Abstract
Background. Determinants of intersubtype differences in human immunodeficiency virus type 1 (HIV-1) clinical disease progression remain unknown.
Methods. HIV-1 subtype was independently determined for 5 separate genomic regions in 396 HIV-1 seroconverters from Rakai, Uganda, using a multiregion hybridization assay. Replication capacities (RC) in samples from a subset of 145 of these subjects were determined. HIV-1 genomic regions and pol RC were examined for association with disease progression. Amino acid polymorphisms were examined for association with pol RC.
Results. In multivariate analyses, the hazard for progression to the composite end point (defined as a CD4+ T-cell count <250 cells/mm3, antiretroviral therapy initiation, or death) among patients with subtype D pol infection was 2.4 times the hazard for those infected with subtype A pol infection (P = .001). Compared with subtype A pol (the reference group), the hazard for progression to the composite end point for subtype D pol infection with a pol RC >67% (ie, the median pol RC) was significantly greater (HR, 4.6; 95% confidence interval [CI], 1.9–11.0; P = .001), whereas the hazard for progression to the composite end point for subtype D pol infection with a pol RC ≤67% was not significantly different (HR, 2.2; 95% CI, 1.0–4.9; P = .051). Amino acid substitutions at protease positions 62 and 64 and at reverse transcriptase position 272 were associated with significant differences in pol RC.
Conclusions. HIV-1 pol gene intersubtype and RC differences are associated with disease progression and may be influenced by amino acid polymorphisms.
Keywords: HIV-1 Subtype, subtype A, subtype D, disease progression, polymerase, replication capacity, amino acid polymorphisms
The global human immunodeficiency virus type 1 (HIV-1) epidemic is genetically diverse [1]. Group M, the predominant group, is divided into 9 distinct subtypes and 58 circulating recombinant forms [2]. Studies from diverse geographic locations including Africa, North America, and Southeast Asia have documented intersubtype differences in disease progression [3–7]. In a prospective cohort from Rakai, Uganda, with known times of seroconversion, the adjusted hazards of death and progression to AIDS for participants infected with subtype D HIV-1 were 5.7 and 2.1 times, respectively, those for participants infected with subtype A [7]. In Mombasa, Kenya, the hazard of death for subtype D–infected commercial sex workers was 2.3 times that for their subtype A–infected counterparts [3].
It is important to understand the virologic factors underlying intersubtype differences in HIV-1 disease progression, which have associated implications for therapeutic and immune targeting, especially among non-B subtypes. In vivo studies examining virologic factors affecting disease progression have mainly involved Western populations, where subtype B predominates [1]. Previously identified virologic factors influencing disease progression include polymerase gene replication capacity (pol RC), coreceptor tropism switch, and deletions in the nef gene [8–11]. Prior studies in cohorts from the United States have demonstrated an association between high pol RC and increased rates of disease progression, independent of HIV-1 load [8, 10, 11]. Studies to determine in vivo virologic factors explaining intersubtype differences in HIV-1 disease progression are best performed in ethnically homogeneous cohorts, with cocirculation of different subtypes, known dates of seroconversion, and long follow-up periods for subjects involved; thus, they are challenging to conduct.
The ethnically homogeneous population in Rakai, Uganda, with a large number of subjects and known dates of seroconversion since 1994, where both HIV-1 subtype A and subtype D cocirculate, provides a unique opportunity to examine the influence of viral factors on intersubtype and intrasubtype differences in disease progression. We examined multiple genomic regions of HIV for association with intersubtype differences in disease progression, tested the influence of phenotypic functionality of a subset of pol genes, and determined whether any amino acid substitutions were correlated with increased pol RC and disease progression in this cohort.
SUBJECTS, MATERIALS, AND METHODS
Subjects
Since 1994, the Rakai Health Sciences Program (RHSP) has conducted annual community-based surveillance of HIV-1 serostatus among persons aged 15–49 years in Rakai, Uganda [12]. From 1997 to 2002, newly identified HIV-1 seroconverters were enrolled into the Molecular Epidemiological Research (MER) study, as previously described [7]. All subjects provided written informed consent, and ethics approvals were obtained from the Uganda Virus Research Institute's Science and Ethics Committee, the Uganda National Council for Science and Technology, the Walter Reed Army Institute of Research, Columbia University, and Johns Hopkins University. The Monogram Biosciences clinical reference laboratory performed genotypic and phenotypic testing.
Briefly, consenting HIV-1 seroconverters were followed up 1, 3, 6, and 12 months after seroconversion and annually thereafter. Study procedures at each visit included completion of a standardized questionnaire, a physical examination, and blood collection. Blood samples were tested to determine CD4+ T-cell count; plasma HIV-1 load; subtype A, C, and D multiple hybridization assay (MHAacd) genotype; and pol sequence and RC. All subjects had samples tested for MHAacd genotype. A subset of 145 subjects was selected for pol sequencing and RC testing [13]. All samples tested for MHAacd genotype, pol sequence, and pol RC were obtained within 1.5 years of the first HIV-1–positive serologic finding. Subjects were followed until 17 October 2007 or were censored at the time of reaching the primary end point or at loss to follow-up (defined as >1 year without clinical review).
Laboratory Analysis
HIV-1 serostatus was determined by Vironostika HV-1 (Organon Teknika, Charlotte, NC) and Cambridge Biotech (Worcester, MA). Discordant enzyme immunoassay results and all seroconversions were confirmed by Western blot (bioMérieux VITEK, St. Louis, MO). Plasma HIV-1 load was determined using the Amplicor HIV-1 Monitor assay, version 1.5 (Roche Molecular Systems). CD4+ T-cell counts were determined using the BD FacsCalibur system until 2003, after which the BD FacsCount system (Becton-Dickinson) was used.
As previously described, the MHAacd targeted 5 separate regions of the HIV-1 genome: gag (HXB2 nucleotides 891–1420), pol (HXB2 nucleotides 1966–2347), vpu (HXB2 nucleotides 5967–6471), env (HXB2 nucleotides 7830–8445), and gp (HXB2 nucleotides 8424–8780) [14]. As a result, each of the 5 genomic regions was separately assigned a subtype. For each genomic region, reactivity to only one of the 3 probes (A, C, or D) indicated a pure subtype, and reactivity to >1 probe indicated mixed infection.
As previously described, GeneSeq HIV and Phenosense HIV assays (Monogram Biosciences, South San Francisco, CA) were used to determine pol genotype (GenBank accession nos. FJ389051-FJ389154) and pol RC, respectively [13, 15]. Briefly, the HIV-1 pol region was amplified following viral extraction from participant samples and then cloned into a test vector. For the GeneSeq HIV assay, sequencing was performed on the vector pools. Subtyping of the pol gene was based on analysis of protease and reverse transcriptase sequences as previously described [13]. For the Phenosense HIV assay, RC of the test vectors containing patient-derived pol was determined and reported as a percentage of the reference, HIV-1 strain NL4-3. In a previous study, replacing the subtype B viral test vector with a subtype C test vector did not result in significant changes in pol RC, suggesting that subtype mismatch between the viral test vector and subject pol gene did not affect pol RC [16].
Study Definitions and Primary and Secondary Study End Points
The time of HIV-1 infection was defined as the midpoint between the latest negative and the first positive HIV-1 test result [7]. The primary study end point was the time from HIV-1 infection to a composite end point, defined as the earliest date of either a CD4+ T-cell count <250 cells/mm3, antiretroviral therapy initiation because of a CD4+ T-cell count <250 cells/mm3, World Health Organization stage III or IV HIV disease, or death. Secondary end points, used in sensitivity analyses, were the time from HIV-1 infection to a CD4+ T-cell count <250 cells/mm3 and the time from HIV-1 infection to death. The subject's HIV-1 load set point was the median of all viral load determinations before a CD4+ T-cell count <250 cells/mm3 was reached.
Statistical Analysis
Baseline characteristics were compared by genomic region, between subjects with reactive and nonreactive probes, and between subjects with subtype A and subtype D, using the Wilcoxon rank sum test for continuous variables and the χ2 test for categorical variables.
To identify HIV-1 genomic regions associated with the intersubtype difference in disease progression, each MHAacd genomic region was separately examined, using Kaplan-Meier analyses and the log-rank test, to compare the time to the primary study end point between subtype A and subtype D. As the study aimed to compare subtypes A and D, subtype C and mixed infection were excluded. To determine whether study inferences would be affected by varying the study end points, sensitivity analyses were performed to examine the times to the primary and secondary end points, using multivariate Cox proportional hazards analyses adjusted for age, sex, and log10 set point HIV-1 load. Age, sex, and log10 set point HIV-1 load were previously identified independent predictors of disease progression in the Rakai population [7, 17]. For primary end point analyses, observations were censored at the last follow-up date or the study cutoff date of 17 October 2007, whichever was earlier. For the secondary end points of a CD4+ T-cell count <250 cells/mm3 and death, observations were also censored at time of initiation of antiretroviral therapy.
The relationship of pol RC and log10-transformed viral load was examined using simple univariate linear regression. To determine the contribution of pol RC to the difference in disease progression between subtype A and subtype D, subjects with the 2 subtypes were stratified into high and low pol RC groups in Kaplan-Meier and Cox proportional hazards analyses of time to composite end point, based on the median RC for pol subtype D–infected subjects.
In the analysis of the association of pol amino acid mutations with pol RC, 42 subtype A and 50 subtype D pol sequences with matched RC data from samples submitted for routine testing to Monogram Biosciences were included for the analysis (MGRM data). Only clinical samples without known drug resistance–associated mutations were selected for inclusion in the MGRM data. To determine whether single pol amino acid mutations correlated with pol RC, we first conducted a feature selection analysis, using several statistical methods. A first univariate analysis compared median pol RC by residue, using a Bonferroni-corrected Wilcoxon rank sum test. For multivariate analyses, amino acid variants were coded as present or absent (if mixtures were present, they counted as full mutations). Regression tree analysis was conducted to find amino acid variants associated with high or low RC, using the median RC for the entire data set as a cut point. Multiple linear regression modeling was used to predict continuous log10 RC, and all amino acid variants with P < .05 were considered to be significant predictors of RC. This first step analysis was limited to amino acid present in at least 5% of the samples, at positions with <90% homogeneity, to allow for a meaningful interpretation of results. Amino acid variants significantly associated with RC in all 3 methods were retained. A second stage analysis, limiting comparisons to pol subtype D, was then performed using the Wilcoxon rank sum test, to control for possible confounding by subtype. All statistical analyses were performed using Stata, version 11 (Stata, College Station, TX) and R [18].
RESULTS
Population Characteristics
Of 491 seroconverters identified from 1997 to 2003, 488 (99.4%) consented to enroll into the Molecular Epidemiology Research study. MHAacd subtyping was successful in at least 1 region in 396 (81.1%) subjects. The percentages with successful MHAacd subtyping, by genomic region, were as follows: gag 332, 83.8%; pol 241, 60.9%; vpu 245, 61.9%; env 237, 59.8%; and gp 275, 69.4% (Table 1). Fifty-one subjects (10.5%) had reactive probes in all 5 genomic regions, of which 2 (3.9%) were subtype A in all 5 genomic regions, 29 (56.9%) were subtype D in all 5 genomic regions, and 20 (39.2%) had probes reactive to subtype A and subtype D, consistent with AD recombinants.
Table 1.
Findings of Subtype A, C, D Multiple Hybridization Assay, by Human Immunodeficiency Virus Type 1 (HIV-1) Genomic Region
| Subtype | HIV-1 Genomic Region, Subjects, No. (%) |
||||
|---|---|---|---|---|---|
| gag (n = 396) | pol (n = 396) | vpu (n = 396) | env (n = 396) | gp (n = 396) | |
| A | 91 (23.0) | 54 (13.6) | 51 (12.9) | 70 (17.7) | 43 (10.9) |
| D | 229 (57.8) | 170 (42.9) | 177 (44.7) | 164 (41.4) | 225 (56.8) |
| C | 4 (1.0) | 9 (2.3) | 14 (3.5) | 0 | 2 (0.5) |
| Mixed | 8 (2.0) | 8 (2.0) | 3 (0.8) | 3 (0.8) | 5 (1.3) |
| Nonreactive | 64 (16.2) | 155 (39.1) | 151 (38.1) | 159 (40.2) | 121 (30.6) |
Comparison of subjects with nonreactive probes to those with reactive probes revealed no significant differences in demographic characteristics or baseline laboratory values for the pol, vpu, and env regions (Table 2). For the gag and gp region, the only difference was that subjects with nonreactive probes had lower median log10-transformed viral loads than those with reactive probes (for gag, 4.3 log10 copies/mL among subjects with nonreactive probes and 4.7 log10 copies/mL among those with reactive probes [P = .042]; for gp, 4.3 and 4.7 log10 copies/mL, respectively [P < .001]).
Table 2.
Characteristics of Study Subjects, Overall and by Findings of Subtype A, C, D Multiple Hybridization Assay of the pol Region
| Characteristic | Overall (n = 396) | Pol Gene Probe Nonreactive (n = 155) | Pol Gene Probe Reactive (n = 241) | Pol Gene Subtype A (n = 54) | Pol Gene Subtype D (n = 170) |
|---|---|---|---|---|---|
| Age at seroconversion, y | |||||
| <25 | 153 (38.8) | 58 (37.7) | 95 (39.6) | 19 (35.8) | 69 (40.6) |
| 25–34 | 161 (40.9) | 59 (38.3) | 102 (42.5) | 23 (43.4) | 73 (42.9) |
| ≥35 | 80 (20.3) | 37 (24.0) | 43 (17.9) | 11 (20.8) | 28 (16.5) |
| Sex | |||||
| Male | 158 (40.2) | 61 (39.6) | 97 (40.6) | 18 (34.0) | 68 (40.2) |
| Female | 235 (59.8) | 93 (60.4) | 142 (59.4) | 35 (66.0) | 101 (59.8) |
| Baseline CD4+ T-cell count, cells/mm3 | 510 (367–717) | 523 (378–701) | 508 (349–725) | 575 (365–801) | 499 (340–691) |
| HIV-1 load, log10 copies/mL | 4.6 (4.1–5.1) | 4.6 (4.0–5.2) | 4.6 (4.1–5.1) | 4.4 (4.2–5.1) | 4.8 (4.1–5.1) |
| Follow-up duration after HIV-1 infection, y | 5.0 (3.5–6.9) | 5.2 (3.5–7.4) | 4.8 (3.5–6.7) | 5.2 (2.8–7.8) | 4.8 (3.6–6.4) |
Data are no. (%) of subjects or median value (interquartile range). Subjects with missing data were excluded from analysis.
In comparing baseline demographic characteristics and laboratory values for subjects with probes reactive for subtype A against those reactive for subtype D, there were no significant differences for the pol, vpu, env, or gp genomic regions (Table 2). In all 5 regions, HIV-1 load did not differ significantly between subjects reactive for subtype A, compared with subjects reactive for subtype D. For the gag genomic region, the only significant difference was that subjects with gag subtype A had a higher median baseline CD4+ T-cell count (586 cells/mm3; interquartile range [IQR], 436–806 cells/mm3) than those with gag subtype D (492 cells/mm3; IQR, 368–680 cells/mm3; P = .006).
The total observation time was 1844 years, with a median follow-up duration from the time of HIV-1 infection to the primary end point or last recorded visit of 5.0 years (IQR, 3.5–6.9 years). A total of 117 subjects (29.5%) were lost to follow-up after a median duration of 3.4 years (IQR, 2.0–4.7 years).
Overall, 208 (52.5%) reached the primary composite end point of either a CD4+ T-cell count of <250 cells/mm3, antiretroviral therapy initiation, or death. One hundred forty-eight subjects (37.4%) progressed to the secondary end point of a CD4+ T-cell count <250 cells/mm3, and 86 (21.7%) progressed to death. For the pol genomic region, higher proportions of subjects with subtype D pol than subjects with subtype A pol reached the composite end point (59.4% vs 35.2%; P = .002), progressed to a CD4+ T-cell count <250 cells/mm3 (41.8% vs 25.9%; P = .037), or died (25.9% vs 9.3%; P = .010). For the gag genomic region, higher proportions of subjects with subtype D gag than subjects with subtype A gag reached the composite end point (56.3% vs 39.6%; P = .007) or died (25.3% vs 11.0%; P = .005). For the gag region, there was no significant difference in progression to CD4+ T-cell count <250 cells/mm3. For the vpu, env, and gp genomic regions, there were no differences in proportions of subjects who reached the composite end point, progressed to a CD4+ T-cell count <250 cells/mm3, or died.
Association of Genomic Region Subtypes With Time to Composite End Point
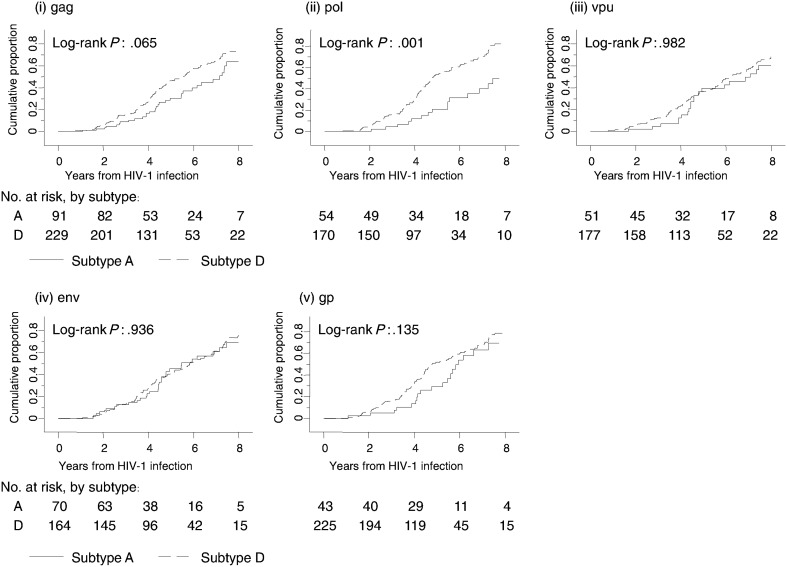
The pol region was the only genomic region with a significant difference in median time from HIV-1 infection to the primary composite end point, comparing subtype D to subtype A (Figure 1). Subjects with a subtype D pol had a median time from HIV-1 infection to the composite end point of 5.2 years (95% confidence interval [CI], 4.7–6.2), compared with 8.4 years (95% CI, 6.9–9.1) for subjects with a subtype A pol (P = .002). The hazard of progression to the composite end point among subjects with subtype D pol was 2.4 times the hazard among those with subtype A (P = .001; Table 3).
Figure 1.
Time from human immunodeficiency virus type 1 (HIV-1) infection to the composite end point, by genomic region. The composite end point was defined as the earliest date of either a CD4+ T-cell count <250 cells/mm3, antiretroviral therapy initiation, or death. The genomic regions are labeled (i) gag, (ii) pol, (iii) vpu, (iv) env, and (v) gp. In each genomic region, subtype A was compared to subtype D.
Table 3.
Primary and Sensitivity Analyses for Time From Human Immunodeficiency Virus Type 1 (HIV-1) Seroconversion to Various Study End Points Among Subjects With pol Subtype D Infection Versus Those With pol Subtype A Infection
| Study End Point | Hazard Ratio (95% CI) | P |
|---|---|---|
| Primary analysis | ||
| Time to composite end point, unadjusted | 2.4 (1.4–3.7) | .001 |
| Sensitivity analysesa | ||
| Time to composite end point | 2.4 (1.4–3.9) | .001 |
| Time to CD4+ T-cell count <250 cells/mm | 2.6 (1.5–4.8) | .001 |
| Time to death | 3.6 (1.4–9.0) | .008 |
a Adjusted for sex, age, and HIV-1 load set point.
Sensitivity Analyses
Table 3 shows the results of sensitivity analyses using different end points, by genomic region, adjusted for sex, age, and set point HIV-1 load. The pol region was the only genomic region with significant differences in times to all end points between subtype D pol and subtype A pol, with hazards of progression for subtype D pol varying from 2.4 to 3.6 times those for subtype A pol (all P < .05). For the gag region, the hazards of progression to the composite end point and to death among subjects with subtype D gag were 1.6 times (95% CI, 1.1–2.3; P = .017) and 2.5 times (95% CI, 1.3–4.8; P = .009), respectively, the hazards for subjects with subtype A gag, but there was no significant difference between subtypes in the risk of progression to CD4+ T-cell count <250 cells/mm3. Multivariate analysis examining the interaction between the pol and gag region was not performed because only 195 subjects (49.2%) had subtype A or D results for both regions, of which only 11 (5.6%) were discordant. The above associations remained unchanged after Bonferroni correction for multiple comparisons.
Relationship of pol RC With pol Gene Subtype, Plasma HIV-1 RNA, and Disease Progression
One hundred of 145 tested samples (67.0%) were successfully sequenced for the pol gene and tested for pol RC. As previously described, failure to obtain results was due to low sample volumes (0.2 mL) [13]. There were no significant differences in the age, sex, or baseline CD4+ T-cell count between individuals with and individuals without pol sequence and RC results (data not shown). Individuals without pol sequence and RC results had lower viral loads than individuals who were successfully tested (median log10-transformed HIV-1 load, 4.36 vs 4.89; P = .004).
Pol subtypes, assigned using the National Center for Biotechnology Information tool, were as follows: 25 subtype A, 65 subtype D, and 10 recombinants. The median RC for subtype D pol was 67% (range, 3%–192%), compared with 28% (range, 6%–57%) for subtype A pol (P < .001). Pol RC were not significantly associated with the log10-transformed set point HIV-1 load (r2 = 0.02; P = .213). Sixty-two subjects had pol subtype data and reactive MHA probes, with concordant results in 53 (85%).
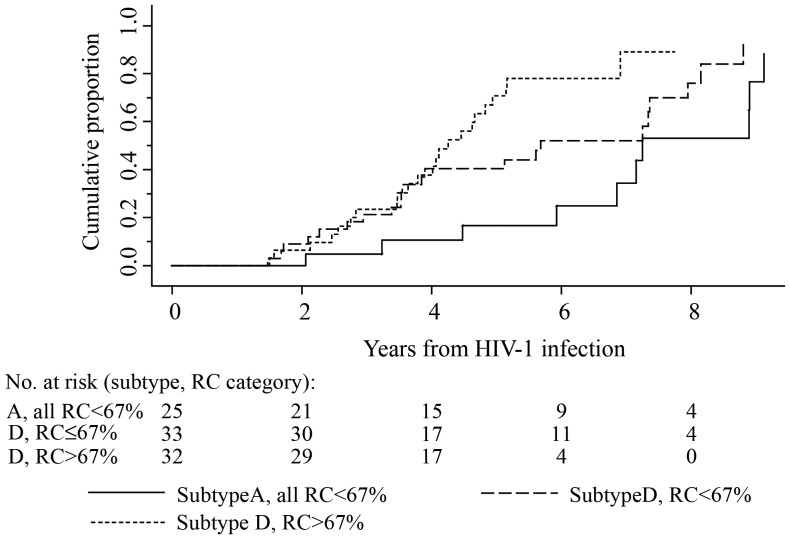
Figure 2 displays the time to the composite end point for subjects divided into 3 categories: (1) subtype A pol, all of which had a pol RC <67%; (2) subtype D pol with a pol RC ≤67%; and (3) subtype D pol with a pol RC >67%. Subjects in the subtype D pol with a pol RC >67% group had the fastest time to the composite end point (median interval, 4.3 years; 95% CI, 3.5–4.9). Subjects in the subtype D pol with a pol RC ≤67% group and those in the subtype A pol group had median times to the composite end point of 5.7 years (95% CI, 3.6–7.4) and 7.3 years (95% CI, 5.9–9.1), respectively (overall log-rank P = .003). In univariate Cox proportional hazards analyses with the hazard of progression to the composite end point among patients with subtype A pol infection as the reference, the hazard of progression for patients with subtype D pol with a pol RC >67% was significantly greater (hazard ratio [HR], 3.8; 95% CI, 1.7–8.5; P = .001), whereas the hazard for progression for patients with subtype D pol with a pol RC ≤67% was not significantly different (HR, 2.0; 95% CI, .9–4.3; P = .078).
Figure 2.
Time from human immunodeficiency virus type 1 (HIV-1) infection to the composite end point, by pol subtype and pol replication capacity (RC). The composite end point was defined as the earliest date of either a CD4+ T-cell count <250 cells/mm3, antiretroviral therapy initiation, or death. Three categories were compared: (i) pol subtype A (all pol subtype A isolates had a RC ≤67%), (ii) pol subtype D with a RC ≤67%, and (iii) pol subtype D with a RC >67%.
In multivariate Cox proportional hazards analyses adjusted for sex, age, and HIV-1 load set point, with the hazard of progression to the composite end point among patients with subtype A pol infection as the reference, the hazard of progression for patients with subtype D pol with a pol RC >67% was significantly greater (HR, 4.6; 95% CI, 1.9–11.0; P = .001), whereas the hazard for those with subtype D pol with a pol RC ≤67% not significantly different (HR, 2.2; 95% CI, 1.0–4.9; P = .051).
Association of pol Amino Acid Polymorphisms With pol RC
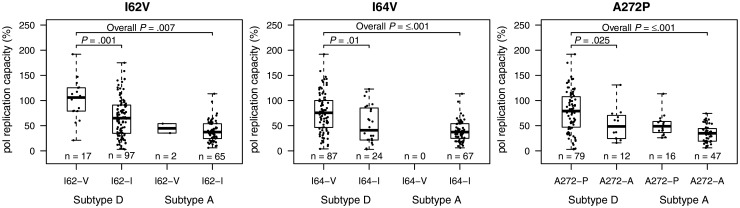
In analyses of 182 samples (90 RHSP, 92 Monogram) from patients with subtype A or D infection who had available pol RC and sequence data, 61 of 404 amino acid positions sequenced had <90% homogeneity. Of these 61 positions, 19 had significantly different median pol RC between residues. Isolates with ambiguous residues were excluded from analysis. Amino acid variants associated with pol RC were protease polymorphisms I62V and I64V and reverse transcriptase polymorphism A272P (Figure 3 and Supplementary Materials).
Figure 3.
Box plots of pol replication capacities (RC), by amino acid polymorphisms and subtype. Results for the 3 amino acid positions (protease polymorphisms I62V and I64V and reverse transcriptase polymorphisms A272P) with significant interresidue differences in median pol RC are shown. The bars represent the highest and lowest pol RC of each category. P values for the first-stage comparison are Bonferroni corrected.
DISCUSSION
In our study, the important role of the pol gene in intersubtype differences in disease progression among patients with HIV-1 subtype A or subtype D infection in Rakai, Uganda was supported by 2 independent lines of evidence. First, among 5 genomic regions subtyped, only the pol region was significantly associated with disease progression in primary and sensitivity analyses comparing subtype D to subtype A. Our second finding, independent of the first, is that subtype D pol isolates had markedly increased pol RC than subtype A pol isolates. In addition, subtype D pol infections with high RC had the highest rates of disease progression, whereas subtype D pol infections with low RC had rates of progression similar to those of subtype A pol infections. Additionally, pol RC was independent of HIV-1 load. Protease polymorphisms I62V and I64V and reverse transcriptase polymorphism A272P were associated with pol RC.
This study extends the evidence linking pol RC with disease progression to include subtype D–infected patients and, additionally, suggests that a high pol RC is a possible biologic mechanism for the increased rates of disease progression seen with subtype D infection, compared to subtype A infection. Prior studies, in predominantly subtype B–infected populations, demonstrated an association between pol RC and disease progression in treatment-nave HIV-1–infected patients, independent of HIV-1 load. Among patients in San Francisco with known dates of infection, a higher pol RC (defined as ≥43%) was associated with increased rates of CD4+ T-cell loss [10]. Similar to our study, pol RC was not associated with HIV-1 load. In a multicenter US study involving HIV-1–infected patients with a baseline CD4+ T-cell count ≥450 cells/mm3, a baseline pol RC <79% was associated with a decreased HR of 0.73 for progression to a composite end point of CD4+ T-cell count ≤350 cells/mm3, treatment initiation, or death, after adjustment for baseline CD4+ T-cell count and HIV-1 load [11].
Interestingly, pol RC was not significantly associated with set point viral load, a finding in agreement with the San Francisco study [10]. This finding is not surprising, given that multiple host and viral factors affect set point viral load, including patient age, race, HLA status, and HIV gag gene epitopes [19, 20]. In a human genome-wide association study, HLA-B*5701 and HLA-C were the 2 polymorphisms significantly associated with viral load set point [21]. Prior studies suggest that viral factors can affect disease progression via pathways independent of HIV-1 load [10, 11, 22].
We identified potential amino acid polymorphisms associated with pol RC. In vitro experiments have demonstrated that single amino acid mutations can result in marked differences in protease catalytic efficiency, a measure known to be correlated with ex vivo replication capacity [23, 24]. In these mutagenesis experiments, the I64V mutation increased protease catalytic efficiency in a subtype B HXB2 strain carrying a deleterious D30N mutation. The hypothesis that amino acid polymorphisms potentially alter pol RC in HIV-1 subtype D could be tested with site-directed forward and backward mutagenesis experiments. Preliminary in vitro evidence suggests that HIV-1 with a higher pol RC is more able to infect thymic progenitor cells and adversely impact immune cells early in development [10].
Our study had limitations. The modest sample size (related to challenges in identifying a large cohort of seroconverters with subtype cocirculation and to sequence heterogeneity challenges in MHAacd subtyping) could have reduced the power to detect true associations between the non-pol genomic regions and disease progression. We could not adjust for coreceptor tropism in our analyses, because of a lack of residual samples. However, as subjects were recruited within 2 years of seroconversion, HIV-1 coreceptor tropism was unlikely to be a significant confounder. From a prior study, phenotypic determinations were available for 20 subjects in this study, of which 18 (90%) had R5-tropic virus [25]. We were unable to study the continuous dose-response association of pol RC and disease progression, because of limited numbers.
We find that the pol gene is an important genomic determinant of the difference in disease progression between subtype D and A infections. The increased pol RC in subtype D pol isolates, compared with subtype A pol isolates, is one possible factor for the observed difference in disease progression, and this association of pol RC with disease progression was independent of viral load. Polymerase gene amino acid polymorphisms potentially influence pol RC. Further in vitro studies examining the pol point mutations, and viral replication in various tissue types would further our understanding of the HIV-1 biology among non-B subtypes.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by a Singapore National Medical Research Training Fellowship Grant (EDG10nov070 to O. T. N.), the National Institutes of Health (NIH; NIDA R01 DA024565 to S. M.), and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–89. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Los Alamos Security. HIV databases. 2012 http://www.hiv.lanl.gov/ Accessed 13 March 2013. [Google Scholar]
- 3.Baeten JM, Chohan B, Lavreys L, et al. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195:1177–80. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 4.Keller M, Lu Y, Lalonde RG, Klein MB. Impact of HIV-1 viral subtype on CD4+ T-cell decline and clinical outcomes in antiretroviral naive patients receiving universal healthcare. AIDS. 2009;23:731–7. doi: 10.1097/QAD.0b013e328326f77f. [DOI] [PubMed] [Google Scholar]
- 5.Ng OT, Lin L, Laeyendecker O, et al. Increased rate of CD4+ T-cell decline and faster time to antiretroviral therapy in HIV-1 subtype CRF01_AE infected seroconverters in Singapore. PLoS One. 2011;6:e15738. doi: 10.1371/journal.pone.0015738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson KE, Costello C, Suriyanon V, Sennun S, Duerr A. Survival of blood donors and their spouses with HIV-1 subtype E (CRF01 A_E) infection in northern Thailand, 1992–2007. AIDS. 2007;21(Suppl 6):S47–54. doi: 10.1097/01.aids.0000299410.37152.17. [DOI] [PubMed] [Google Scholar]
- 7.Kiwanuka N, Laeyendecker O, Robb M, et al. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197:707–13. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 8.Daar ES, Kesler KL, Petropoulos CJ, et al. Baseline HIV type 1 coreceptor tropism predicts disease progression. Clin Infect Dis. 2007;45:643–9. doi: 10.1086/520650. [DOI] [PubMed] [Google Scholar]
- 9.Learmont JC, Geczy AF, Mills J, et al. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340:1715–22. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- 10.Barbour JD, Hecht FM, Wrin T, et al. Higher CD4+ T cell counts associated with low viral pol replication capacity among treatment-naive adults in early HIV-1 infection. J Infect Dis. 2004;190:251–6. doi: 10.1086/422036. [DOI] [PubMed] [Google Scholar]
- 11.Goetz MB, Leduc R, Wyman N, et al. HIV replication capacity is an independent predictor of disease progression in persons with untreated chronic HIV infection. J Acquir Immune Defic Syndr. 2010;53:472–9. doi: 10.1097/QAI.0b013e3181cae480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiwanuka N, Robb M, Kigozi G, et al. Knowledge about vaccines and willingness to participate in preventive HIV vaccine trials: a population-based study, Rakai, Uganda. J Acquir Immune Defic Syndr. 2004;36:721–5. doi: 10.1097/00126334-200406010-00009. [DOI] [PubMed] [Google Scholar]
- 13.Eshleman SH, Laeyendecker O, Parkin N, et al. Antiretroviral drug susceptibility among drug-naive adults with recent HIV infection in Rakai, Uganda. AIDS. 2009;23:845–52. doi: 10.1097/QAD.0b013e328327957a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoelscher M, Dowling WE, Sanders-Buell E, et al. Detection of HIV-1 subtypes, recombinants, and dual infections in east Africa by a multi-region hybridization assay. AIDS. 2002;16:2055–64. doi: 10.1097/00002030-200210180-00011. [DOI] [PubMed] [Google Scholar]
- 15.Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–8. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe SS, Stawiski E, Parkin NT. Interpretation of drug-susceptibility and replication-capacity results from subtype C HIV-1 Protease/RT is not influenced by the subtype of the resistance test vector. Antivir Ther. 2007;12:S118. [Google Scholar]
- 17.Kiwanuka N, Robb M, Laeyendecker O, et al. HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J Acquir Immune Defic Syndr. 2010;54:180–4. doi: 10.1097/QAI.0b013e3181c98fc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Institute for Statistic and Mathematics. The R project for statistical computing. 2012 http://www.r-project.org. Accessed 4 February 2013. [Google Scholar]
- 19.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–30. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue L, Prentice HA, Farmer P, et al. Cumulative impact of host and viral factors on HIV-1 viral load control during early infection. J Virol. 2013;87:708–15. doi: 10.1128/JVI.02118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prince JL, Claiborne DT, Carlson JM, et al. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 2012;8:e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parera M, Fernandez G, Clotet B, Martinez MA. HIV-1 protease catalytic efficiency effects caused by random single amino acid substitutions. Mol Biol Evol. 2007;24:382–7. doi: 10.1093/molbev/msl168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabana M, Fernandez G, Parera M, Clotet B, Martinez MA. Catalytic efficiency and phenotype of HIV-1 proteases encoding single critical resistance substitutions. Virology. 2002;300:71–8. doi: 10.1006/viro.2002.1520. [DOI] [PubMed] [Google Scholar]
- 25.Redd AD, Laeyendecker O, Kong X, et al. Efficiency of CCR5 coreceptor utilization by the HIV quasispecies increases over time, but is not associated with disease progression. AIDS Res Hum Retroviruses. 2012;28:289–94. doi: 10.1089/aid.2011.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.