LMTK3 at both protein and mRNA levels, suppression of S phase ratio and cell proliferation in vitro. Inhibition of miR-34a with its specific inhibitor almost restored the suppression effect caused by miR-34a. Finally, re-introduction of LMTK3 in MCF-7-miR-34a cells can partially rescue the suppressing effect in proliferation and LMTK3 expression at both mRNA and protein level. These results clearly indicate that LMTK3 is a novel functional target of miR-34a and may play an essential role in ERα positive breast cancer suppression in vitro and in vivo.
More than two-thirds of breast tumors express ERα (Thomas and Gustafsson, 2011) and patients with ERα-positive disease exhibit endocrine resistance (Thomas and Gustafsson, 2011). LMTK3 was first identified to regulate ERα activity by a whole human kinome siRNA screen using expression of the estrogen-responsive TFF1 gene, and silencing of LMTK3 reduced tumor volume in an orthotopic mouse model and abrogated proliferation of ERα positive but not ERα negative cells, indicative of its role in regulating ERα activity (Giamas et al., 2011). It has already been shown that LMTK3 is over-expressed in more aggressive forms of breast cancer, and its levels correlate with disease free survival and overall survival as well as predicting response or resistance to hormonal treatment. LMTK3 expressions were also shown to directly correlate with the tumor proliferation marker Ki67 (p<0.002) (Stebbing et al., 2012), indicating its broad function including proliferation in breast cancer. In this study, we confirmed that LMTK3 was indeed a functional target of miR-34a and found that the expression of LMTK3 was negatively regulated by miR-34a. It should be noted that the mature miR-34a sequence (5′-UGGCAGUGUCUUAGCUG GUUGU-3′) is highly conserved among mammalian species. The potential miR-34a binding site (5′-CACUGCC-3′) is also presented in the known LMTK3 3′UTR of chimpanzee, dog, pig, bush baby, and cow, suggesting that miR-34a might regulate LMTK3 expression of these species in a similar manner.
The activity of ERα was dependent on its two major phosphorylation sites (Ser118 and Ser167). Ser118 was phosphorylated by mitogen-activated protein kinase-3 (MAPK3) (Zoubir et al., 2008) and Ser167 was phosphorylated by AKT (Campbell et al., 2001) and p90RSK (Joel et al., 1998). There is a discrepancy in the regulation of ERα expression by LMTK3. Having shown that LMTK3 stabilize ERα stability at mRNA and protein (Giamas et al., 2011; Stebbing et al., 2012) other reports have shown that overexpression of LMTK3 was negatively associated with the expression of hormone receptors, ERa and PgR (Stebbing et al., 2013). However, the concurrent results in above two articles are the positive correlation of LMTK3 expression level with high tumor grade and clinical outcome including overall survival. In agreement with these results, our study have shown that ERα protein level was slightly reduced to 0.88-fold but the p-ERαSer167 was significantly upregulated to 1.45-fold in miR-34a group while LMTK3 was significantly downregulated to 0.45-fold. Rescue experiment by re-introducing LMTK3 mediated by pcDNA3.1(+)-LMTK3-CDS could restore both LMTK3 protein and the p-ERαSer167 levels, indicating that LMTK3 regulate the phosphorylation of ERα at Ser167 in a negative correlation manner. This result is consistent with the article reported previously (Giamas et al., 2011). Phosphorylation of ERα at Ser167 was confirmed to be associated with being longer disease-free and overall survival in breast cancer patients (Jiang et al., 2007). In miR-34a group, p-AKTSer473 was strikingly downregulated to 0.65-fold even though the total AKT had no change. The phosphorylation of GSK3β at Ser9, the major component downstream of PI3K/AKT signaling pathway, was also downregulated to 0.68-fold, indicating the downregulation of PI3K/AKT signaling pathway. The reduced expression of Ki67 and cyclin D1, the cell proliferation markers, were observed in this group. P-AKTSer473 participates in phosphorylation of ERα at Ser167, but p90RSK may be more important than AKT for Ser167 phosphorylation (Jiang et al., 2007). P-AKTSer473 was shown to be associated with a two- to three-fold reduction in overall survival. The negative association between the p-AKTSer473 and p-ERαSer167 probably is because p-AKTSer473 and p90RSK are competitive in phosphorylation of ERα at Ser167. Rescue experiments are carried out in miR-34a group by transient transfection of LMTK3 coding region without 3′UTR; the expression of Ki67, cyclin D1, p-AKTSer473, and p-GSK3βSer9 were almost restored compared with miR-34a group transfected with pcDNA 3.1 vector. However, there is a little discrepancy between cell proliferation and the mechanism alteration in EV group re-introducing LMTK3. The proliferation in this group is faster than other groups, but the effect of LMTK3 on AKT signal pathway is not very consistent. These may be because of the high basal expression levels of cyclinD1 and Ki67, which make the cells not sensitive to LMTK3 overexpression but sensitive to low level of LMTK3 expression. It is also possible that other signaling pathways such as apoptosis may involve in. Taken together, it is reasonable to hypothesize that post-transcriptional regulation of LMTK3 by miR-34a may provide a new insight into breast cancer progression and therapeutic target in breast cancer treatment.
Conclusion
We developed an miR-34a overexpression model in ERα-positive breast cancer cell line MCF-7 and showed that miR-34a suppresses MCF-7 cell proliferation in vitro and in vivo by targeting a new functional gene-LMTK3 in an E2-dependent manner. Further studies are needed to identify the exact mechanisms of miR-34a involved in breast cancer progression.
Acknowledgments
This research was supported by the National Nature Scientific Foundation of China (NO.81241093), the Fundamental Research Funds for Jilin University (NO.450060445246), the High-tech Industrial Development Project of Jilin Province (NO.20090633), the Specialized Research Fund for the Doctoral Program of Higher Education (NO.20100061120028), the Scientific Research Foundation of Jilin Province (NO. 20130206001YY, 20120713, and 200905169), and the Scientific Research Foundation of Changchun (NO.12SF29).
Disclosure Statement
The authors have declared that no competing interests exist.
References
- Bader A.G. miR-34- a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.A. Bhat-Nakshatri P. Patel N.M. Constantinidou D. Ali S. Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- Eulalio A. Huntzinger E. Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Giamas G. Filipovic A. Jacob J. Messier W. Zhang H. Yang D. Zhang W. Shifa B.A. Photiou A. Tralau-Stewart C., et al. Kinome screening for regulators of the estrogen receptor identifies LMTK3 as a new therapeutic target in breast cancer. Nat Med. 2011;17:715–719. doi: 10.1038/nm.2351. [DOI] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Jia C.Y. Li H.H. Zhu X.C. Dong Y.W. Fu D. Zhao Q.L. Wu W. Wu X.Z. MiR-223 suppresses cell proliferation by targeting IGF-1R. PLoS One. 2011;6:e27008. doi: 10.1371/journal.pone.0027008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. Sarwar N. Peston D. Kulinskaya E. Shousha S. Coombes R.C. Ali S. Phosphorylation of estrogen receptor-alpha at Ser167 is indicative of longer disease-free and overall survival in breast cancer patients. Clin Cancer Res. 2007;13:5769–5776. doi: 10.1158/1078-0432.CCR-07-0822. [DOI] [PubMed] [Google Scholar]
- Joel P.B. Smith J. Sturgill T.W. Fisher T.L. Blenis J. Lannigan D.A. pp90rsk1 regulates estrogen receptor-mediated transcription through phosphorylation of Ser-167. Mol Cell Biol. 1998;18:1978–1984. doi: 10.1128/mcb.18.4.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.H. Kim H.S. Li X.Y. Lee I. Choi H.S. Kang S.E. Cha S.Y. Ryu J.K. Yoon D. Fearon E.R., et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Day E. Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenouda S.K. Alahari S.K. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- Stebbing J. Filipovic A. Ellis I.O. Green A.R. D'Silva T.R. Lenz H.J. Coombes R.C. Wang T. Lee S.C. Giamas G. LMTK3 expression in breast cancer: association with tumor phenotype and clinical outcome. Breast Cancer Res Treat. 2012;132:537–544. doi: 10.1007/s10549-011-1622-z. [DOI] [PubMed] [Google Scholar]
- Stebbing J. Filipovic A. Lit L.C. Blighe K. Grothey A. Xu Y. Miki Y. Chow L.W. Coombes R.C. Sasano H., et al. LMTK3 is implicated in endocrine resistance via multiple signaling pathways. Oncogene. 2013;32:3371–3380. doi: 10.1038/onc.2012.343. [DOI] [PubMed] [Google Scholar]
- Svoboda M. Sana J. Redova M. Navratil J. Palacova M. Fabian P. Slaby O. Vyzula R. MiR-34b is associated with clinical outcome in triple-negative breast cancer patients. Diagn Pathol. 2012;7:31. doi: 10.1186/1746-1596-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. Gustafsson J.A. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011;11:597–608. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- Winter J. Jung S. Keller S. Gregory R.I. Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wu R.C. Qin J. Yi P. Wong J. Tsai S.Y. Tsai M.J. O'Malley B.W. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Yang S. Li Y. Gao J. Zhang T. Li S. Luo A. Chen H. Ding F. Wang X. Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2012 doi: 10.1038/onc.2012.432. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Zoubir M. Mathieu M.C. Mazouni C. Liedtke C. Corley L. Geha S. Bouaziz J. Spielmann M. Drusche F. Symmans W.F., et al. Modulation of ER phosphorylation on serine 118 by endocrine therapy: a new surrogate marker for efficacy. Ann Oncol. 2008;19:1402–1406. doi: 10.1093/annonc/mdn151. [DOI] [PubMed] [Google Scholar]