Summary
Inducible gene expression based upon Tet repressor (tet regulation) is a broadly applied tool in molecular genetics. In its original environment, Tet repressor (TetR) negatively controls tetracycline (tc) resistance in bacteria. In the presence of tc, TetR is induced and detaches from its cognate DNA sequence tetO, so that a tc antiporter protein is expressed. In this article, we provide a comprehensive overview about tet regulation in bacteria and illustrate the parameters of different regulatory architectures. While some of these set‐ups rely on natural tet‐control regions like those found on transposon Tn10, highly efficient variations of this system have recently been adapted to different Gram‐negative and Gram‐positive bacteria. Novel tet‐controllable artificial or hybrid promoters were employed for target gene expression. They are controlled by regulators expressed at different levels either in a constitutive or in an autoregulated manner. The resulting tetsystems have been used for various purposes. We discuss integrative elements vested with tc‐sensitive promoters, as well as tet regulation in Gram‐negative and Gram‐positive bacteria for analytical purposes and for protein overproduction. Also the use of TetR as an in vivo biosensor for tetracyclines or as a regulatory device in synthetic biology constructs is outlined. Technical specifications underlying different regulatory set‐ups are highlighted, and finally recent developments concerning variations of TetR are presented, which may expand the use of prokaryotic tet systems in the future.
Introduction
Genome sequencing of microorganisms has become a routinely used procedure and resulted in more than 300 completely sequenced bacterial genomes up to date (http://www.tigr.org). It is one of the major challenges of the post‐genomic era to extract useable information from this vast amount of sequence data by determining the function of proteins that have no ortho‐ or paralogues with known activities. Indeed, approximately one‐fourth of the open reading frames (ORFs) of the best‐studied bacterium Escherichia coli encode proteins whose function is still unknown (Richmond et al., 1999). In general, this is the case for about 30–40% of the ORFs found in bacterial genomes. A widely used approach for delineating gene–function relationships is to create a deletion mutant and to determine the resulting phenotype. Obviously, this method is restricted to non‐essential genes and, more importantly, also fails for the analysis of genes that are critical under specific growth conditions, e.g. for intracellular growth. On the other hand, both essential genes and genes that need to be expressed for pathogenicity are preferred targets for developing novel anti‐infectives because it is assumed that bacteria cannot easily develop target modification‐based resistance against antibiotics acting on such vital functions. It is anticipated that some genes with unknown functions encode factors that could eventually be attacked by new classes of antibiotics.
In fact, there is an increasing need to combat bacterial infections with newly developed drugs, because the commonly used ones are more and more rapidly incapacitated by resistance development in pathogenic bacteria. These circumstances have generated substantial efforts directed at the elaboration of new high‐throughput methods to determine gene function, e.g. by conditional gene silencing so that the impact of the encoded proteins can be studied under various conditions. In general, there is no shortage of bacterial gene regulation systems that have successfully been exploited for that purpose. However, the optimal inducible regulation system for targetvalidation and evaluation should be able to reveal much more information than just whether a gene product is essential or not. A regulatory system allowing graded expression will yield information about the threshold amount of an essential protein necessary for survival, thereby defining the minimal inhibitory activity of a potential drug acting on this protein. This information might be a pivotal advantage for selecting a target for drug development. A regulatory system allowing graded expression in in vivo models, e.g. mice infected with pathogenic bacteria, could mimic drug activity by adjusting target protein expression to various levels. Thereby the effect of a potential drug on disease symptoms can be revealed or possible side‐effects originating from the physiological response of the pathogen to a drug, or the survival properties of the pathogen under treatment conditions can be monitored.
Regulation of bacterial genes within the infected host or host cells can only be accomplished if the effector for the regulatory system is readily applicable in mammals, is not or only slowly metabolized by the host and is taken up by the mammalian cells to reach intracellular pathogens. Tetracycline‐dependent gene regulatory systems fulfil these requirements. Tetracycline can slowly diffuse across natural and artificial membranes and hence can passively penetrate most cells (reviewed in Berens and Hillen, 2003). Furthermore, as tc is being widely used as a drug since the mid‐1950s, its pharmacokinetics and slow metabolization rate in mammals are well established (Chopra and Roberts, 2001). For an efficient gene regulation system, it would additionally be desirable if one could turn a gene on or off at will within a large regulatory window as some proteins may only be needed in minute amounts while others must be highly expressed to fully exert their biological function. All of these advantageous properties are combined in tc‐dependent gene regulation (tet regulation).
The widespread use of tet regulation in the eukaryotic kingdom, in particular for the control of mammalian genes by means of TetR fused to mammalian transcription factor domains like VP16 for transactivation (Gossen and Bujard, 1992) or KRAB for transsilencing (Deuschle et al., 1995), is an impressive demonstration of the versatility of this gene regulation system (summarized in Berens and Hillen, 2003). In this review we describe the properties of tet regulation pertinent to the above outlined special tasks, present an overview of various applications of TetR in prokaryotes and finally review the attempts to expand the potential applications by engineering regulator–effector pairs with novel properties.
The origin of tet regulation
Resistance to the antibiotic tc in Gram‐negative bacteria is mostly brought about by proton‐dependent antiport of the drug accomplished by the membrane‐residing TetA protein (reviewed in Chopra and Roberts, 2001). The conserved genetic organization and sequence variants of this regulatory system have been summarized in Hillen and Berens (1994). Currently, there are 14 sequence variants known which are widespread among the eubacteria, occurring in 35 genera covering five of 24 phyla (Berens and Hillen, 2004; Agerso and Guardabassi, 2005; Thompson et al., 2007). The common genetic structure consists of the tetA gene encoding the membrane‐spanning tc antiporter which is under transcriptional control of the tc‐responsive Tet repressor TetR encoded by the divergently oriented tetR gene. TetR is an all α‐helical protein and is active as a homodimer both in its DNA‐bound and in its induced state (Fig. 1); detailed information on the TetR structures has been reviewed (Saenger et al., 2000). As tc blocks protein biosynthesis, its inducer potency must clearly surpass its inhibitory capacity, hence induction for these determinants has had to acquire a remarkable sensitivity. Owing to the fact that even low level expression of the membrane protein TetA is disadvantageous for bacterial cells as demonstrated for E. coli (Nguyen et al., 1989), repression of this gene is very efficient. This rarely found combination of low basal expression with efficient induction is the result of the thermodynamic properties of the underlying regulatory reactions. The specificity and efficiency of regulation result from a high binding constant of TetR to tetO, while at the same time the affinity for non‐operator sequences is rather low compared with other repressors (Berens and Hillen, 2003). This favourable affinity ratio for binding of specific over non‐specific DNA sequences implies that TetR can exert efficient regulation in organisms with much larger genomes than E. coli. Furthermore, these properties lead to a high occupancy of tetO and, thus, contribute to efficient repression. In addition, the distinct genetic organization of these determinants as studied extensively for the tc resistance determinant tet(B) found on transposon Tn10 contributes to the efficiency of regulation. The detailed genetic arrangement is displayed in Fig. 2 and indicates that tetR and tetA are separated by a total of three promoters and two tet operators (tetO1 and tetO2) in their intergenic region. The functional analysis of the two operators revealed that both of them can be bound independently by TetR, and that occupation of tetO1 inhibits tetR and tetA transcription, whereas the occupation of tetO2 represses only the expression of the exporter protein (Meier et al., 1988). Notably, the affinity of tetO2 for TetR is about twice larger than that for tetO1. TetR‐mediated regulation of tc export has been reviewed (Grkovic et al., 2002).
Figure 1.
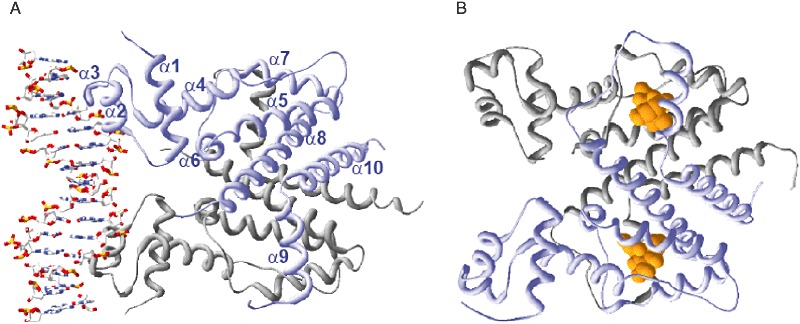
A. Structure of the TetR–tetO complex as determined by Orth and colleagues (2000). TetR is depicted in a ribbon representation with one monomer coloured grey and the other coloured blue. The numbers of helices are given in the blue monomer. Note that the linker sequence between helices α8 and α9 of both monomers is not resolved. The bound DNA is depicted as a ball‐and‐stick model. B. Structure of TetR in the tc‐bound form according to Hinrichs and colleagues (1994). Representations are as in (A). Tetracycline is given as orange spheres, with two molecules bound to the two inducer‐binding pockets of TetR.
Figure 2.

Sequence of the tetR–tetA intergenic region of Tn10 according to Chalmers and colleagues (2000). The three promoters are depicted as thin arrows with the –35 and the –10 regions symbolized by black boxes. The arrow tips mark the transcription start points. Squares indicate tetO sequences, and the black or grey filled arrows indicate the start codons of tetR or tetA respectively.
tet regulation of Tn10 obeys a closed‐loop logic of continuous control (Batchelor et al., 2004) and it has been previously assumed that autoregulation of TetR serves the purpose of preventing induction when the TetR level may be diluted by cell division. While this is still a valid consideration, more recent data from artificially constructed regulatory circuits indicate that negative autoregulation results in a large decrease of the response time after administration of the inducer (Rosenfeld et al., 2002). In the light of these results, it is very well possible that autoregulation of tetR also contributes to the high sensitivity of induction by small amounts of drug and to less heterogeneity in tetA expression in the affected cell population (Golding et al., 2005). The tetO2operator exclusively controlling tetA expression would then be important for keeping the induced expression of tetA at a moderate level so that toxicity of this protein would not counteract protection from tetracyclines. Thus, shaped by the outlined evolutionary pressure, gene regulation mediated by TetR combines the seemingly contradictory features of tight repression and sensitive induction.
The use of Tn10 and transposon‐derived elements to obtain tc‐regulatable transcriptional fusions
Creating gene disruptions in bacteria is often achieved using transposons (Hayes, 2003), which also frequently exert polar effects on adjacent genes. For Tn10, it had been realized early on that, while inactivating the gene located at the insertion site, this element also confers tc‐inducible transcription of regions located upstream or downstream of that site (Kleckner et al., 1978). The induced expression levels, however, are usually quite low, thus limiting the use of full‐length Tn10 to generate mutants with tc‐dependent phenotypes. Deletion variants of Tn10 have been constructed and resulted in much higher efficiency of tc‐inducible expression. A mini‐Tn10, basically consisting only of the tetR/tetA divergon, and the terminal sequences of the flanking IS10 elements (Way et al., 1984), was used to obtain tc‐inducible auxotrophs in E. coli (Takiff et al., 1992). An improved version of this Tn10deletion mutant, called T‐POP (Rappleye and Roth, 1997), found use in E. coli and Salmonella for gene analysis. A T‐POP‐controlled flagella master operon in Salmonella enterica serovar Typhi served to analyse the temporal expression of components of the flagella motor from that operon after tc induction. A general search for conditional auxotrophic mutants in E. coli, using a random insertion library of T‐POP, has resulted in four strains with the desired phenotype (Hidalgo et al., 2004), while an E. coli strain deleted for the porin‐encoding tsx gene has been randomly mutagenized by T‐POP to identify tc‐controllable suppressors (Bucarey et al., 2005). Taken together, these Tn10 derivatives with enhanced tc‐dependent outward transcription have proven to be useful in E. coli and its close relative Salmonella. Both elements do not bear transposase genes, but are mobilized by transposase encoded within the bacteria.
A completely different approach towards insertional mutagenesis was pioneered by Reznikoff and colleagues. Their method involves the in vitro assembly of a purified hyperactive mutant transposase derived from transposon Tn5 and DNA fragments containing optimized transposase recognition sites at both ends. A ternary complex formed by two molecules of transposase and one linear DNA fragment (called a transposome) can efficiently insert into target DNA in vitro or be transferred via electroporation into E. coli (Goryshin et al., 2000). Upon integration of the DNA, genetically stable insertion mutants are obtained because the transposase is encoded neither on the integrative element nor elsewhere in the target cell and hence cannot remove the integrated DNA from the insertion locus at a later time point. This approach has been utilized to generate mutants with tc‐dependent phenotypes by placing a modified tetA promoter on an insertion element termed InsTetG–1 (Köstner et al., 2006). Electroporation of InsTetG–1 transposomes into E. coli or S. enterica serovar Typhimurium that express TetR resulted in tc‐inducible auxotrophic or lethal strains. A similar transposome approach has also been used to obtain Bacillus subtilisstrains with such phenotypes by random insertion. To this end, different variants of integrative elements, designated InsTetG+, were constructed which harbour an outward‐directed tc‐responsive promoter suitable for B. subtilis. TetR was encoded either on the elements themselves, on a plasmid or on the chromosome (Bertram et al., 2005).
Utilization of tet expression systems in Gram‐negative bacteria
The first plasmid‐based tc‐controlled expression system used a tetA–lacZ translational fusion as reporter (de la Torre et al., 1984). While this group exploited the native tetA promoter of Tn10, Lutz and Bujard (1997) developed a synthetic tet‐sensitive promoter, termed PLtetO‐1, composed of the phage λ PL promoter with tetO sequences replacing λ cI repressor binding sites. The efficiency of this set‐up was compared with two differently regulated expression systems in E. coli, and the excellent regulatory properties of tet controlled gene expression were quantitatively established. Among the regulatory systems tested, the tet set‐up produced the largest regulatory window with about 5000‐fold induction of gene expression using anhydrotetracycline (atc) as an inducer and the lowest basal expression level estimated to amount to only one mRNA molecule per three E. coli cells in the repressed state. These results demonstrated clearly that tet‐based regulatory systems would have the potential to control the expression of genes encoding poisonous products and proteins which exert their biological function with only a few copies per cell. It should be noted, however, that accomplishing such low expression levels in the reduced state requires careful adjustment of the regulatory system with respect to the ratio of TetR molecules to the copy number of regulated promoter(s) in the cell (see below). This work also highlighted the use of luciferase as a reporter enzyme which is useful for determining the repressed state expression level, because very few copies of the protein already lead to a detectable enzymatic activity.
These results paved the way for several applications specifically requiring low expression levels of a desired target protein. In particular, the property of tight repression achievable by tet regulation has been exploited for conditional expression of Flp recombinase or the meganuclease I‐Sce, to obtain E. coli strains in which site‐specific recombination, or DNA restriction events, respectively, depended strictly on the addition of an inducer for TetR (Pósfai et al., 1994; 1997; 1999). These studies also clearly demonstrated that tet regulation is able to repress the expression of enzymes mediating irreversible genetic modifications to levels completely alleviating their activities. This is further highlighted by a tet controlled two‐stage expression system in which a cloned gene needs to be inverted by Flp recombinase so that the reading frame becomes attached to the promoter (Sektas and Szybalski, 1998). Flp recombinase in turn is provided under tet control resulting in a well‐regulated all‐or‐none expression system. The tight repression observed in the work described above is also due to the high affinity and specificity of TetR for tetO. The latter characteristic is very much supported by the remarkably low affinity of TetR for non‐cognate DNA, thereby reducing loss of regulator molecules by binding to unspecific, non‐tetO sites on chromosomal DNA (reviewed in Hillen and Berens, 1994). This highly specific DNA binding was exploited for the construction of E. coli strains containing multiple tetO repeats in various sections of the chromosome and expressing a TetR–eYfp fusion protein retaining binding activity for tetO, so that the assembly of this fusion protein on the chromosome became observable by confocal microscopy (Possoz et al., 2006). These authors demonstrated clearly that binding of this repressor–reporter fusion to DNA leads to a block of replication at several sites of the chromosome, which could be relieved by administering an inducer for TetR.
tet controlled expression of rpoH was used to determine the σ32 regulon and to examine the degradation characteristics of this alternative sigma factor of E. coli (Zhao et al., 2005). To this end an E. coli strain with tetR ectopically integrated into the chromosome was used. A similar strain has been exploited for expression of inhibitory single‐stranded DNA (ssDNA) to block ftsZ expression (Tan et al., 2004). It turned out that induction of ssDNA synthesis led not only to the loss of FtsZ activity but also stopped cell growth. Hence, the authors concluded that FtsZ would be a validated target for the development of anti‐infectives. This work was followed up by expressing a randomly constructed ssDNA library under tet control to screen for essential genes in E. coli. This approach was validated by the fact that inhibition of RNA polymerase activity via induction of a respective ssDNA fragment resulted in growth arrest (Tan and Chen, 2005). A tet‐regulation system has been used to control expression of an antigen from Plasmodium falciparum in S. enterica serovar Typhi for human vaccine production (Qian and Pan, 2002). Remarkably, it has also been demonstrated that a respective Salmonella strain can be induced in the liver and spleen of infected mice, where the majority of the bacteria retained the expression plasmid for at least 14 days. Recently, a shuttle vector with a tet controlled expression system has been constructed for E. coli and the recently identified bacterium Laribacter hongkongensis which is associated with gastroenteritis (Woo et al., 2005). Using two common reporters, Gfp and glutathione S‐transferase, it has been demonstrated clearly that tet regulation was efficiently possible in both bacteria using one common set‐up. Finally, tet regulation was established in Vibrio cholerae to identify genes which are differentially expressed dependent on growth in vitro or during infection of mice. This was achieved by random chromosomal integration of promoterless tetR which, upon insertion downstream of an active promoter, led to repression of a tet controlled gfp gene (Hsiao et al., 2006).
tet regulation for phage display systems and production of secreted proteins
tet controlled transcriptional regulation was exploited for the expression of secreted Fab fragments of antibodies by constructing a generally applicable multicopy expression vector with tight repression exerted by TetR (Skerra, 1994). The results of this study showed very low background expression levels in the uninduced state and efficient induction of Fab fragment expression in the presence of atc and subsequent secretion of the product into the periplasm of several E. coli hosts. This approach has been applied to the expression of several antibody fragments, which gave rise to high yields of the desired proteins (Schiweck et al., 1997; Griep et al., 1999). Accordingly, the expression of so‐called ‘anticalin’ mutant pools of lipocalin‐like folded proteins with variable ligand‐binding specificities can efficiently be controlled by a similar tet system (Beste et al., 1999).
Indeed, tet control showed a distinct general advantage for the construction of mutant libraries for screening purposes, e.g. in phage display (Daugherty et al., 1999; Zahn et al., 1999), where the choice of expression system is crucial for the maintenance of the diversity in mutant libraries in E. coli because the property of the expressed protein often has a pronounced effect on the growth rate of the respective cell. The fastest growth rate is usually exhibited by cells that do not produce any protein from the library. Hence, they would outgrow candidate cells unless repression is tight enough to avoid all library imposed growth effects. This is an important point because any screening protocol will only enrich positive candidates, may it be done by SELEX, cell sorting or any other method. The following expansion of the candidate pool suffers from potential growth effects resulting in the worst case in elimination of slow‐growing candidates. Hence, the complexity in tightly controlled mutant banks remains much larger as compared with less tightly controlled ones. Tightly regulated tet expression systems were promoted for the isolation of mutants with desired properties from degenerated peptide libraries by phage display (Paschke et al., 2001; Vogt and Skerra, 2004). A general comparison of tet regulation with other systems for heterologous protein expression can be found in a recent review (Terpe, 2006).
tet regulation in Gram‐positive bacteria
While export‐mediated tc resistance is always tightly regulated by TetR in Gram‐negative bacteria, the frequently occurring tet(K) determinant in Staphylococcus aureus lacks a TetR variant (reviewed in Chopra and Roberts, 2001). TetR‐mediated transgene regulation has been established in Gram‐positive bacteria by meeting the different promoter requirements of these organisms to ensure both proper expression of the regulator and adequate regulation of a target gene. Although the conserved –10 and –35 regions of σA promoters match with the ones of their σ70 counterparts of E. coli (Helmann, 1995), the B. subtilis RNA polymerase shows stricter discrimination. Furthermore, B. subtilis promoters often need so‐called –16 regions to function efficiently (Voskuil and Chambliss, 1998; Camacho and Salas, 1999). Accordingly, an unmodified Tn10‐based regulatory system was not functional in B. subtilis. To circumvent this obstacle, the B. subtilis xylA promoter was equipped with one or two tetoperator sequences in a construct termed Pxyl/tet, which could be regulated in B. subtilis by tetR placed in divergent orientation, driven by a modified autoregulated promoter termed P* (Geissendörfer and Hillen, 1990). This novel plasmid‐based expression system marked the beginning of tet regulation in Gram‐positive bacteria. The need for efficient and reliable target validation methods in pathogenic Gram‐positive bacteria provided the motivation for applying this system in S. aureus and different streptococci. A comparison between Pxyl/tetand two other inducible systems in S. aureus (Zhang et al., 2000) was followed by a thorough characterization of tet regulation using Pxyl/tet controlled gfp expression. These studies established that tc control is effective not only when used in bacteria cultured in vitro, but can also be applied to eukaryotic cell cultures for intracellular bacteria, and even worked well with bacteria in animal models for infectious diseases (Bateman et al., 2001). Pxyl/tet has also been used to overexpress a small untranslated RNA, SR1, to gain insights into its function (Licht et al., 2005).
An improved tet‐regulation system was recently established, which produced much higher regulation factors in B. subtilis. This was achieved via enhanced, constitutive expression of TetR (Kamionka et al., 2005). It is expected that this strategy would also improve tetregulation in other Gram‐positive bacteria of low G+C content.
A different mode of tet regulation has often been used in S. aureus, in which a target gene is conditionally downregulated by dosed expression of antisense RNA employing tc‐sensitive promoters. The result of this approach is the tc‐induced silencing of a gene, as opposed to induction of expression. The same goal can nowadays also be accomplished using TetR variants, called revTetR, which require tc to bind to tetO (see below for details). The antisense method is particularly well suited for regulation of essential genes which are left untouched in their original genetic locus, but can thus nevertheless be regulated in trans. The efficiency of tet controlled antisense RNA regulation has been demonstrated for the hla gene, assumed to play an important role in pathogenicity (Ji et al., 1999). The tc‐induced knockdown of hla expression completely eliminated lethality of an S. aureus infection in mice, thereby also establishing that tet regulation can be effectively used to control the expression of genes from staphylococci in mammalian infection models. This study was soon followed by the comprehensive analysis of essential genes in S. aureus using a tet controlled random pool of antisense RNA, where essential genes for infection have also been characterized in infected mice (Ji et al., 2001).
tet‐regulated antisense approaches have been applied for further studies of S. aureus genetics: the importance of a two‐component regulatory system has been established, which turned out to be essential and, when expressed at lower levels, increased the organism's susceptibility for phosphomycin (Sun et al., 2005). An essential glycoprotease has been identified (Zheng et al., 2005), which has recently been characterized (Zheng et al., 2007), and tet controlled antisense‐mediated knockdown of a polypeptide deformylase resulted in an increased sensitivity for an antibacterial compound targeting this enzyme (Yang et al., 2006). It is noteworthy that in B. subtilis direct and antisense control exerted by Pxyl/tet leads to similar repressed expression levels as demonstrated by Western blots directed against HPr kinase/phosphorylase (HPrK/P), a central player in carbon catabolite regulation in B. subtilis, that was regulated using both approaches (Bertram et al., 2006). Even the lowest accomplishable expression level did not yield the phenotype of an HPrK/P knockout mutant. This result indicates that some genes, presumably especially the ones encoding regulatory functions, can exert their phenotypes at very low expression levels. The tet controlled antisense RNA strategy for target identification, validation and mechanism of action has been thoroughly reviewed (Yin and Ji, 2002).
Pxyl/tetregulation has also been utilized for the construction of genetic tools for strain construction in S. aureus. Two methods were introduced, one allowing for allelic replacement, thereby yielding regulated expression of essential genes (Fan et al., 2001), and another enabling selection against the episomal state of integrative plasmids (Bae and Schneewind, 2006). Targets from the bacterial fatty acid biosynthesis enzymes have been expressed in a tet controlled manner in S. aureus, and these strains have been used to adjust so‐called sensitized phenotypes for the evaluation of drug candidates. A sensitized phenotype results from adjusting the expression level of the target protein below the wild‐type level which increases the effect of a potential drug directed against this target, thereby increasing the number of lead compounds derived from such a screen. This approach has been extensively used to characterize a variety of inhibitors of bacterial fatty acid synthesis (Ling et al., 2004) and to confirm the mode of action of novel compounds in whole‐cell assays (Ji et al., 2004). In addition to S. aureus, streptococcal species have been the preferred bacteria for application of the tet system. The effect of an eukaryotic‐type Ser/Thr kinase has been evaluated in pathogenic group B streptococci (Rajagopal et al., 2005) by using tet controlled expression of that kinase for complementation of knockouts. tet regulation has been found to be superior to other regulatory systems for setting up a conditional knockout strategy for identifying essential genes in Streptococcus mutans(Wang and Kuramitsu, 2005). While these two studies made use of the fact that Pxyl/tetis also active in streptococci, two different tc‐regulatable promoters have been developed for Streptococcus pneumoniae(Stieger et al., 1999). These constructs were subsequently used to adjust the expression levels of target genes in that species by tet regulation (Ulijasz et al., 2004).
The last years have also seen the successful adaptation of tc‐dependent expression systems for the analysis of genes in mycobacteria including the pathogenic strain Mycobacterium tuberculosis. The application in this organism included antisense control in free‐living and intracellular bacteria (Blokpoel et al., 2005), the construction of conditional lethal knockouts of ftsZ by a tc‐dependent promoter leading to a more than 100‐fold induction (Ehrt et al., 2005), and the construction of a tc‐addressable conditional auxotroph containing trpD under tet control in single copy, which resulted in tight control of expression (Carroll et al., 2005). Furthermore, tet‐regulated expression constructs for use in Streptomycetes have been described and yielded about 270‐fold induction in members of that genus (Rodriguez‐Garcia et al., 2005).
Use of TetR for bacterial biosensors monitoring tc
The high affinity of tc and its derivatives for TetR combined with the sensitive induction of reporter gene expression has been used to construct bacterial biosensor strains to detect tc in various environments. Whole‐cell biosensors based on tet‐dependent expression of Gfp in E. coli have been used for detection and quantification of tetracyclines in the rat intestine (Bahl et al., 2004). When the tet(M) resistance determinant against tetracyclines was also introduced into that reporter strain, it resulted in a remarkable expansion of the concentration range of tetracyclines that could be detected (Bahl et al., 2005). In a similar approach, an E. coli strain expressing Gfp in a tet‐dependent manner was used to track oxytetracycline synthesis by Streptomyces rimosus in sterile soil samples (Hansen et al., 2001). This whole‐cell biosensor may also be equipped with different reporter genes encoding β‐galactosidase (β‐gal) or luciferase. Such constructs can be transferred to other Gram‐negative bacteria by conjugation, and respective E. coli strains were useful for determining oxytetracycline in milk or pork serum (Kurittu et al., 2000).
Use of TetR for synthetic biology
The rapidly developing area of synthetic biology attempts to develop quantitative models for complex regulatory circuits in living cells on the basis of quantitative interpretations of simple building blocks from which such complex regulation may be constructed in the future. Currently, artificial genetic circuits are constructed mostly in E. coli or yeast, using well‐established transcription control systems in various combinations. The most often used bacterial promoters in such circuits are the lambda pl/pr promoters which are supplied with either their natural lambda operators, or lac operator or tetoperator, so that they respond to various inducers. TetR is frequently used in these constructs for the reproducible adjustment of different expression levels of other repressors or activators. It was found that TetR with a C‐terminal tag enforcing degradation can be well suited for synthetic biology approaches involving time‐resolved experiments (Elowitz and Leibler, 2000; Guet et al., 2002), while in a different study a TetR single‐amino‐acid exchange mutant for decreased operator binding, which was furthermore tagged by eGfp at the C‐terminus, was used (Becskei and Serrano, 2000). In fact, several further Gfp proteins were fused to TetR, while both portions of the fusion construct retained their activity (Michaelis et al., 1997; Rosenfeld et al., 2002; Lau et al., 2003). Detailed information about engineered gene regulatory networks in general and synthetic biological circuits involving TetR can be found in some excellent reviews (Kaern et al., 2003; McDaniel and Weiss, 2005; Sprinzak and Elowitz, 2005).
Overview of bacterial tet regulation architectures and parameters of induction
tetregulation in bacteria requires expression of a TetR variant and a promoter equipped with tetO. The native arrangement in Tn10, in which the tetR and tetA promoters PR1, PR2 and PA overlap, share tetO sequences (Fig. 2) and, hence, mutually influence each other (Hillen and Berens, 1994), has been exploited in several studies (de la Torre et al., 1984; Way et al., 1984; Chopra et al., 1990; Takiff et al., 1992; Pósfai et al., 1994; 1997; 1999; Rappleye and Roth, 1997; Sektas and Szybalski, 1998; Sektas et al., 1999; Hansen and Sorensen, 2000; Karlinsey et al., 2000; Hansen et al., 2001; Bahl et al., 2004; 2005; Hidalgo et al., 2004; Bucarey et al., 2005). One study has made use of the tet(Z) determinant found in Corynebacterium glutamicum (Blokpoel et al., 2005).
The P*/Pxyl/tetsystem is an adaptation of this intertwined bidirectional promoter/operator arrangement to accommodate the stricter sequence requirements for promoters in Gram‐positive bacteria (Geissendörfer and Hillen, 1990). This set‐up involves autoregulation of tetR expression and has also been applied in Gram‐positive species of low G+C content from the genera Bacillus, Staphylococcus and Streptococcus (Ji et al., 1999; 2001; 2004; Zhang et al., 2000; Bateman et al., 2001; Fan et al., 2001; Bertram et al., 2005; 2006; Carroll et al., 2005; Licht et al., 2005; Rajagopal et al., 2005; Sun et al., 2005; Wang and Kuramitsu, 2005; Zheng et al., 2005; Bae and Schneewind, 2006; Yang et al., 2006), as well as for the Gram‐negative bacterium Laribacter hongkongensis (Woo et al., 2005). Using the strong PxylA of B. subtilis or synthetic promoters to drive tetRexpression increased the regulatory window obtained with Pxyl/tet(Bertram et al., 2005; Kamionka et al., 2005). Several examples indicate that tetR autoregulation does not generally offer an advantage for regulation efficiency in transgenic set‐ups; for example, the tet(Z) determinant used in mycobacteria displayed considerable leakiness in the repressed state (Blokpoel et al., 2005) and, accordingly, P*/Pxyl/tet controlled complementation of a serine/threonine kinase occurred even in the absence of inducer (Rajagopal et al., 2005). Compounds like atc combine a reduced antibiotic activity with increased induction and hence can be administered in higher concentrations to obtain full induction even with high TetR amounts. In fact, uncoupling tetR expression from the regulated promoter has yielded some of the largest regulatory windows with tetR expressed from a transcriptional fusion with β‐lactamase (Skerra, 1994) or by phage promoters (Lutz and Bujard, 1997; Qian and Pan, 2002; Tan et al., 2004; Tan and Chen, 2005; Rodriguez‐Garcia et al., 2005).
Numerous tet controlled promoters have been described, which differ in the number and positioning of tetO sequences. Bacillus subtilis promoters with two tetO sites can be repressed stronger than those with one operator (Kamionka et al., 2005), albeit at the cost of incomplete induction in case of P*/Pxyl/tet (Geissendörfer and Hillen, 1990; Bertram et al., 2005). The only promoter with three tetO sites has been used in streptomycetes (Rodriguez‐Garcia et al., 2005). In the often used PLtetO‐1 promoter, the two tet operators flank the –35 site (Lutz and Bujard, 1997), while they bracket the –10 region in PtetA and Pxyl/tet. Figure 3 summarizes the arrangements of tetR and the respective target gene with respect to vicinity and episomal or chromosomal location and shows which ones were used in various studies. In addition, Fig. 4 schematically depicts some most efficient and/or most often applied promoters for target gene expression with corresponding induction factors that have been determined.
Figure 3.
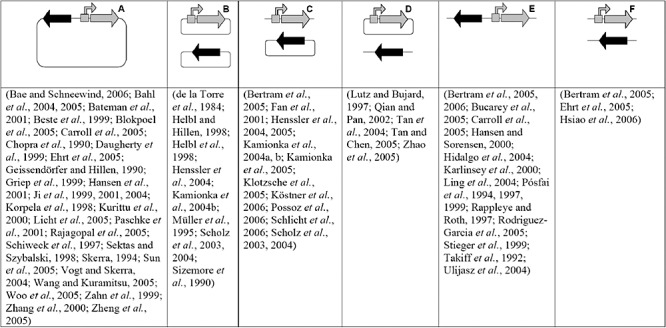
Different architectures of tet‐regulation system are shown. Depicted are tetR (black arrow) and the target gene (grey arrow), controlled by a tc‐sensitive promoter (grey bent arrow with grey box symbolizing tetO). Situations are: (A) target gene and tetR on one plasmid; (B) target gene and tetR on two distinct plasmids; (C) target gene and tetR on chromosome; (D) target gene on plasmid, tetR on chromosome; (E) target gene and tetR on chromosome, adjacent; (F) target gene and tetR on chromosome, distinct loci. Remarks: Several studies applied more than one architecture. The articles by Pósfai and colleagues (1994; 1997; 1999) describe the use of plasmids which were subsequently integrated into the genome. Possoz and colleagues (2006) applied a plasmid expressing TetR for non‐transcriptional regulation. Bae and Schneewind (2006) used tet regulation to counterselect for the episomal state of the DNA. In contrast to the drawing in the head of the figure, Rodriguez‐Garcia and colleagues (2005) placed tetR collinear to, and hence uncoupled from, the target gene, which is also true for the construct of Skerra (1994) and derivatives thereof. The tet‐regulation vector developed by Woo and colleagues (2005) has only approximately one copy per Laribacter cell.
Figure 4.
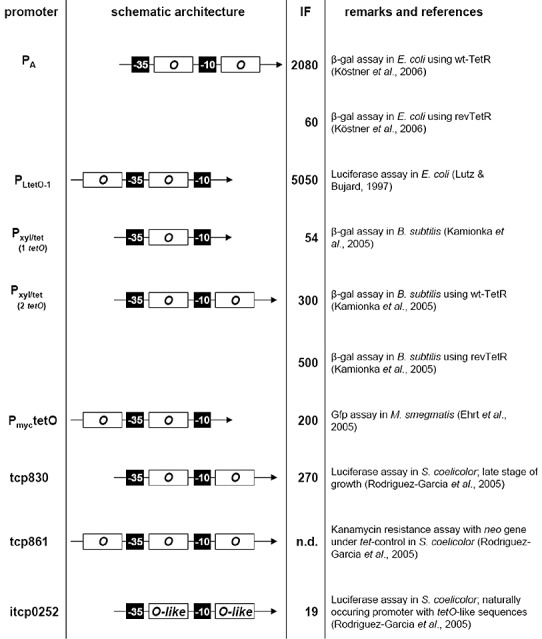
Representation of selected promoters for tetregulation. The architecture is schematically depicted and not drawn to scale. –35 and –10 denote the respective base‐pair hexamers of the promoters. ‘O’ designates tet operator. The indicated approximated induction factors (IF) were achieved under circumstances briefly outlined at the right side. n.d., not determined.
For relief of repression, the predominant inducer of tet‐regulated genes in literature is tc, followed by atc, while also further tc derivatives were assayed for TetR induction in various bacteria (Chopra et al., 1990; Korpela et al., 1998; Kurittu et al., 2000; Blokpoel et al., 2005). Doxycycline (dox), the preferred compound for mammalian tet systems, is rarely the compound of choice (Wang and Kuramitsu, 2005), probably due to antibiotic activity. One means to overcome growth inhibition imposed by tc is the expression of suitable resistance factors which allow for administration of otherwise harmful concentrations of the drug (Takiff et al., 1992; Karlinsey et al., 2000; Bahl et al., 2005; Yang et al., 2006). In turn, it then takes much higher tc concentrations for maximal expression of a tet‐controlled gene (Chopra et al., 1990). Some early applications of tet regulation in E. coliused a heat‐inactivated chlortetracycline stock solution for induction (Pósfai et al., 1994; 1997; 1999; Sektas and Szybalski, 1998; Sektas et al., 1999). This treatment probably leads to rapid degradation of the drug, and we suspect that trace amounts are converted to atc, which has a decreased antibiotic activity and is a much more potent inducer of TetR (Degenkolb et al., 1991). These properties have made atc the preferred inducer for tet controlled bacterial expression systems nowadays. It must be kept in mind, however, that atc decomposes upon irradiation with light leading to rapid changes in inducer concentration at longer exposure times.
A commonly applied concentration of tc as inducers is 200 ng ml–1 (equalling ~0.5 µM); however, tet regulation offers doseable expression dependent on inducer concentration, as demonstrated by Chopra and colleagues (1990), Griep and colleagues (1999) and Tan and colleagues (2004) in E. coli. Similar analyses confirming doseability of tet regulation were conducted with staphylococci (Zhang et al., 2000; Bateman et al., 2001; Fan et al., 2001; Sun et al., 2005; Zheng et al., 2005) and mycobacteria (Blokpoel et al., 2005; Carroll et al., 2005; Ehrt et al., 2005). The advantageous pharmacokinetic properties of tc also enable the induction of intracellular bacteria within eukaryotic cell cultures (Bateman et al., 2001; Blokpoel et al., 2005; Ehrt et al., 2005), or even in infected mice and rats (Ji et al., 1999; 2001; Bateman et al., 2001; Qian and Pan, 2002; Bahl et al., 2004) (see above), where the drug is injected or given orally via drinking water. Although the inducer concentration which actually reaches the bacterial cells within the hosts cannot easily be determined, relative dose–response correlations have been established (Bateman et al., 2001; Ji et al., 2001; Bahl et al., 2004).
Concerning induction time and the duration of the drug's exposure, Skerra (1994) found an increase in Fab fragment production expressed via a tet system 1 h after induction. A similar response time was determined with S. enterica serovar Typhimurium cells harbouring a tet‐controlled flagellar master operon, which gained motility approximately 45 min after addition of tc (Karlinsey et al., 2000), which, according to the timescale, is consistent with early findings by de la Torre and colleagues (1984). Various studies have confirmed a graded time response dependence of tet‐inducible transcription (Ji et al., 1999; Zhang et al., 2000; Bateman et al., 2001; Tan et al., 2004; Ehrt et al., 2005; Sun et al., 2005; Tan and Chen, 2005; Wang and Kuramitsu, 2005; Woo et al., 2005; Zhao et al., 2005; Zheng et al., 2005). Interestingly, Possoz and colleagues (2006) could demonstrate impressively that as early as 5 min after addition of atc, cells with TetR bound to tetO arrays on the chromosome were relieved from stalling in a non‐replicative state.
Contrasting to the induction time, a reversal of an induced phenotype was obtained 4 h after the inducer was washed out of the cells (Hidalgo et al., 2004). To quickly achieve both on‐ and off‐states, it would hence be desirable to have a toggle‐switch architecture at hand. Such a set‐up has been described, exploiting TetR and LacI that mutually control each other and a target gene (Gardner et al., 2000). It is also conceivable to express both wt‐TetR and one of some recently constructed revTetR variants, which display a stringently different inducer preference (see below), to rapidly switch between the on‐ and the off‐state of a target gene by administering different tc derivatives.
Modifications of tet regulation: revTetR and TetR mutants employing different effectors and operators
Most applications of tet regulation in bacteria have used the wt‐TetR variant from Tn10, called TetR(B). However, genetic and thermodynamic analyses have revealed that a chimera, designated TetR(BD) containing the DNA reading head of TetR(B) and the protein core of TetR(D) (encoded by the Salmonella plasmid RA1), exhibits enhanced stability and better regulatory properties (Schnappinger et al., 1998). The stability of TetR is further enhanced in a monomeric single‐chain TetR protein called scTetR, which has been constructed by fusing the C‐terminus of the first monomer to the N‐terminus of the second monomer by means of a flexible polypeptide linker. This variant retains all activities in E. coli (Kamionka et al., 2006), and fused to a eukaryotic read‐out domain also in eukaryotes (Krueger et al., 2003). Genetic stability of the sctetR gene is assured by employing alternative codons in one‐half of the gene thereby reducing the similarity between the two tetR sequences.
Much effort has been put into the understanding and engineering of TetR interactions with DNA or effectors. tetO sequences changed in one base pair of each palindromic half‐side (Fig. 5A) were used to develop two TetR(B) variants with altered operator preferences (Helbl and Hillen, 1998; Helbl et al., 1998). Recently, another TetR mutant has been developed, which specifically recognizes an operator with two altered positions in each palindromic half‐site (M. Krueger, O. Scholz, S. Wisshak, and W. Hillen, unpubl. results). As a result, TetR variants can bind to promoters with mutant tet operators to repress different downstream genes in the same cell. To accomplish independent regulation of these genes, the respective TetR variants must furthermore be specific for distinct inducers. TetR is not induced by tc derivatives lacking the 4‐dimethylamino grouping like 4‐de‐dimethylamino‐atc (Fig. 5B); however, in vitro evolution has yielded a triple mutant of TetR, which is induced exclusively by this compound and not by atc, tc or dox (Henssler et al., 2004).
Figure 5.

A. Depiction of tetO2 and two derived operator variants with single‐base‐pair exchanges in each half‐side. B. Chemical structure of tc (with key carbon atoms numbered), atc and 4‐de‐dimethylamino‐atc. C. Regulation principle of wt‐TetR (top) and revTetR (bottom). wt‐TetR is depicted with black ovals representing the protein core and grey ovals symbolizing DNA reading heads. revTetR is depicted accordingly with hatched fillings. A tet controlled target gene is repressed (grey arrow) or induced (white arrow), dependent on the absence or presence of effector molecules (white triangles) bound to TetR.
Independent tet regulation of two genes in one prokaryotic cell has been accomplished by employing TetR variants with such different characteristics. To this end, two TetR variants differing (i) in their dimerization domain sequence to prevent heterodimerization [classes (BD) versus (B) (Schnappinger et al., 1998)], (ii) in their DNA binding site specificity (tetO versus tetO‐4C) and (iii) in their inducer specificity (4‐de‐dimethylamino‐atc versus atc) were expressed in the same E. coli cell, yielding selective regulation of two reporter genes (Kamionka et al., 2004b). Furthermore, a novel gain of function scTetR variant with two different inducer‐binding pockets requires both of these compounds for induction, indicating that TetR detaches from tetO only when two inducer molecules are bound (Kamionka et al., 2006).
More than 100 revTetR variants display enhanced tetO binding in the presence of tetracyclines (Fig. 5C). Some of these novel regulators have regulatory properties of similar efficiencies as wt‐TetR (Kamionka et al., 2004a; Scholz et al., 2004); however, their exploitation in prokaryotes is only beginning. revTetR variants have been used to regulate the InsTetG–1 element in E. coli (Köstner et al., 2006), InsTetG+ variants in B. subtilis(Bertram et al., 2005), and in another B. subtilis architecture revTetR displayed at least the same regulatory efficiency as wt‐TetR (Kamionka et al., 2005). It was recently demonstrated that the different phenotypic alterations of shifted operator and inducer preference and reverse behaviour of TetR variants can functionally be combined in novel regulators, which, e.g. only in the presence of 4‐de‐dimethylamino‐atc bind to tetO (Bertram et al., 2004; Henssler et al., 2005). A dodecapeptide, termed Tip (transcription‐inducing peptide), isolated by phage display has pronounced inducing capacities for TetR(B) (Klotzsche et al., 2005). This is the first known inducer for TetR belonging to a different class of substances. A properly constructed insertion element created random translational fusions with Tip in E. coli. The expression levels of the Tip fusion proteins could be quantified by tet‐regulated reporter gene expression (Schlicht et al., 2006).
Conclusions
tet regulation has proven very useful for inducible transgene expression in both eukaryotes and prokaryotes. Its usefulness concerning the latter group of organisms is reflected not only by spanning a considerable repertoire of species, ranging from E. coli to high G+C Gram‐positive bacteria, but also by the published and ongoing development of tet‐regulation systems for diverse applications. It seems likely that we will witness both the elaboration of new and even more efficient set‐ups as well as the adaptation of established systems to further bacteria, such as Chlamydiae (Dugan et al., 2004). Exploiting Tet repressors with different modes of allostery or ligand‐binding specificities (or both) will pave the way for multigene regulation set‐ups, and, indeed, first steps have already been taken in this direction (Kamionka et al., 2004b). Further developments are under way and one can imagine that novel tet systems will be tailored for any given task in any prokaryotic organism of choice.
Acknowledgments
We thank Drs Eva‐Maria Henßler for preparation of Fig. 1 and Chris Berens for fruitful discussions and critically reading of the manuscript. Work in the authors’ lab was supported by the Deutsche Forschungsgemeinschaft through SFB 473, by the VolkswagenStiftung and by the Fonds der Chemischen Industrie.
Note added in proof
While finishing this article, we became aware of further recent studies employing tetR in bacteria. Wright et al. (Cell Microbiol, 2007, doi:10.1111/j.1462‐5822.2007.00952.x) used the tet‐regulatory region of a commercially available E. coli strain to construct a conditional fim mutant and to analyse the effects of pili on biofilm formation of a uropathogenic E. coli variant. Also Da Re et al. (Appl Environ Microbiol, 2007, 73, 3391–3403) studied the effects of regulated genes promoting biofilm formation in E. coli applying integrative sequences, comparable to InsTet elements described above. According to personal communication with Y. Zhang, tet‐regulation has been introduced into the Gram‐negative bacterium Photorhabdus luminescens for induction of Red/ET recombineering. Gründling and Schneewind integrated an autoregulated tetR gene and a divergently oriented Pxyl/tet promoter into the S. aureus chromosome to express genes involved in lipoteichoic acid synthesis (J Bacteriol, 2007, 189, 2521–2530 and Proc Natl Acad Sci USA, 2007, 104, 8478–8483). Tahlan et al. (Mol Microbiol, 2007, 64, 951–961) made use of synthetic promoters carrying tetO to drive expression of lux genes in an E. coli tc‐biosensor strain. Guo et al. (J Bacteriol, 2007, 189, 4614–4623) established gene regulation in Mycobacterium smegmatis via revTetR, as proven by conditional expression of secA1, essential for in vitro growth.
References
- Agerso Y., Guardabassi L. Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J Antimicrob Chemother. 2005;55:566–569. doi: 10.1093/jac/dki051. [DOI] [PubMed] [Google Scholar]
- Bae T., Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter‐selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Bahl M.I., Hansen L.H., Licht T.R., Sorensen S.J. In vivo detection and quantification of tetracycline by use of a whole‐cell biosensor in the rat intestine. Antimicrob Agents Chemother. 2004;48:1112–1117. doi: 10.1128/AAC.48.4.1112-1117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl M.I., Hansen L.H., Sorensen S.J. Construction of an extended range whole‐cell tetracycline biosensor by use of the tet(M) resistance gene. FEMS Microbiol Lett. 2005;253:201–205. doi: 10.1016/j.femsle.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Batchelor E., Silhavy T.J., Goulian M. Continuous control in bacterial regulatory circuits. J Bacteriol. 2004;186:7618–7625. doi: 10.1128/JB.186.22.7618-7625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman B.T., Donegan N.P., Jarry T.M., Palma M., Cheung A.L. Evaluation of a tetracycline‐inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect Immun. 2001;69:7851–7857. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A., Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Berens C., Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur J Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- Berens C., Hillen W. Gene regulation by tetracyclines. Genet Eng (N Y) 2004;26:255–277. doi: 10.1007/978-0-306-48573-2_13. [DOI] [PubMed] [Google Scholar]
- Bertram R., Kraft C., Wisshak S., Mueller J., Scholz O., Hillen W. Phenotypes of combined tet repressor mutants for effector and operator recognition and allostery. J Mol Microbiol Biotechnol. 2004;8:104–110. doi: 10.1159/000084565. [DOI] [PubMed] [Google Scholar]
- Bertram R., Köstner M., Müller J., Vazquez Ramos J., Hillen W. Integrative elements for Bacillus subtilis yielding tetracycline‐dependent growth phenotypes. Nucleic Acids Res. 2005;33:e153. doi: 10.1093/nar/gni154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram R., Wünsche A., Sprehe M., Hillen W. Regulated expression of HPrK/P does not affect carbon catabolite repression of the xyn operon and of rocG in Bacillus subtilis. FEMS Microbiol Lett. 2006;259:147–152. doi: 10.1111/j.1574-6968.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- Beste G., Schmidt F.S., Stibora T., Skerra A. Small antibody‐like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc Natl Acad Sci USA. 1999;96:1898–1903. doi: 10.1073/pnas.96.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokpoel M.C., Murphy H.N., O'Toole R., Wiles S., Runn E.S., Stewart G.R. Tetracycline‐inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005;33:e22. doi: 10.1093/nar/gni023. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucarey S.A., Villagra N.A., Martinic M.P., Trombert A.N., Santiviago C.A., Maulen N.P. The Salmonella enterica serovar Typhi tsx gene, encoding a nucleoside‐specific porin, is essential for prototrophic growth in the absence of nucleosides. Infect Immun. 2005;73:6210–6219. doi: 10.1128/IAI.73.10.6210-6219.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho A., Salas M. Effect of mutations in the ‘extended –10’ motif of three Bacillus subtilis sigmaA‐RNA polymerase‐dependent promoters. J Mol Biol. 1999;286:683–693. doi: 10.1006/jmbi.1998.2526. [DOI] [PubMed] [Google Scholar]
- Carroll P., Muttucumaru D.G., Parish T. Use of a tetracycline‐inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl Environ Microbiol. 2005;71:3077–3084. doi: 10.1128/AEM.71.6.3077-3084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R., Sewitz S., Lipkow K., Crellin P. Complete nucleotide sequence of Tn10. J Bacteriol. 2000;182:2970–2972. doi: 10.1128/jb.182.10.2970-2972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Hacker K., Misulovin Z., Rothstein D.M. Sensitive biological detection method for tetracyclines using a tetA–lacZ fusion system. Antimicrob Agents Chemother. 1990;34:111–116. doi: 10.1128/aac.34.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty P.S., Olsen M.J., Iverson B.L., Georgiou G. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. 1999;12:613–621. doi: 10.1093/protein/12.7.613. [DOI] [PubMed] [Google Scholar]
- Degenkolb J., Takahashi M., Ellestad G.A., Hillen W. Structural requirements of tetracycline–Tet repressor interaction: determination of equilibrium binding constants for tetracycline analogs with the Tet repressor. Antimicrob Agents Chemother. 1991;35:1591–1595. doi: 10.1128/aac.35.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Meyer W.K., Thiesen H.J. Tetracycline‐reversible silencing of eukaryotic promoters. Mol Cell Biol. 1995;15:1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan J., Rockey D.D., Jones L., Andersen A.A. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv‐like gene. Antimicrob Agents Chemother. 2004;48:3989–3995. doi: 10.1128/AAC.48.10.3989-3995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S., Guo X.V., Hickey C.M., Ryou M., Monteleone M., Riley L.W., Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M.B., Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Fan F., Lunsford R.D., Sylvester D., Fan J., Celesnik H., Iordanescu S. Regulated ectopic expression and allelic‐replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus. Plasmid. 2001;46:71–75. doi: 10.1006/plas.2001.1526. et al. [DOI] [PubMed] [Google Scholar]
- Gardner T.S., Cantor C.R., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Geissendörfer M., Hillen W. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl Microbiol Biotechnol. 1990;33:657–663. doi: 10.1007/BF00604933. [DOI] [PubMed] [Google Scholar]
- Golding I., Paulsson J., Zawilski S.M., Cox E.C. Real‐time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Goryshin I.Y., Jendrisak J., Hoffman L.M., Meis R., Reznikoff W.S. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat Biotechnol. 2000;18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline‐responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep R.A., Van Twisk C., Kerschbaumer R.J., Harper K., Torrance L., Himmler G. pSKAP/S: an expression vector for the production of single‐chain Fv alkaline phosphatase fusion proteins. Protein Expr Purif. 1999;16:63–69. doi: 10.1006/prep.1999.1041. et al. [DOI] [PubMed] [Google Scholar]
- Grkovic S., Brown M.H., Skurray R.A. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guet C.C., Elowitz M.B., Hsing W., Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- Hansen L.H., Sorensen S.J. Detection and quantification of tetracyclines by whole cell biosensors. FEMS Microbiol Lett. 2000;190:273–278. doi: 10.1111/j.1574-6968.2000.tb09298.x. [DOI] [PubMed] [Google Scholar]
- Hansen L.H., Ferrari B., Sorensen A.H., Veal D., Sorensen S.J. Detection of oxytetracycline production by Streptomyces rimosus in soil microcosms by combining whole‐cell biosensors and flow cytometry. Appl Environ Microbiol. 2001;67:239–244. doi: 10.1128/AEM.67.1.239-244.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. Transposon‐based strategies for microbial functional genomics and proteomics. Annu Rev Genet. 2003;37:3–29. doi: 10.1146/annurev.genet.37.110801.142807. [DOI] [PubMed] [Google Scholar]
- Helbl V., Hillen W. Stepwise selection of TetR variants recognizing tet operator 4C with high affinity and specificity. J Mol Biol. 1998;276:313–318. doi: 10.1006/jmbi.1997.1540. [DOI] [PubMed] [Google Scholar]
- Helbl V., Tiebel B., Hillen W. Stepwise selection of TetR variants recognizing tet operator 6C with high affinity and specificity. J Mol Biol. 1998;276:319–324. doi: 10.1006/jmbi.1997.1539. [DOI] [PubMed] [Google Scholar]
- Helmann J.D. Compilation and analysis of Bacillus subtilis sigma A‐dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssler E.M., Scholz O., Lochner S., Gmeiner P., Hillen W. Structure‐based design of Tet repressor to optimize a new inducer specificity. Biochemistry. 2004;43:9512–9518. doi: 10.1021/bi049682j. [DOI] [PubMed] [Google Scholar]
- Henssler E.M., Bertram R., Wisshak S., Hillen W. Tet repressor mutants with altered effector binding and allostery. FEBS J. 2005;272:4487–4496. doi: 10.1111/j.1742-4658.2005.04868.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo A.A., Trombert A.N., Castro‐Alonso J.C., Santiviago C.A., Tesser B.R., Youderian P., Mora G.C. Insertions of mini‐Tn10 transposon T‐POP in Salmonella enterica sv. typhi. Genetics. 2004;167:1069–1077. doi: 10.1534/genetics.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen W., Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- Hinrichs W., Kisker C., Düvel M., Müller A., Tovar K., Hillen W., Saenger W. Structure of the Tet repressor–tetracycline complex and regulation of antibiotic resistance. Science. 1994;264:418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- Hsiao A., Liu Z., Joelsson A., Zhu J. Vibrio cholerae virulence regulator‐coordinated evasion of host immunity. Proc Natl Acad Sci USA. 2006;103:14542–14547. doi: 10.1073/pnas.0604650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Marra A., Rosenberg M., Woodnutt G. Regulated antisense RNA eliminates alpha‐toxin virulence in Staphylococcus aureus infection. J Bacteriol. 1999;181:6585–6590. doi: 10.1128/jb.181.21.6585-6590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Zhang B., Van S.F., Horn W., Woodnutt P., Burnham G. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. et al. [DOI] [PubMed] [Google Scholar]
- Ji Y., Yin D., Fox B., Holmes D.J., Payne D., Rosenberg M. Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol Lett. 2004;231:177–184. doi: 10.1016/S0378-1097(03)00931-5. [DOI] [PubMed] [Google Scholar]
- Kaern M., Blake W.J., Collins J.J. The engineering of gene regulatory networks. Annu Rev Biomed Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- Kamionka A., Bogdanska‐Urbaniak J., Scholz O., Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004a;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamionka A., Sehnal M., Scholz O., Hillen W. Independent regulation of two genes in Escherichia coli by tetracyclines and Tet repressor variants. J Bacteriol. 2004b;186:4399–4401. doi: 10.1128/JB.186.13.4399-4401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamionka A., Bertram R., Hillen W. Tetracycline‐dependent conditional gene knockout in Bacillus subtilis. Appl Environ Microbiol. 2005;71:728–733. doi: 10.1128/AEM.71.2.728-733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamionka A., Majewski M., Roth K., Bertram R., Kraft C., Hillen W. Induction of single chain tetracycline repressor requires the binding of two inducers. Nucleic Acids Res. 2006;34:3834–3841. doi: 10.1093/nar/gkl316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey J.E., Tanaka S., Bettenworth V., Yamaguchi S., Boos W., Aizawa S.I., Hughes K.T. Completion of the hook‐basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Barker D.F., Ross D.G., Botstein D. Properties of the translocatable tetracycline‐resistance element Tn10 in Escherichia coli and bacteriophage lambda. Genetics. 1978;90:427–461. doi: 10.1093/genetics/90.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzsche M., Berens C., Hillen W. A peptide triggers allostery in tet repressor by binding to a unique site. J Biol Chem. 2005;280:24591–24599. doi: 10.1074/jbc.M501872200. [DOI] [PubMed] [Google Scholar]
- Korpela M.T., Kurittu J.S., Karvinen J.T., Karp M.T. A recombinant Escherichia coli sensor strain for the detection of tetracyclines. Anal Chem. 1998;70:4457–4462. doi: 10.1021/ac980740e. [DOI] [PubMed] [Google Scholar]
- Köstner M., Schmidt B., Bertram R., Hillen W. Generating tetracycline‐inducible auxotrophy in Escherichia coli and Salmonella enterica serovar Typhimurium by using an insertion element and a hyperactive transposase. Appl Environ Microbiol. 2006;72:4717–4725. doi: 10.1128/AEM.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger C., Berens C., Schmidt A., Schnappinger D., Hillen W. Single‐chain Tet transregulators. Nucleic Acids Res. 2003;31:3050–3056. doi: 10.1093/nar/gkg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurittu J., Karp M., Korpela M. Detection of tetracyclines with luminescent bacterial strains. Luminescence. 2000;15:291–297. doi: 10.1002/1522-7243(200009/10)15:5<291::AID-BIO596>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lau I.F., Filipe S.R., Soballe B., Okstad O.A., Barre F.X., Sherratt D.J. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Licht A., Preis S., Brantl S. Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in Bacillus subtilis. Mol Microbiol. 2005;58:189–206. doi: 10.1111/j.1365-2958.2005.04810.x. [DOI] [PubMed] [Google Scholar]
- Ling L.L., Xian J., Ali S., Geng B., Fan J., Mills D.M. Identification and characterization of inhibitors of bacterial enoyl‐acyl carrier protein reductase. Antimicrob Agents Chemother. 2004;48:1541–1547. doi: 10.1128/AAC.48.5.1541-1547.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R., Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1‐I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel R., Weiss R. Advances in synthetic biology: on the path from prototypes to applications. Curr Opin Biotechnol. 2005;16:476–483. doi: 10.1016/j.copbio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Meier I., Wray L.V., Hillen W. Differential regulation of the Tn10‐encoded tetracycline resistance genes tetA and tetR by the tandem tet operators O1 and O2. EMBO J. 1988;7:567–572. doi: 10.1002/j.1460-2075.1988.tb02846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Müller G., Hecht B., Helbl V., Hinrichs W., Saenger W., Hillen W. Characterization of non‐inducible Tet repressor mutants suggests conformational changes necessary for induction. Nat Struct Biol. 1995;2:693–703. doi: 10.1038/nsb0895-693. [DOI] [PubMed] [Google Scholar]
- Nguyen T.N., Phan Q.G., Duong L.P., Bertrand K.P., Lenski R.E. Effects of carriage and expression of the Tn10 tetracycline‐resistance operon on the fitness of Escherichia coli K12. Mol Biol Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- Orth P., Schnappinger D., Hillen W., Saenger W., Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor‐operator system. Nat Struct Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- Paschke M., Zahn G., Warsinke A., Höhne W. New series of vectors for phage display and prokaryotic expression of proteins. Biotechniques. 2001;30:720–724, 726. [PubMed] [Google Scholar]
- Pósfai G., Koob M., Hradecná Z., Hasan N., Filutowicz M., Szybalski W. In vivo excision and amplification of large segments of the Escherichia coli genome. Nucleic Acids Res. 1994;22:2392–2398. doi: 10.1093/nar/22.12.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Koob M.D., Kirkpatrick H.A., Blattner F.R. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Kolisnychenko V., Bereczki Z., Blattner F.R. Markerless gene replacement in Escherichia coli stimulated by a double‐strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possoz C., Filipe S.R., Grainge I., Sherratt D.J. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor‐bound DNA in vivo. EMBO J. 2006;25:2596–2604. doi: 10.1038/sj.emboj.7601155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian F., Pan W. Construction of a tetR‐integrated Salmonella enterica serovar Typhi CVD908 strain that tightly controls expression of the major merozoite surface protein of Plasmodium falciparum for applications in human vaccine production. Infect Immun. 2002;70:2029–2038. doi: 10.1128/IAI.70.4.2029-2038.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal L., Vo A., Silvestroni A., Rubens C.E. Regulation of purine biosynthesis by a eukaryotic‐type kinase in Streptococcus agalactiae. Mol Microbiol. 2005;56:1329–1346. doi: 10.1111/j.1365-2958.2005.04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye C.A., Roth J.R. A Tn10 derivative (T‐POP) for isolation of insertions with conditional (tetracycline‐dependent) phenotypes. J Bacteriol. 1997;179:5827–5834. doi: 10.1128/jb.179.18.5827-5834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond C.S., Glasner J.D., Mau R., Jin H., Blattner F.R. Genome‐wide expression profiling in Escherichia coli K‐12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Garcia A., Combes P., Perez‐Redondo R., Smith M.C. Natural and synthetic tetracycline‐inducible promoters for use in the antibiotic‐producing bacteria Streptomyces. Nucleic Acids Res. 2005;33:e87. doi: 10.1093/nar/gni086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N., Elowitz M.B., Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Saenger W., Orth P., Kisker C., Hillen W., Hinrichs W. The tetracycline repressor – a paradigm for a biological switch. Angew Chem Int Ed Engl. 2000;39:2042–2052. doi: 10.1002/1521-3773(20000616)39:12<2042::aid-anie2042>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Schiweck W., Buxbaum B., Schätzlein C., Neiss H.G., Skerra A. Sequence analysis and bacterial production of the anti‐c‐myc antibody 9E10: the V(H) domain has an extended CDR‐H3 and exhibits unusual solubility. FEBS Lett. 1997;414:33–38. doi: 10.1016/s0014-5793(97)00983-6. [DOI] [PubMed] [Google Scholar]
- Schlicht M., Berens C., Daam J., Hillen W. Random insertion of a TetR‐inducing peptide tag into Escherichia coli proteins allows analysis of protein levels by induction of reporter gene expression. Appl Environ Microbiol. 2006;72:5637–5642. doi: 10.1128/AEM.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D., Schubert P., Pfleiderer K., Hillen W. Determinants of protein–protein recognition by four helix bundles: changing the dimerization specificity of Tet repressor. EMBO J. 1998;17:535–543. doi: 10.1093/emboj/17.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz O., Köstner M., Reich M., Gastiger S., Hillen W. Teaching TetR to recognize a new inducer. J Mol Biol. 2003;329:217–227. doi: 10.1016/s0022-2836(03)00427-3. [DOI] [PubMed] [Google Scholar]
- Scholz O., Henssler E.M., Bail J., Schubert P., Bogdanska‐Urbaniak J., Sopp S. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. et al. [DOI] [PubMed] [Google Scholar]
- Sektas M., Szybalski W. Tightly controlled two‐stage expression vectors employing the Flp/FRT‐mediated inversion of cloned genes. Mol Biotechnol. 1998;9:17–24. doi: 10.1007/BF02752694. [DOI] [PubMed] [Google Scholar]
- Sektas M., Gregorowicz M., Szybalski W. Transient conversion to RecA+ phenotype to permit P1 transduction in any Escherichia coli recA– strains. Biotechniques. 1999;27:911–914. doi: 10.2144/99275bm07. [DOI] [PubMed] [Google Scholar]
- Sizemore C., Wissmann A., Gülland U., Hillen W. Quantitative analysis of Tn10 Tet repressor binding to a complete set of tet operator mutants. Nucleic Acids Res. 1990;18:2875–2880. doi: 10.1093/nar/18.10.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene. 1994;151:131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- Sprinzak D., Elowitz M.B. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- Stieger M., Wohlgensinger B., Kamber M., Lutz R., Keck W. Integrational plasmids for the tetracycline‐regulated expression of genes in Streptococcus pneumoniae. Gene. 1999;226:243–251. doi: 10.1016/s0378-1119(98)00561-7. [DOI] [PubMed] [Google Scholar]
- Sun J., Zheng L., Landwehr C., Yang J., Ji Y. Identification of a novel essential two‐component signal transduction system, YhcSR, in Staphylococcus aureus. J Bacteriol. 2005;187:7876–7880. doi: 10.1128/JB.187.22.7876-7880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiff H.E., Baker T., Copeland T., Chen S.M., Court D.L. Locating essential Escherichia coli genes by using mini‐Tn10 transposons: the pdxJ operon. J Bacteriol. 1992;174:1544–1553. doi: 10.1128/jb.174.5.1544-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X.X., Chen Y. A novel genomic approach identifies bacterial DNA‐dependent RNA polymerase as the target of an antibacterial oligodeoxynucleotide, RBL1. Biochemistry. 2005;44:6708–6714. doi: 10.1021/bi0475626. [DOI] [PubMed] [Google Scholar]
- Tan X.X., Rose K., Margolin W., Chen Y. DNA enzyme generated by a novel single‐stranded DNA expression vector inhibits expression of the essential bacterial cell division gene ftsZ. Biochemistry. 2004;43:1111–1117. doi: 10.1021/bi035164h. [DOI] [PubMed] [Google Scholar]
- Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- Thompson S.A., Maani E.V., Lindell A.H., King C.J., McArthur J.V. A novel tetracycline resistance determinant isolated from an environmental strain of Serratia marcescens. Appl Environ Microbiol. 2007;73:2199–2206. doi: 10.1128/AEM.02511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre J.C., Ortín J., Domingo E., Delamarter J., Allet B., Davies J. Plasmid vectors based on Tn10 DNA: gene expression regulated by tetracycline. Plasmid. 1984;12:103–110. doi: 10.1016/0147-619x(84)90056-8. et al. [DOI] [PubMed] [Google Scholar]
- Ulijasz A.T., Andes D.R., Glasner J.D., Weisblum B. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J Bacteriol. 2004;186:8123–8136. doi: 10.1128/JB.186.23.8123-8136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M., Skerra A. Construction of an artificial receptor protein (‘anticalin’) based on the human apolipoprotein D. Chembiochem. 2004;5:191–199. doi: 10.1002/cbic.200300703. [DOI] [PubMed] [Google Scholar]
- Voskuil M.I., Chambliss G.H. The –16 region of Bacillus subtilis and other gram‐positive bacterial promoters. Nucleic Acids Res. 1998;26:3584–3590. doi: 10.1093/nar/26.15.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Kuramitsu H.K. Inducible antisense RNA expression in the characterization of gene functions in Streptococcus mutans. Infect Immun. 2005;73:3568–3576. doi: 10.1128/IAI.73.6.3568-3576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J.C., Davis M.A., Morisato D., Roberts D.E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Ma S.S., Teng J.L., Li M.W., Kao R.Y., Lau S.K., Yuen K.Y. Construction of an inducible expression shuttle vector for Laribacter hongkongensis, a novel bacterium associated with gastroenteritis. FEMS Microbiol Lett. 2005;252:57–65. doi: 10.1016/j.femsle.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Yang J., Zheng L., Ji Y. Confirmation of the mode of action of an antibacterial inhibitor using regulated antisense RNA. World J Microbiol Biotechnol. 2006;22:299–303. [Google Scholar]
- Yin D., Ji Y. Genomic analysis using conditional phenotypes generated by antisense RNA. Curr Opin Microbiol. 2002;5:330–333. doi: 10.1016/s1369-5274(02)00315-6. [DOI] [PubMed] [Google Scholar]
- Zahn G., Skerra A., Hohne W. Investigation of a tetracycline‐regulated phage display system. Protein Eng. 1999;12:1031–1034. doi: 10.1093/protein/12.12.1031. [DOI] [PubMed] [Google Scholar]
- Zhang L., Fan F., Palmer L.M., Lonetto M.A., Petit C., Voelker L.L. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene. 2000;255:297–305. doi: 10.1016/s0378-1119(00)00325-5. et al. [DOI] [PubMed] [Google Scholar]
- Zhao K., Liu M., Burgess R.R. The global transcriptional response of Escherichia coli to induced sigma 32 protein involves sigma 32 regulon activation followed by inactivation and degradation of sigma 32 in vivo. J Biol Chem. 2005;280:17758–17768. doi: 10.1074/jbc.M500393200. [DOI] [PubMed] [Google Scholar]
- Zheng L., Yang J., Landwehr C., Fan F., Ji Y. Identification of an essential glycoprotease in Staphylococcus aureus. FEMS Microbiol Lett. 2005;245:279–285. doi: 10.1016/j.femsle.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Zheng L., Yu C., Bayles K., Lasa I., Ji Y. Conditional mutation of an essential putative glycoprotease eliminates autolysis in Staphylococcus aureus. J Bacteriol. 2007;189:2734–2742. doi: 10.1128/JB.01806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


