Summary
Whole‐transcriptome analysis was used here for the first time in the rhizosphere to discern the genes involved in the pathogenic response of Pseudomonas aeruginosa PAO1 as well as to discern the response of the poplar tree. Differential gene expression shows that 185 genes of the bacterium and 753 genes of the poplar tree were induced in the rhizosphere. Using the P. aeruginosatranscriptome analysis, isogenic knockout mutants, and two novel plant assays (poplar and barley), seven novel PAO1 virulence genes were identified (PA1385, PA2146, PA2462, PA2463, PA2663, PA4150 and PA4295). The uncharacterized putative haemolysin repressor, PA2463, upon inactivation, resulted in greater poplar virulence and elevated haemolysis while this mutant remained competitive in the rhizosphere. In addition, disruption of the haemolysin gene itself (PA2462) reduced the haemolytic activity of P. aeruginosa, caused less cytotoxicity and reduced barley virulence, as expected. Inactivating PA1385, a putative glycosyl transferase, reduced both poplar and barley virulence. Furthermore, disrupting PA2663, a putative membrane protein, reduced biofilm formation by 20‐fold. Inactivation of PA3476 (rhlI) increased virulence with barley as well as haemolytic activity and cytotoxicity, so quorum sensing is important in plant pathogenesis. Hence, this strategy is capable of elucidating virulence genes for an important pathogen.
Introduction
Greater than 99% of all bacteria are found in biofilms (Sauer et al., 2004), and 80% of bacterial infections are caused by bacteria living in biofilms (Costerton, 2004); hence, discerning the genetic basis of disease caused by biofilms is important. However, discovering pathogenic genes with animal models can be expensive, time consuming and tedious (Prithiviraj et al., 2005). In addition, animal models often do not fully resemble all aspects of human disease caused by bacteria like Pseudomonas aeruginosa (Rahme et al., 2000) whereas the interactions of bacteria with plants are often similar to the interactions of bacteria with eukaryotes (Lugtenberg et al., 2002). Therefore, plant models can provide a fast, inexpensive and high‐throughput method for discovering bacterial virulence factors (Prithiviraj et al., 2005; Filiatrault et al., 2006).
Pseudomonas aeruginosa with its 6.3 million base pairs and 5570 open reading frames (ORFs) is one of the largest of the sequenced bacterial genomes (Stover et al., 2000). Pseudomonas aeruginosa is an opportunistic pathogen and causes urinary tract, respiratory tract and skin infections (Stover et al., 2000). Pseudomonas aeruginosa primarily causes nosocomial infections, and it is frequently resistant to commonly used antibiotics and disinfectants (Stover et al., 2000). Although this bacterium is well studied, roughly one‐fourth of its ORFs are uncharacterized (Lewenza et al., 2005).
Populus trichocarpa is a model woody plant due to its small genome size (520 Mb) (Tuskan et al., 2004) and routine transformation system mediated by Agrobacterium tumefaciens (Fillatti et al., 1987). The poplar genome encodes more than 45 000 putative protein‐coding genes (Tuskan et al., 2006), and the poplar genome project is expected to help identify tree‐specific characteristics such as those for wood formation, perennial crown development and distribution of water/nutrients over long distances (Tuskan et al., 2004). However, there is little known about bacterial pathogens in the poplar rhizosphere. Injury and tumour formation on poplar stems are caused by the bacterial pathogens Xanthomonas spp. and Agrobacterium spp., and enhanced resistance of poplar to these bacteria was achieved by engineering expression of the antimicrobial peptide D4E1 (Mentag et al., 2003). Although determinants of disease resistance are not well studied in woody plants, poplar has diverse disease resistance proteins that typically have nucleotidebinding sites and/or leucine‐rich repeats (LRR) in contrast to those in the model grass plants, Arabidopsis and rice (Tuskan et al., 2006). Hence, poplar is a good model for pathogenesis research of woody plants.
The pathogenicity of P. aeruginosais dependent on more than biofilms; other virulence factors include adhesins, haemagglutinin, protein toxins, phenazine and cyanide (Gallagher and Manoil, 2001). Pseudomonas aeruginosais a known colonizer of respiratory tract of cystic fibrosis patients (Potera, 1999) and causes disease in humans, animals, nematodes and insects (Lewenza et al., 2005); yet, it is not recognized as an important rhizosphere bacterium although it is frequently found in soils (Filiatrault et al., 2006) and has been found to colonize cucumber roots (Lugtenberg and Dekkers, 1999), lettuce leaves (Filiatrault et al., 2006), sweet basil roots (Walker et al., 2004), sugar beet roots (Mark et al., 2005), wheat roots (Lugtenberg and Dekkers, 1999) and Arabidopsis roots (Walker et al., 2004). We hypothesized that not only could P. aeruginosa survive in the rhizosphere but that it may be pathogenic to some trees. Here we show P. aeruginosa is virulent to poplars (it effectively kills the tree in 48 h) and is virulent to barley (prevents seed germination); hence, we used poplar trees and barley as model organisms to identify P. aeruginosapathogenic genes (two novel virulence assays were developed). Using DNA microarrays, the whole‐transcriptome response of the tree contacted both with and without P. aeruginosafor 12 h and the bacterium contacted with and without poplar tree roots for 2 days was determined. In addition, isogenic mutants were used to identify seven previously uncharacterized genes as pathogenic determinants.
Results
Pseudomonas aeruginosa is a poplar pathogen
As P. aeruginosais a pathogen for Arabidopsis(Walker et al., 2004), sweet basil (Walker et al., 2004) and lettuce (Filiatrault et al., 2006), and given our interest in rhizoremediation (Yee et al., 1998; Shim et al., 2000), we investigated whether this strain was a pathogen for poplar trees and then developed a wilting poplar tree assay to quantify PAO1 pathogenesis. As shown in Fig. 1, after 48 h, P. aeruginosaPAO1 killed poplar trees and caused a 16 ± 12‐fold increase in branch wilting. Poplar trees were contacted with both the wild‐type P. aeruginosaPAO1 from the University of British Columbia (P. aeruginosaPAO1‐UBC) and the wild‐type strain from the University of Washington (P. aeruginosaPAO1‐UW) as mutants from both universities were utilized (Fig. 1C and E respectively). However, colonization of poplar trees with the indigenous bacterium Pseudomonassp. strain Pb3‐1 (Shim et al., 2000) did not cause wilting in 2 days (Fig. 1B). In addition, poplar trees that were not inoculated with bacteria were not affected for up to 10 days (a poplar tree contacted in HRP medium without bacteria after 7 days is shown in Fig. 1A). Confocal microscopy was used to confirm the presence of P. aeruginosa on the growing poplar root (Fig. 1G). Evidence of a small number of GFP‐tagged pseudomonads was also found inside root sections after 48 h. We also investigated whether P. aeruginosaPAO1 or its metabolites kill the poplar trees by filter sterilizing the liquid from a flask that contained a poplar tree contacted with P. aeruginosaPAO1 for 2 days. The cell‐free supernatant did not kill the poplar trees after 3 days; therefore, the P. aeruginosa PAO1 cells themselves kill poplar trees.
Figure 1.

Poplar tree wilting after exposure to bacteria: (A) no bacteria after 7 days, (B) indigenous Pseudomonassp. strain Pb3‐1 after 48 h, (C) P. aeruginosaPAO1‐UBC after 48 h contact, (D) P. aeruginosaPAO1 PA2463‐UBC after 48 h contact, (E) P. aeruginosaPAO1‐UW after 48 h contact, (F) P. aeruginosaPAO1 PA1385‐UW after 48 h contact and (G) P. aeruginosaPAO1/pMRP9‐1 on poplar roots after 24 h visualized with confocal microscopy (scale bar represents 50 µm).
Differentially expressed P. aeruginosa genes upon infection
To determine the global transcriptome response to colonization of poplar trees by P. aeruginosa and to identify the genes required for poplar pathogenesis, we compared differential gene expression at 48 h for the pseudomonad on poplar roots versus the genes required for colonization of glass wool. In this way, only the genes required for pathogenesis and interaction with poplar trees are differentially expressed while the genes required for biofilm formation are not identified as the biofilm state was used for the pseudomonad both on poplar roots and on glass wool. Colonization of P. aeruginosa induced 185 genes greater than twofold change including genes for carbon compound metabolism, a colicin immunity protein, energy metabolism, membrane protein, fatty acid, phospholipid metabolism, motility and attachment genes (partial gene list shown in Table S1). Similarly, colonization of P. aeruginosa on poplar roots repressed 419 genes greater than twofold including those for adaptation, protection, amino acid biosynthesis, biosynthesis of cofactors, cell division, chemotaxis, flagella, motility, attachment, energy metabolism, membrane proteins, fatty acid, phospholipid metabolism, protein secretion, transcription and translation regulation (partial gene list shown in Table S2).
In comparison with glass wool, colonization on poplar roots induced expression of components of the type III secretion system in P. aeruginosa including PA1691 (pscT, a homologue of yscTin Yersinia pseudotuberculosis) (Bergman et al., 1994) and PA1718 (pscE) (Quinaud et al., 2005) (Table S1). Additional virulence genes were upregulated including PA1712 (exsB, an exoenzyme S synthesis protein secreted by the type III secretion system) (Yahr et al., 1996), PA1432 (lasI, an autoinducer synthesis protein) (Passador et al., 1993) and PA4540 (a putative haemolysin activator and homologue of hxuB in Haemophilus influenzae) (Cope et al., 1995). Furthermore, PA4295 was induced in our microarray data; this gene is close to the PA4296 virulence gene that was identified in the lettuce leaf model (Wagner et al., 2006) (Table S1). Hence, activation of these virulent factors is necessary for infection of the poplar tree by P. aeruginosa.
Exploration of pathogenesis via knockout mutants using poplar wilting and barley germination
To corroborate the DNA microarray results and to explore the proteins related to the differentially expressed genes, a series of P. aeruginosaisogenic knockout mutants were utilized: strains with mutations in genes PA0513, PA0984, PA1385, PA2146, PA2462, PA2463, PA2663, PA3278 (transposon mutation was inserted in the promoter region of PA3278 gene, hence, PA3278 function may not be completely disrupted), PA4150, PA4151, PA4153, PA4295 and PA4549. Two of the most induced genes (PA2461 and PA2459) are part of a putative haemolysin operon; hence, we investigated pathogenesis with isogenic mutations in PA2462 (putative haemolysin) and PA2463 (putative haemolysin regulator); note that the Affymetrix chip does not contain probes for the PA2463 gene. The other knockout mutants were investigated as they were part of the top 20 induced genes of P. aeruginosaPAO1 contacted with poplar trees and mutants were readily available. Quorum sensing has a significant role in bacterial colonization and virulence in bacteria (Gallagher et al., 2002; Chun et al., 2004); hence, strains with the PA0996 (pqsA), PA2587 (pqsH) and PA3476 (rhlI) inactivations were also evaluated. As shown in Table S3, knockout mutants were obtained from both the collections of the University of British Columbia (UBC) (Lewenza et al., 2005) and the University of Washington (UW) (Jacobs et al., 2003); each mutation was verified using four polymerase chain reactions (PCR) for each mutant and the wild‐type strain, and for all of the assays, each mutant was compared with its respective wild‐type strain.
The PA2463 mutant showed over twofold more wilting than the wild‐type strain in 48 h (Figs 1D and 2A); hence, it is more pathogenic than the wild‐type strain and appears to encode a haemolysin repressor. Conversely, the PA1385 mutant showed threefold less wilting in 48 h (Fig. 1F) and inactivation of PA0996 caused 2.5‐fold less wilting (Fig. 2A).
Figure 2.
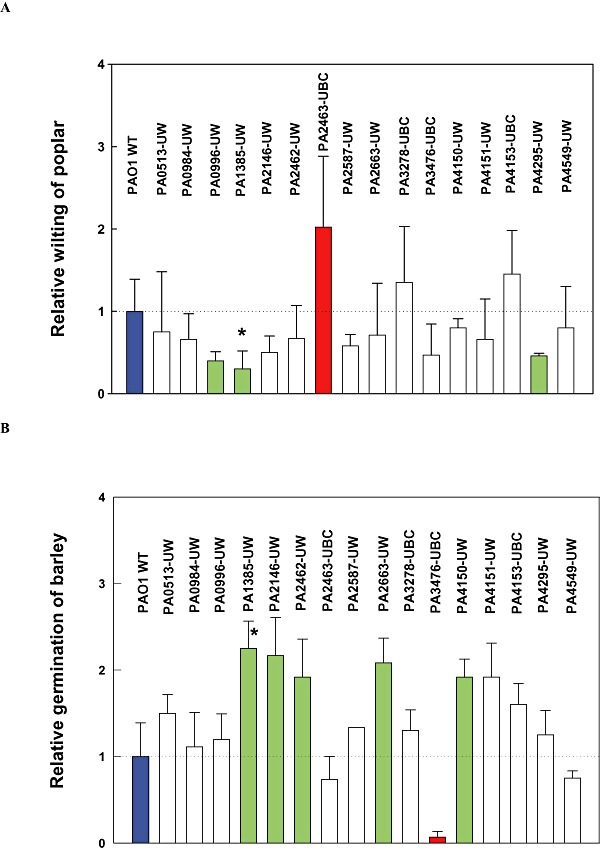
Pseudomonas aeruginosavirulence as indicated by poplar tree wilting (A) and inhibition of barley seed germination (B). Red indicates more virulence, green indicates less virulence and white indicates similar virulence compared with the wild‐type strain based on a t‐test. Poplars were grown at 22°C, and barley seeds were germinated at 25°C for 3 days (at least 45 seeds). Error bars indicate one standard deviation. Asterisk indicates the mutant showing similar trends in virulence for both poplar and barley.
To corroborate the poplar wilting assay, we tested the ability of P. aeruginosato prevent germination of barley seeds (also a novel assay for P. aeruginosa). Wild‐type PAO1 significantly reduced the germination of barley seeds to 20% (PAO1‐UW) and 33% (PAO1‐UBC), whereas 87% of the seeds germinated in the absence of P. aeruginosa, and Pb3‐1 did not inhibit germination. Hence, wild‐type P. aeruginosa is virulent not only to woody plants (e.g. poplar) but also is virulent to a grass (e.g. barley). The effect of the isogenic mutations on barley seed germination is shown in Fig. 2B. After 3 days, isogenic mutations that caused statistically less virulence included PA1385 (225% more germination), PA2146 (217%), PA2462 (192%), PA2663 (208%) and PA4150 (192%); therefore, the proteins encoded by these genes are important for plant pathogenesis. Conversely, quorum‐sensing mutant PA3476 (rhlI) was significantly more virulent to barley because only 7% of seeds could germinate after infection with this strain (Fig. 2B).
Rhizosphere competition of the knockout mutants
The knockout mutants were also tested for their ability to compete simultaneously with the wild‐type strain in the poplar rhizosphere. The more virulent PA2463 mutant and the PA2587 and PA4295 mutants showed statistically almost the same ability to compete with the wild type in the rhizosphere whereas the PA4153 mutant was 100‐fold less competitive (Table 1); hence, PA4153 encodes a protein that is very important for competition in the rhizosphere. As a control, the wild‐type strain (P. aeruginosaPAO1‐UW) with and without pMRP9‐1 was inoculated on four poplar trees with two different ratios of bacteria, and after 2 days, the ratio of bacteria found on the roots matched the starting ratio.
Table 1.
Competition (percentage of mutant cells on poplar tree roots versus the wild‐type strain), biofilm formation (relative to wild type), swimming motility and specific growth rate in LB medium for the P. aeruginosa PAO1 mutants.
| Strains | Competition (%) | Relative biofilm (LB) | Relative biofilm (LB glu) | Swimming motility (cm, 5 h) | Swimming motility (cm, 18 h) | Growth rate |
|---|---|---|---|---|---|---|
| Wild‐type‐UBC | NA | 1.0 ± 0.3 | 1.0 ± 0.2 | 0.24 ± 0.00 | 1.55 ± 0.06 | 0.89 ± 0.04 |
| PA2463 | 90 ± 30 | 1.0 ± 0.4 | 0.9 ± 0.6 | 0.16 ± 0.01 | 1.19 ± 0.08 | 1.03 ± 0.04 |
| PA3278 | 60 ± 6 | 1.1 ± 0.3 | 1.6 ± 0.3 | 0.19 ± 0.01 | 1.1 ± 0.3 | 0.89 ± 0.01 |
| PA3476 | 53 ± 14 | 1.8 ± 0.7 | 2.7 ± 0.7 | 0.19 ± 0.02 | 1.5 ± 0.1 | 0.93 ± 0.01 |
| PA4153 | 0.9 ± 0.4 | 1.1 ± 0.3 | 1.3 ± 0.4 | 0.20 ± 0.03 | 1.72 ± 0.07 | 0.89 ± 0.02 |
| Wild‐type‐UW | NA | 1.0 ± 0.2 | 1.0 ± 0.3 | 0.2 ± 0.1 | 1.2 ± 0.2 | 1.02 ± 0.05 |
| PA0513 | 42 ± 26 | 0.8 ± 0.1 | 0.8 ± 0.2 | 0.5 ± 0.1 | 2.1 ± 0.1 | 1.04 ± 0.01 |
| PA0984 | 21 ± 7 | 0.5 ± 0.1 | 0.7 ± 0.1 | 0.32 ± 0.05 | 1.72 ± 0.08 | 1.02 ± 0.06 |
| PA0996 | 67 ± 21 | 0.6 ± 0.1 | 0.7 ± 0.2 | 0.27 ±0.05 | 1.5 ± 0.2 | 1.02 ± 0.00 |
| PA1385 | 37 ± 19 | 0.7 ± 0.1 | 0.7 ± 0.2 | 0.28 ± 0.07 | 1.8 ± 0.2 | 1.07 ± 0.07 |
| PA2146 | 62 ± 21 | 0.7 ± 0.1 | 0.7 ± 0.2 | 0.36 ± 0.03 | 1.93 ± 0.03 | 1.0 ± 0.1 |
| PA2462 | 39 ± 5 | 0.5 ± 0.1 | 0.6 ± 0.1 | 0.30 ± 0.05 | 1.6 ± 0.2 | 0.99 ± 0.01 |
| PA2587 | 88 ± 36 | 0.6 ± 0.1 | 0.7 ± 0.2 | 0.34 ± 0.03 | 1.8 ± 0.2 | 1.14 ± 0.02 |
| PA2663 | 29 ± 12 | 0.05 ± 0.01 | 0.09 ± 0.03 | 0.29 ± 0.08 | 1.8 ± 0.2 | 0.98 ± 0.01 |
| PA4150 | 42 ± 11 | 0.6 ± 0.1 | 0.7 ± 0.1 | 0.4 ± 0.1 | 1.7 ± 0.5 | 1.15 ± 0.06 |
| PA4151 | 18 ± 4 | 0.6 ± 0.1 | 0.6 ± 0.2 | 0.37 ± 0.04 | 1.8 ± 0.2 | 1.11 ± 0.00 |
| PA4295 | 84 ± 74 | 0.5 ± 0.1 | 0.7 ± 0.2 | 0.42 ± 0.07 | 1.9 ± 0.2 | 1.10 ± 0.04 |
| PA4549 | 45 ± 21 | 0.7 ± 0.1 | 0.8 ± 0.2 | 0.5 ± 0.1 | 2.2 ± 0.1 | 1.11 ± 0.07 |
UBC indicates wild‐type P. aeruginosafrom the University of British Columbia and UW indicates wild‐type P. aeruginosafrom the University of Washington. Data indicate the mean ± one standard deviation. NA, not applicable.
Swimming motility, biofilm formation and growth rates
Martínez‐Granero and colleagues (2006) showed that the most severely impaired Pseudomonascolonization mutants appeared to be non‐motile mutants, and colonization of plant tissue is a significant step in plant–bacteria interaction for pathogenesis (Lugtenberg et al., 2002). Hence, swimming motility and biofilm formation were evaluated here for the isogenic mutants (Table 1). The PA2463 and PA3278 knockouts exhibited 23 to 29% less motility than the wild‐type strain in 18 h, respectively, whereas the PA0513, PA0984, PA1385, PA2146, PA2587, PA4151, PA4153, PA4295 and PA4549 mutants showed 50–83% more motility. The PA3476 mutation did not cause a difference in swimming motility which agrees with a previous report (Overhage et al., 2007).
In LB glu medium at 30°C, mutations in PA3278 and PA3476 consistently enhanced biofilm formation, whereas mutations in PA0984, PA2462, PA2663, PA4151 and PA4295 decreased biofilm formation (Table 1). Of these, the PA2663 mutation was most significant as it reduced biofilm formation in both medium by a remarkable 15‐fold (Table 1). The PA1385 gene has been shown to encode polysaccharide biosynthesis enzymes that are important for biofilm development (Jackson et al., 2004); this mutant had about 30% less biofilm formation in LB glu medium.
The knockout mutations were not deleterious for the strains and most of the transposon insertions increased the specific growth rate slightly (Table 1); hence, the changes in phenotypes are not due to growth rate differences. Notably, the PA2463 mutation caused a 16 ± 7% increase in growth rate compared with the wild‐type strain but this increase in growth rate is probably not the cause of its enhanced virulence as other strains (e.g. PA4150) grew more rapidly but were less virulent (Fig. 2).
Haemolysis and cytotoxicity
As PA2463 is a putative regulator of haemolytic activity (Winsor et al., 2005), we measured haemolysis with this knockout mutant compared with the wild‐type strain. Pseudomonas aeruginosaPAO1‐UBC showed around 15% haemolysis, whereas P. aeruginosaPAO1‐UW showed around 45% haemolysis. The PA2463 mutant showed 4.7 ± 0.1‐fold more haemolytic activity than the wild‐type cells; hence, the increased pathogenesis from this strain may be due to its increased haemolytic activity. Inactivation of the PA2462 putative haemolysin gene (Winsor et al., 2005) itself reduced haemolysis by 47% and cytotoxicity by 70%. Furthermore, inactivation of the PA0984 immunity protein (He et al., 2004) reduced haemolysis by 41% and cytotoxicity by 67%. In contrast, inactivation of PA2663, which caused a large decrease in biofilm formation, did not affect the haemolytic activity and cytotoxicity of P. aeruginosa. Inactivation of PA3476 increased P. aeruginosavirulence for barley, and it was found to increase both haemolytic activity by 219% and cytotoxicity by 68%. Inactivation of PA1385, a putative glycosyl transferase (Winsor et al., 2005), decreased haemolytic activity by 50%. However, the inactivation of PA1385 did not change cytotoxicity. Overall, there was good agreement with changes in haemolysis and cytotoxicity (Fig. 3) and virulence (Fig. 2).
Figure 3.
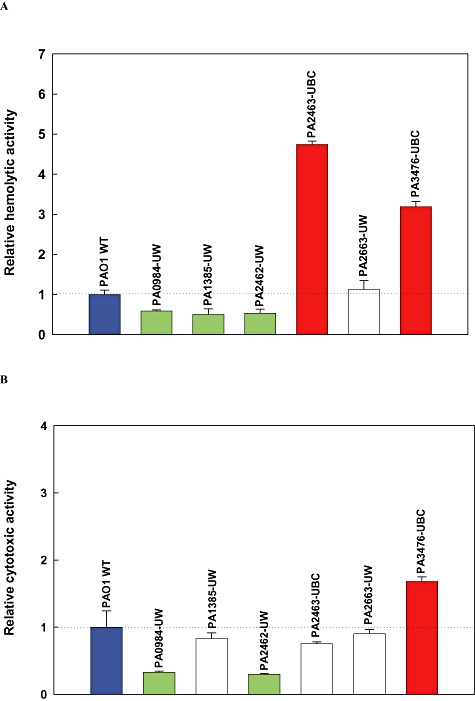
Pseudomonas aeruginosahaemolytic activity (A) and cytotoxic activity (B) in whole cells. Red indicates more virulence, green indicates less virulence and white indicates similar virulence compared with the wild‐type strain based on a t‐test. The data for the mutants are normalized by the values for the respective wild‐type strains. Error bars indicate standard deviations.
Poplar genetic response to P. aeruginosa infection
To complement our investigation of the genes required for P. aeruginosaPAO1 pathogenesis of poplar, we investigated the response of the poplar roots to P. aeruginosainfection using an Affymetrix oligo‐array that contains DNA elements for 56 055 poplar transcripts. In poplar roots, 753 genes were induced by PAO1 (1.3% of the genome, partial list in Table S4) and 1017 genes were repressed (1.8% of the genome, partial list in Table S5) after 12 h of P. aeruginosainfection. These genes participate in a wide range of biological functions including signal transduction, pathogenesis, primary and secondary metabolism, transport, chaperoning and transcription regulation.
Induction of expression of pathogenesis‐related proteins is one of the best‐known responses for grass plants and pathogenic microbes (Pinto and Ricardo, 1995; Glazebrook et al., 1996). Upon P. aeruginosainfection in the current study, transcription of pathogenesis‐related protein 5 (Glazebrook et al., 1996) and PR10 (Pinto and Ricardo, 1995) increased 74‐fold and 34‐fold in poplar roots respectively (Table S4). Transcription of the genes related to secondary metabolism was also activated, such as cytochrome P450 (37‐fold), anthocyanin 5‐aromatic acyltransferase (32‐fold), flavonoid 3‐hydroxylase (16‐fold) and berberine bridge enzyme (52‐fold) (Table S4).Berberine bridge enzyme catalyses the reaction from reticuline to scoulerine, which is the precursor of berberine (Dittrich and Kutchan, 1991).
We also found differentially regulated genes encoding the components of signal transduction and transcription factors upon P. aeruginosainfection, such as nucleotide binding site‐LRR disease resistance genes, mybtranscriptional factors, AP2 domain transcription factors, Zn‐finger type transcription factors, SCARECROW and the NO APICAL MERISTEM/ATAF/CUP‐SHAPED COTYLEDON(NAC) transcription factors in poplar roots (Tables S4 and S5). Differential regulation was also observed in plant hormone‐responsive genes upon P. aeruginosainfection, such as induction of ethylene‐induced transcription factors, auxin‐responsive factor 1 (ARF1) and ARF10, as well as downregulation of small auxin up RNA. In addition to plant hormone‐responsive genes, the amount of phytosulfokine transcript, encoding a small peptide‐growth factor in plants (Matsubayashi and Sakagami, 1996), was increased to 52‐fold in poplar roots upon infection of P. aeruginosa.
Discussion
In this study, we demonstrate that P. aeruginosaPAO1 is a pathogen for poplar trees and barley. In addition, through microarrays and mutagenesis, we identified seven novel genes involved in plant pathogenesis for either the poplar or barley plant models (PA1385, PA2146, PA2462, PA2463, PA2663, PA4150 and PA4295). Mutation in PA2463 increased virulence for poplar trees (Fig. 2); to our knowledge, the PA2463 gene has not been studied previously. As the function of PA2463 is predicted to be a haemolysin regulator (Winsor et al., 2005), we tested this mutant and found it has more than four times elevated haemolytic activity. Given that disruption of the adjacent putative haemolysin gene PA2462 reduced by half haemolysis activity (Fig. 3) and reduced virulence with barley (Fig. 2) and given that haemolysin is a known virulence factor (Gallagher and Manoil, 2001), our findings suggest that PA2463 acts as a negative regulator of haemolysin formation, and this haemolysin is important for P. aeruginosaplant pathogenesis. Furthermore, disruption of PA1385 (induced 3.2‐fold in the poplar rhizosphere) decreased virulence in both plant models by threefold; PA1385 has not been reported in previous P. aeruginosa DNA microarray studies (Hentzer et al., 2003; Schuster et al., 2003; Wagner et al., 2003; Mark et al., 2005; Filiatrault et al., 2006; Wagner et al., 2006). Moreover, the PA1385 mutant decreased haemolytic activity by half. PA1385 encodes a probable glycosyl transferase involved in polysaccharide formation (Jackson et al., 2004).Forquin and colleagues (2007) recently showed a direct relationship between the glycosyl transferase genes and haemolysin and virulence in Streptococcus agalactiae.
Bacteria with disruptions in the PA2663, PA4150 and PA4295 genes were also less virulent in either the barley or poplar rhizosphere; these genes have not been identified previously in P. aeruginosaDNA microarray studies (Hentzer et al., 2003; Schuster et al., 2003; Wagner et al., 2003; Mark et al., 2005; Filiatrault et al., 2006; Wagner et al., 2006). PA2663 encodes a hypothetical membrane protein (Winsor et al., 2005), and inactivation of this gene decreased biofilm formation significantly, but did not affect cytotoxicity or haemolysis. PA4150 encodes a possible dehydrogenase (Winsor et al., 2005). PA4296 is adjacent to PA4295 (identified here as induced in the poplar rhizosphere) and was shown to influence twitching, swarming, and cause virulence in the lettuce model (Wagner et al., 2006).
PA0984 encodes an immunity protein (He et al., 2004), and disrupting PA0984 reduced haemolytic activity and cytotoxicity (Fig. 3); this gene was identified previously as induced by root exudates from sugar beet (Mark et al., 2005). Similarly, the PA2146 knockout decreased virulence here with the barley model and was also identified previously as induced by sugar beet root exudates (Mark et al., 2005); however, this is the first report of its influence on virulence. PA2146 has 92% similarity with yciGthat controls swarming in Escherichia coli(Inoue et al., 2007). In addition, PA0764 (mucB, negative regulator of alginate synthesis), PA1458 (cheA, response regulator) and PA1092 (fliC, flagellin type B) were found here to be downregulated in the poplar rhizosphere as seen before with sugar beet root exudates (Mark et al. 2005); Mark and colleagues (2005) also showed 6 h of root exudate treatment suppressed expression of the alg genes involved in alginate synthesis that is one of the essential activities for biofilm‐forming bacteria. However, in our research, expression of most of the alg genes was induced slightly during infection of the poplar roots.
The lasand rhlquorum‐sensing systems control around 10% of the P. aeruginosa genome (Wagner et al., 2006), and PA0996 (pqsA) and PA2587 (pqsH) encode quorum‐sensing genes (Gallagher et al., 2002; Hentzer et al., 2003; Schuster et al., 2003; Wagner et al., 2003) that are upregulated in the presence of acyl‐homoserine lactone (Schuster et al., 2003). In the current study, the mutation in PA0996 decreased competitiveness during poplar root colonization and decreased virulence with poplar trees. In contrast, the rhlI mutation (PA3476) caused a large increase in virulence with barley and increased both cytotoxicity and haemolytic activity. This is the first report that quorum sensing as mediated by PA0996 and PA3476 is significant for virulence of P. aeruginosa in plants whereas loss of one of these quorum‐sensing genes has been shown to decrease virulence with Bacillus subtilis(Park et al., 2005), nematodes (Gallagher et al., 2002) and mice (Rumbaugh et al., 1999; Gallagher et al., 2002) respectively. The rhlI mutant (PA3476) also increased biofilm formation in LB glu media. In contrast, Davies et al. (1998) reported that the rhlImutant formed similar biofilms to the wild type in flow cells after 2 weeks in ERPI medium, and we observed the similar behaviour in this medium (data not shown); hence, the differences in biofilm formation are due to the differences in experimental design (temperature and medium). The pqsA mutant (PA0996) also formed less biofilm both in LB and in LB glu medium and was less competitive in the rhizosphere. Of the 16 knockout genes, all but except PA2463, PA2587 and PA4295 showed less competitiveness than the wild‐type strain on poplar roots. Strains that lacked PA0984, PA4151 or PA4153 were 5–100 times less competitive than the wild‐type strain respectively.
The approach here was successful in identifying novel virulence genes, rhizosphere competition genes and biofilm genes for P. aeruginosaPAO1 colonizing poplar roots. Nonetheless, one limitation is that the gene expression of the bacteria has been examined only once (48 h after infection). Another limitation is that the gene expression of both bacteria and plants were examined on the plant roots rather than inside plant tissue; however, it is difficult to obtain sufficient bacteria for microarray experiments from bacteria inside the plant.
Although poplar is widely used as a model woody plant, global changes in its transcriptome are little known in response to bacterial infection. Here, P. aeruginosa infection increased expression of pathogenesis‐related proteins, indicating that induction of these proteins is also a common response in woody plants like that seen in Arabidopsis (Glazebrook et al., 1996) and Lupinus albus (Pinto and Ricardo, 1995). PR5 protein (induced 74‐fold) is a homologue of tobacco osmotin that has antifungal activity (Yun et al., 1997). Osmotin is a putative apoptosis inducer through PHO36, an osmotin receptor, in budding yeast (Narasimhan et al., 2005). Although antibacterial activity of osmotin is not well known, it might play a defensive role in poplar roots against P. aeruginosainfection. Moreover, we found upregulation of the gene encoding the berberine bridge enzyme. Berberine treatment triggers apoptosis‐like cell death and enhances generation of reactive oxygen species in human cells (Jantova et al., 2006). Taken together with osmotin, production of berberine may also be one of the defence responses in poplar against pathogenic attack through induction of apoptosis cell death for cells infected with P. aeruginosa.
We also identified some poplar genes for transcriptional regulators that were differentially regulated during P. aeruginosa infection such as the LRR disease resistance gene, the NAC transcriptional factor and the myb transcriptional factor. A typical nucleotide binding site‐LRR disease resistance gene, FLS2, functions in recognition of flagellin in Arabidopsisand participates in activation of the defence response in the innate immune system (Gómez‐Gómez and Boller, 2000). In poplar, expression of the LRR genes indicates a diverse response to P. aeruginosainfection, and three of the LRRs were highly induced and seven were suppressed. Besides FLS2, some classes of NAC transcription factors in rice were controlled by flagellin perception (Fujiwara et al., 2004). Furthermore, flg22, a peptide containing the most conserved domain of flagellin, induces some LRRs and the myb transcriptional factor in Arabidopsis (Navarro et al., 2004). These results suggest that flagellin‐mediated signal transduction may play an essential part in the early defence mechanism in poplar.
Altered expression of auxin‐related genes has been observed often during plant–microbe interactions (Wang et al., 2005a), which suggests auxin‐mediated morphological changes in plants. Auxin‐responsive transcriptional factors mediate auxin‐dependent transcriptional activation or repression, and ARF10 participates in the development of root cap cells (Wang et al., 2005b). Auxin has diverse effects on plant growth, including tissue development and tropism to light and gravity (Perrot‐Rechenmann and Napier, 2005). When the pathogenic bacterium A. tumefaciensinfects a plant, an auxin‐mediated tumour is formed (Ooms et al., 1981). Pseudomonas aeruginosaalso synthesizes auxin, and this may affect the auxin‐mediated response in plants. It is interesting that auxin induces expression of SCARECROW(Gao et al., 2004), which regulates cell differentiation in the root cortex/endodermis daughter cells (Di Laurenzio et al., 1996). Thus, developmental regulation is often caused by auxin‐related signalling in plants, and it may be one of the adaptive responses to bacterial infection.
Investigations of plant–microbe interactions in the rhizosphere are often complicated by the need to separate plant and microbe samples from soil without introducing artefacts. In this research, we used a hydroponic system to study poplar and P. aeruginosa interactions in the rhizosphere, and it enabled us to obtain samples from the rhizosphere without soil effects. We note that the term rhizosphere includes hydroponic growth conditions (Cramer et al., 1999; Jauert et al., 2002; Morgan et al., 2005; Soda et al., 2007), even though it was originally defined as the soil compartment influenced by the roots (Hinsinger and Marschener, 2006). Thus, our hydroponic system is also useful for rhizosphere research, although some differences may occur in results obtained in a soil rhizosphere versus a hydroponic rhizosphere.
Previously, P. aeruginosaPA14 was shown to infect disparate species by using the same virulence factors (Rahme et al., 1995; Rahme et al., 1997; Mahajan‐Miklos et al., 1999; Tan et al., 1999; Rahme et al., 2000; Hendrickson et al., 2001). Analogously, as some of the genes we identified through the Pseudomonas transcriptome analysis are significant for virulence with both plants and human models (e.g. PA1385, PA2462, PA2463 and PA3476), some virulence‐related functions of P. aeruginosaare not limited to specific hosts. It is also clear that virulence gene expression in biofilms requires a live poplar tree as P. aeruginosagenes were clearly differentially expressed relative to the inert solid (glass wool). Overall, this work broadens our understanding of the genetic basis of pathogenesis in the rhizosphere (as well as biofilm formation) with P. aeruginosaand increases our understanding of the response of poplar trees to pathogenic bacteria; it may also help to define the interaction of this pathogenic bacterium (and others) with other eukaryotic hosts (e.g. humans).
Experimental procedures
Bacterial strains and growth
Strains and plasmids are listed in Table S3. LB medium (Sambrook et al., 1989) was used to grow the bacterial strains, and HRP minimal medium [contains (per litre) 5.5 g of KH2PO4, 1.5 g of K2HPO4, 1 g of (NH4)2SO4, 0.735 g of MgCl2·6H2O and 0.1 g of NaCl] (Huynh et al., 1989) with 0.25 wt% sucrose was used during contact of bacteria with plants. The sequenced P. aeruginosaPAO1 Holloway strain (Stover et al., 2000) was used with the DNA microarrays. Colonization of P. aeruginosa PAO1 on poplar tree roots was visualized using a TCS SP5 scanning confocal laser microscope with a 63× HCX PL FLUOTAR L dry objective with correction collar and numerical aperture of 0.7 (Leica Microsystems, Mannheim, Germany) using the constitutive green fluorescent protein plasmid pMRP9‐1 (Davies et al., 1998). To select for the knockout mutants, 50 µg ml−1 tetracycline was used.
Poplar trees growth
To facilitate rapid growth of plants and removal of soil without damaging fragile plant roots, poplar cuttings (DN‐34 Imperial hybrid poplar, Segal Ranch, Grandview, WA) were surface sterilized with 3% hydrogen peroxide for 10 min and planted in polyethylene autoclavable bags (Fisher Scientific, Pittsburg, PA) containing 3.5 kg of autoclaved sand. Poplar trees were placed under 60 W Spot‐Gro plant light bulbs (Sylvania, St. Marys, PA), illuminated for 16 h each day and irrigated with 10% sterile Hoagland's solution every day (Shim et al., 2000).
Bacterial RNA isolation and microarray analysis
After 5 weeks of growth, 25 poplar trees were gently removed from the autoclave bags by cutting the bag with scissors, and the fragile roots were exposed by gently rinsing with distilled water in a manner that preserved the fine root filaments. Overnight P. aeruginosaPAO1 cultures were re‐suspended in 1.5 l of 1× HRP minimal medium with 0.25% sucrose at a turbidity of 0.6 at 600 nm, and the poplar trees were dipped in a 3 l plastic beaker containing the re‐suspended PAO1 culture. As sucrose is the main carbohydrate in the young poplars (Bonicel et al., 1987), it was used as a carbon source to obtain more bacteria. Furthermore, sucrose does not affect plant root colonization (Lugtenberg et al., 1999). The bacterial culture and poplar trees were shaken at 150 r.p.m. (KS250BS1 shaker, IKA, Germany) at room temperature for 48 h and illuminated for 16 h per day. Total wet weight of poplar trees roots was approximately 20 g. In the glass wool experiment, cells were prepared with the same manner except 20 g of glass wool was used instead of poplar trees. After 48 h of contacting PAO1 with poplar roots or glass wool, the poplar roots and glass wool were washed in 200 ml of 0°C 0.85% NaCl buffer for 30 s, and the biofilm cells were removed from the poplar roots or glass wool by sonicating at 22 W (FS3 sonicator, Fisher Scientific, Pittsburg, PA) in 200 ml of 0°C 0.85% NaCl buffer. The buffer was centrifuged at 10 000 g for 2 min at 4°C (J2‐HS centrifuge, Beckman, Palo Alto, CA). RNA isolation from P. aeruginosaPAO1, cDNA synthesis, fragmentation and hybridizations were as described previously (Domka et al., 2007). The absence of contamination was verified by streaking the culture contacted with poplar trees on LB agar plates and confirming the presence of green colonies as P. aeruginosaPAO1.
The Affymetrix Genechip P. aeruginosaGenome Array (Affymetrix, P/N 900339) contains 5500 of the 5570 ORFs of P. aeruginosa(Whiteley et al., 2001). The reliability of induced and repressed genes was ensured with a P‐value less than 0.05. The intensities of polyadenosine RNA control were used to monitor the labelling process. The total signal intensity was scaled to an average value of 500. Genes were identified as differentially expressed if the expression ratio was greater than twofold change based on standard deviations of the genes of 1.7−1.9 (Domka et al., 2007). The expression data have been submitted to the NCBI Gene Expression Omnibus (GSE5887).
Poplar RNA isolation and microarray analysis
Overnight P. aeruginosaPAO1 cultures were re‐suspended in 1.5 l of 1× HRP + 0.25% sucrose at turbidity at 600 nm of 0.6. Two 5‐week‐old poplar trees with roots were gently taken from sand and washed with distilled water in a manner that preserved the fine root filaments. One poplar tree was dipped in the culture containing P. aeruginosaPAO1 and the other one was dipped in medium lacking P. aeruginosaPAO1. The trees were shaken in 250 ml Erlenmeyer flasks at 150 r.p.m. for 12 h at room temperature, then the poplar roots were dipped in 100 ml of 0°C 0.85% NaCl buffer, the roots of the poplar tree were cut with sterile scissors and the roots were sonicated for 2 min to remove bacteria. To avoid the effect of sonication on gene expression, sonication was also performed for the poplar tree not contacted with P. aeruginosaPAO1. RNA was isolated from poplar roots as described previously (Brunner et al., 2004) with minor modifications by grinding roots to a powder with a pestle in a mortar containing liquid nitrogen. The remaining steps of RNA isolation were performed according to the Qiagen RNeasy Plant Mini kit protocol. Subsequent steps consisting of cDNA synthesis, biotin labelling of cDNA, fragmentation, hybridization and scanning were performed at the Center for Functional Genomics at the State University of New York, University at Albany.
The GeneChip Poplar Genome Array (Affymetrix, P/N 900728) contains 61 251 poplar probe sets representing 56 055 transcripts. Induced and repressed genes were identified as differentially expressed if the P‐value was less than 0.05 and the expression ratio was greater than 4 based on the standard deviations of 4.2 for both arrays (Domka et al., 2007). Data quality was assessed by the hybridization controls and scaling factors. The expression data have been deposited in the NCBI Gene Expression Omnibus (GSE5887).
Verification of knockouts
The P. aeruginosa transposon mutants were obtained from UBC (Lewenza et al., 2005) or UW (Jacobs et al., 2003); the UBC library was constructed with a mini‐Tn5‐luxCDABE (promoter trap), and the UW library was constructed using ISphoA/hah or ISlacZ/hah (contain internal promoters). Insertion of mini‐Tn5‐luxCDABE transposon in PA2463 locus was confirmed by a PCR‐based method with chromosomal DNA purified from both wild‐type PAO1 and PA2463 mutant (McPhee et al., 2003) (see Table S6 for primer sequences). The PA2463‐F primer and PA2463‐R primer were used to confirm the presence of the transposon insertion in PA2463 locus by amplifying 450 bp of the partial PA2463 gene. In addition, the Tn5‐out primer and PA2463‐R were used to amplify 700 bp fragment corresponding to the end of the transposon and its flanking region of PA2463 gene. Similarly, all of the knockout mutants used in this study were verified by PCR.
Poplar pathogenicity wilting assay
The degree of wilting of poplar tree branches was devised by us to indicate poplar tree health. To minimize contamination, poplar trees were dipped in bacterial cultures in 1× HRP medium containing kanamycin 50 (µg ml−1); kanamycin at this concentration does not alter tree viability (poplar trees were viable for more than 7 days in 1× HRP medium with kanamycin without bacteria). The change in branch angle was measured during the poplar tree–bacteria contact. To measure pathogenicity of mutants and wild‐type strain, angles were measured every 24 h from the vertical axis (clockwise) and the stem of poplar was considered to be origin. For each strain, at least three separate trees were used. The data for wilting experiments were analysed with a Student's t‐test, and those with the P‐value less than or equal to 0.05 were chosen as significant (Ross, 2004).
Root microscopy
To observe P. aeruginosa cells inside roots, poplar trees were infected with P. aeruginosa PAO1‐UW tagged with pMRP9‐1 by placing trees in 250 ml flasks. After 48 h, poplar root tips were embedded in optimum cutting temperature compound (Tissue‐Tek, SAKURA Finetechnical, Tokyo, Japan), sectioned at 20 µm thickness by a cryostat microtome (JUNG CM 1800, Leica) and observed using scanning confocal laser microscope.
Barley seed pathogenicity assay
Barley seeds (cultivar Belford) were purchased from Stover Seed Company (Los Angels, CA) and were surface‐sterilized in 1% sodium hypochlorite solution for 30 min, then washed with sterilized distilled water 10 times. Pseudomonas aeruginosa, grown in LB medium at 30°C, was harvested at a turbidity at 600 nm of 1. Pseudomonas aeruginosa cells were washed once with sterilized distilled water and twice with 1× Hoagland solution, and then re‐suspended to turbidity at 600 nm of 1.00 ± 0.03. Fifteen barley seeds were germinated in 10 ml of 1× Hoagland solution without (control), with PAO1 or with each mutant at 25°C with gentle shaking. After 3 days, the number of germinated seeds was counted compared with that of wild‐type PAO1 treatment. All experiments were repeated at least three times (45 seeds). The datafor barley germination experiments were analysed with a Student's t‐test and those with a P‐value less than or equal to 0.05 were chosen as significant (Ross, 2004).
Crystal violet biofilm assay
Biofilm formation was quantified in 96‐well polystyrene plates as described previously (Ren et al., 2005). Overnight P. aeruginosaPAO1 cultures were diluted in LB and LB supplemented with 0.2 wt% glucose to a turbidity of 0.05 of 600 nm. Diluted cultures were inoculated into the plates and were grown at 30°C without shaking. Before measuring the biofilm mass, the growth of the cells was quantified using turbidity at 620 nm. Ten replicate wells were averaged to obtain each data point. Two independent cultures were used.
Swimming motility assay
Agar plates containing 1% tryptone, 0.25% NaCl and 0.3% agar were used to assay motility in plates as described previously (González Barrios et al., 2006). Motility halos were measured at 5 and 18 h. Six plates were used to evaluate motility in each strain. Two independent cultures were tested for each strain.
Bacterial rhizosphere competition assay
One millilitre of overnight‐grown P. aeruginosaPAO1 and the knockout mutants were inoculated each into 25 ml of LB and were grown to a turbidity of 1.6 at 600 nm. Once the cells reached this optical density (OD), they were washed with HRP minimal media at 5500 g for 5 min at 4°C, and re‐suspended in 7.5 ml of HRP minimal media. The OD of the mutants and wild‐type were measured after re‐suspension. The same amount of mutant and wild‐type cells was added to a total of 150 ml of HRP minimal media in a 250 ml Erlenmeyer flask. One poplar tree was placed into the culture containing P. aeruginosaPAO1 and the mutant was contacted for 48 h; the root tip was cut with sterilized scissors, sonicated in 100 ml of 0.85% NaCl buffer, then the supernatant was plated with appropriate dilutions on LB agar containing kanamycin 50 µg ml−1 to determine the total number of P. aeruginosacells as well as LB containing kanamycin 50 µg ml−1 and tetracycline 50 µg ml−1 to determine the number of P. aeruginosaknockout cells. Bacterial concentrations were expressed as colony‐forming units per gram of dry root weight. All experiments were repeated at least twice. The data for the crystal violet biofilm assay, swimming motility and bacterial rhizosphere competition experiments were analysed with a Student's t‐test, and those with a P‐value less than or equal to 0.05 were chosen as significant (Ross, 2004).
Haemolysis and cytotoxicity assays
Haemolysis assays to determine the haemolytic activity were carried out as described previously (Blocker et al., 1999) with some modifications. Basically, bacteria were grown overnight at 37°C and inoculated into fresh medium until they reached turbidity at 600 nm of approximately 1. Bacteria were then pelleted at 1000 g for 10 min and suspended at equal concentrations in 2 ml of saline. The whole cells were then aliquoted into 96‐well plates in 200 µl final volume at a concentration equivalent to 108 colony‐forming units and diluted 1:2 in saline six times. Eighty microlitres of 2% human red blood cells and 80 µl of each bacterial preparation were then added to a fresh 96‐well plate and incubated at 37°C for 2 h. The plate was centrifuged at 1000 g for 10 min at 4°C. The supernatant was transferred to a fresh 96‐well plate and the OD450 was determined. Controls include saline (negative) and 0.02% Tween‐20 (positive) in all experiments. The positive control was set as 100% haemolysis, and the negative control was set as 0% haemolysis. The value for red blood cells without Tween and bacteria was used to subtract the background in the spectrophotometer readings. Each sample was prepared in quadruplicate.
Standard lactate dehydrogenase release cytotoxicity assay was used in these studies (Brander et al., 1993; Behl et al., 1994) as described previously (Cirillo et al., 2001). The procedure used was essentially as recommended by the manufacturer of the CytoTox96 Non‐Radioactive Cytotoxicity Assay system (Promega). Serial dilutions were made of each bacterial strain at multiplicity of infections of 500, 250, 100 and 10 in a final volume of 100 µl for each assay using 4 × 104 human peripheral blood monocytic cells (PBMCs). Appropriate numbers of cells for CytoTox96 assays were determined as suggested by the manufacturer (Promega). As a positive control for 100% cytotoxicity, the cells are lysed with 9% v/v Triton X‐100 (Promega). The cells were incubated with the bacteria for 4 h at 37°C + 5% CO2. Cytotoxicity readings were taken using an ELISA plate reader at 450 nm. Percent cytotoxicity was calculated as recommended by the manufacturer and corrected for small differences in the inocula used. PBMCs were isolated from 50 ml of human blood obtained from healthy volunteers. The mononuclear cell fraction was purified by centrifugation in Ficoll at 700 g for 30 min at room temperature. The PBMCs containing band was removed, washed twice in Hanks balanced salt solution (Gibco) and suspended in Roswell Park Memorial Institute medium with 0.1% heat‐inactivated human serum to a concentration of 106 cells ml−1.
Acknowledgments
We thank Professor Marvin Whiteley for the pMRP9‐1 plasmid, Professor Tim McDermott for P. aeruginosaPAO1, and Professor Robert E.W. Hancock and Professor Colin Manoil for providing knockout mutants. This research was supported by the National Institutes of Health (EB003872‐01A1) and the National Science Foundation (BES‐0331416).
Supplementary material
The following supplementary material is available for this article online:
Partial list of P. aeruginosa PAO1 genes induced more than 2.5-fold after 48 hours of poplar root contact versus contact with glass wool.
Partial list of P. aeruginosa PAO1 genes repressed more than 3-fold with after 48 hours of poplar root contact versus contact with glass wool.
Strains and plasmids used.
Partial list of poplar tree root genes induced more than 15-fold after 12 hours of P. aeruginosa PAO1 contact versus no bacteria.
Partial list of poplar tree root genes repressed more than 20-fold after 12 hours of P. aeruginosa PAO1 contact versus no bacteria.
Primers used for verifying the P. aeruginosa knockout mutations.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Behl C., Davis J.B., Lesley R., Schubert D. Hydrogen peroxide mediates amyloid beta protein toxicity. Cell. 1994;77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Bergman T., Erickson K., Galyov E., Persson C., Wolf‐Watz H. The lcrByscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994;176:2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A., Gounon P., Larquet E., Niebuhr K., Cabiaux V., Parsot C., Sansonetti P. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonicel A., Haddad G., Gagnaire J. Seasonal‐variations of starch and major soluble sugars in the different organs of young poplars. Plant Physiol Biochem. 1987;25:451–459. [Google Scholar]
- Brander C., Wyss‐Coray T., Mauri D., Bettens F., Pichler W.J. Carrier‐mediated uptake and presentation of a major histocompatibility complex class I‐restricted peptide. Eur J Immunol. 1993;23:3217–3223. doi: 10.1002/eji.1830231226. [DOI] [PubMed] [Google Scholar]
- Brunner A.M., Yakovlev I.A., Strauss S.H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004;4:14. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun C.K., Ozer E.A., Welsh M.J., Zabner J., Greenberg E.P. Inactivation of a Pseudomonas aeruginosa quorum‐sensing signal by human airway epithelia. Proc Natl Acad Sci USA. 2004;101:3587–3590. doi: 10.1073/pnas.0308750101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo S.L., Bermudez L.E., El‐Etr S.H., Duhamel G.E., Cirillo J.D. Legionella pneumophila entry gene rtxA is involved in virulence. Infect Immun. 2001;69:508–517. doi: 10.1128/IAI.69.1.508-517.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope L.D., Yogev R., Ursula M.E., Hansen E.J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton B. Microbial ecology comes of age and joins the general ecology community. Proc Natl Acad Sci USA. 2004;101:16983–16984. doi: 10.1073/pnas.0407886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer M.D., Gao Z.F., Lips S.H. The influence of dissolved inorganic carbon in the rhizosphere on carbon and nitrogen metabolism in salinity‐treated tomato plants. New Phytol. 1999;142:441–450. [Google Scholar]
- Davies D.G., Parsek M.R., Pearson J.P., Iglewski B.H., Costerton J.W., Greenberg E.P. The involvement of cell‐to‐cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka‐Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. et al. [DOI] [PubMed] [Google Scholar]
- Dittrich H., Kutchan T.M. Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci USA. 1991;88:9969–9973. doi: 10.1073/pnas.88.22.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J., Lee J., Bansal T., Wood T.K. Temporal gene‐expression in Escherichia coli K‐12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Filiatrault M.J., Picardo K.F., Ngai H., Passador L., Iglewski B.H. Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect Immun. 2006;74:4237–4245. doi: 10.1128/IAI.02014-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti J.J., Sellmer J., McCown B., Haissig B., Comai L. Agrobacterium mediated transformation and regeneration of Populus. Mol Gen Genet. 1987;206:192–199. [Google Scholar]
- Forquin M.P., Tazi A., Rosa‐Fraile M., Poyart C., Trieu‐Cuot P., Dramsi S. The putative glycosyltransferase‐encoding gene cylJ and the group B Streptococcus (GBS)‐specific gene cylK modulate hemolysin production and virulence of GBS. Infect Immun. 2007;75:2063–2066. doi: 10.1128/IAI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Tanaka N., Kaneda T., Takayama S., Isogai A., Che F.S. Rice cDNA microarray‐based gene expression profiling of the response to flagellin perception in cultured rice cells. Mol Plant Microbe Interact. 2004;17:986–998. doi: 10.1094/MPMI.2004.17.9.986. [DOI] [PubMed] [Google Scholar]
- Gallagher L.A., Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L.A., McKnight S.L., Kuznetsova M.S., Pesci E.C., Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M.J., Parkin I., Lydiate D., Hannoufa A. An auxin‐responsive SCARECROW‐like transcriptional activator interacts with histone deacetylase. Plant Mol Biol. 2004;55:417–431. doi: 10.1007/s11103-004-0892-9. [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E.E., Ausubel F.M. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Barrios A.F., Zuo R., Hashimoto Y., Yang L., Bentley W.E., Wood T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gómez L., Boller T. FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- He J., Baldini R.L., Déziel E., Saucier M., Zhang Q., Liberati N.T. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci USA. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson E.L., Plotnikova J., Mahajan‐Miklos S., Rahme L.G., Ausubel F.M. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J Bacteriol. 2001;183:7126–7134. doi: 10.1128/JB.183.24.7126-7134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M., Wu H., Anderson J.B., Riedel K., Rasmussen T.B., Bagge N. Attenuation of Pseudomonas aeruginosa biofilm virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsinger P., Marschener P. Rhizosphere‐perspectives and challenges – a tribute to Lorenz Hiltner 12–17 September 2004 – Munich, Germany. Plant Soil. 2006;283:7–8. [Google Scholar]
- Huynh T.V., Dahlbeck D., Staskawicz B.J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- Inoue T., Shingaki R., Hirose S., Waki K., Mori H., Fukui K. Genome‐wide screening of genes required for swarming motility in Escherichia coli K‐12. J Bacteriol. 2007;189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K.D., Starkey M., Kremer S., Parsek M.R., Wozniak D.J. Identification of psl, a locus encoding a potential exopolysaccharide that is essential forPseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M.A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantova S., Cipak L., Letasiova S. Berberine induces apoptosis through a mitochondrial/caspase pathway in human promonocytic U937 cells. Toxicol in Vitro. 2006;21:25–31. doi: 10.1016/j.tiv.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Jauert P., Schumacher T.E., Boe A., Reese R.N. Rhizosphere acidification and cadmium uptake by strawberry clover. J Environ Qual. 2002;31:627–633. [PubMed] [Google Scholar]
- Lewenza S., Falsafi R.K., Winsor G., Gooderham W.J., McPhee J.B., Brinkman F.S., Hancock R.E.W. Construction of a mini‐Tn5‐luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 2005;15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B.J., Dekkers L.C. What makes Pseudomonas bacteria rhizosphere competent? Environ Microbiol. 1999;1:9–13. doi: 10.1046/j.1462-2920.1999.00005.x. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B.J., Kravchenko L.V., Simons M. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol. 1999;1:439–446. doi: 10.1046/j.1462-2920.1999.00054.x. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B.J.J., Chin A.W.T.F., Bloemberg G.V. Microbe–plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek. 2002;81:373–383. doi: 10.1023/a:1020596903142. [DOI] [PubMed] [Google Scholar]
- McPhee J.B., Lewenza S., Hancock R.E. Cationic antimicrobial peptides activate a two‐component regulatory system, PmrA‐PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- Mahajan‐Miklos S., Tan M.W., Rahme L.G., and, Ausubel F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- Mark G.L., Dow J.M., Kiely P.D., Higgins H., Haynes J., Baysse C. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe‐plant interactions. Proc Natl Acad Sci USA. 2005;102:17454–17459. doi: 10.1073/pnas.0506407102. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Granero F., Rivilla R., Martín M. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl Environ Microbiol. 2006;72:3429–3434. doi: 10.1128/AEM.72.5.3429-3434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentag R., Luckevich M., Morency M.J., Seguin A. Bacterial disease resistance of transgenic hybrid poplar expressing the synthetic antimicrobial peptide D4E1. Tree Physiol. 2003;23:405–411. doi: 10.1093/treephys/23.6.405. [DOI] [PubMed] [Google Scholar]
- Morgan J.A., Bending G.D., White P.J. Biological costs and benefits to plant–microbe interactions in the rhizosphere. J Exp Bot. 2005;56:1729–1739. doi: 10.1093/jxb/eri205. [DOI] [PubMed] [Google Scholar]
- Narasimhan M.L., Coca M.A., Jin J., Yamauchi T., Ito Y., Kadowaki T. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol Cell. 2005;17:171–180. doi: 10.1016/j.molcel.2004.11.050. et al. [DOI] [PubMed] [Google Scholar]
- Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J.D.G. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene‐dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135:1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms G., Hooykaas P.J., Moolenaar G., Schilperoort R.A. Grown gall plant tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti plasmids; analysis of T‐DNA functions. Gene. 1981;14:33–50. doi: 10.1016/0378-1119(81)90146-3. [DOI] [PubMed] [Google Scholar]
- Overhage J., Lewenza S., Marr A.K., Hancock R.E.W. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini‐Tn5‐lux mutant library. J Bacteriol. 2007;189:2164–2169. doi: 10.1128/JB.01623-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Heo Y.J., Choi Y.S., Deziel E., Cho Y.H. Conserved virulence factors of Pseudomonas aeruginosa are required for killing Bacillus subtilis. J Microbiol. 2005;43:443–450. [PubMed] [Google Scholar]
- Passador L., Cook J.M., Gambello M.J., Rust L., Iglewski B.H. Expression of Pseudomonas aeruginosa virulence genes requires cell‐to‐cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- Perrot‐Rechenmann C., Napier R.M. Auxins. Vitam Horm. 2005;72:203–233. doi: 10.1016/S0083-6729(04)72006-3. [DOI] [PubMed] [Google Scholar]
- Pinto M.P., Ricardo C.P. Lupinus albus L. pathogenesis‐related proteins that show similarity to PR‐10 proteins. Plant Physiol. 1995;109:1345–1351. doi: 10.1104/pp.109.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potera C. Forging a link between biofilms and disease. Science. 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- Prithiviraj B., Weir T., Bais H.P., Schweizer H.P., Vivanco J.M. Plant models for animal pathogenesis. Cell Microbiol. 2005;7:315–324. doi: 10.1111/j.1462-5822.2005.00494.x. [DOI] [PubMed] [Google Scholar]
- Quinaud M., Chabert J., Faudry E., Neumann E., Lemaire D., Pastor A. The PscE–PscF–PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J Biol Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. et al. [DOI] [PubMed] [Google Scholar]
- Rahme L.G., Stevens E.J., Wolfort S.F., Shao J., Tompkins R.G., Ausubel F.M. Common virulence factors for bacterial pathogenecity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Rahme L.G., Tan M.W., Le L., Wong S.M., Tompkins R.G., Calderwood S.B., Ausubel F.M. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L.G., Ausubel F.M., Cao H., Drenkard E., Goumnerov B.C., Lau G.W. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci USA. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Zuo R., Gonzalez Barrios A.F., Bedzyk L.A., Eldridge G.R., Pasmore M.E., Wood T.K. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl Environ Microbiol. 2005;71:4022–4034. doi: 10.1128/AEM.71.7.4022-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S.M. Elsevier Academic Press; 2004. [Google Scholar]
- Rumbaugh K.P., Griswold J.A., Iglewski B.H., Hamood A.N. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sauer K., Cullen M.C., Rickard A.H., Zeef L.A., Davies D.G., Gilbert P. Characterization of nutrient‐induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Lostroh C.P., Ogi T., Greenberg E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum‐controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Chauhan S., Ryoo D., Bowers K., Thomas S.M., Canada K.A. Rhizosphere competitiveness of trichloroethylene‐degrading, poplar‐colonizing recombinant bacteria. Appl Environ Microbiol. 2000;66:4673–4678. doi: 10.1128/aem.66.11.4673-4678.2000. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda S., Ike M., Ogasawara Y., Yoshinaka M., Mishima D., Fujita M. Effects of light intensity and water temperature on oxygen release from roots into water lettuce rhizosphere. Water Res. 2007;41:487–491. doi: 10.1016/j.watres.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. et al. [DOI] [PubMed] [Google Scholar]
- Tan M.W., Rahme L.G., Sternberg J.A., Tompkins R.G., Ausubel F.M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., DiFazio S.P., Teichmann T. Poplar genomics is getting popular: the impact of the poplar genome project on tree research. Plant Biol (Stuttg) 2004;6:2–4. doi: 10.1055/s-2003-44715. [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. et al. [DOI] [PubMed] [Google Scholar]
- Wagner V.E., Bushnell D., Passador L., Brooks A.I., Iglewski B.H. Microarray analysis of Pseudomonas aeruginosa quorum‐sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V.E., Li L.L., Isabella V.M., Iglewski B.H. Analysis of the hierarchy of quorum‐sensing regulation in Pseudomonas aeruginosa. Anal Bioanal Chem. 2006;387:469–479. doi: 10.1007/s00216-006-0964-6. [DOI] [PubMed] [Google Scholar]
- Walker T.S., Bais H.P., Déziel E., Schweizer H.P., Rahme L.G., Fall R., Vivanco J.M. Pseudomonas aeruginosa–plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004;134:320–331. doi: 10.1104/pp.103.027888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ohara Y., Nakayashiki H., Tosa Y., Mayama S. Microarray analysis of the gene expression profile induced by the endophytic plant growth‐promoting rhizobacteria, Pseudomonas fluorescens FPT9601‐T5 in Arabidopsis. Mol Plant Microbe Interact. 2005a;18:385–396. doi: 10.1094/MPMI-18-0385. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Wang L.J., Mao Y.B., Cai W.J., Xue H.W., Chen X.Y. Control of root cap formation by MicroRNA‐targeted auxin response factors in Arabidopsis. Plant Cell. 2005b;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Bangera M.G., Bumgarner R.E., Parsek M.R., Teitzel G.M., Lory S., Greenberg E.P. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Winsor G.L., Lo R., Ho Sui S.J., Ung K.S.E., Huang S., Cheng D. Pseudomonas aeruginosa genome database and PseudoCAP: facilitating community‐based, continually updated, genome annotation. Nucleic Acids Res. 2005;33:D338–343. doi: 10.1093/nar/gki047. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr T.L., Goranson J., Frank D.W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- Yee D.C., Maynard J.A., Wood T.K. Rhizoremediation of trichloroethylene by a recombinant, root‐colonizing Pseudomonas fluorescens strain expressing toluene ortho‐monooxygenase constitutively. Appl Environ Microbiol. 1998;64:112–118. doi: 10.1128/aem.64.1.112-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D.J., Zhao Y., Pardo J.M., Narasimhan M.L., Damsz B., Lee H. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc Natl Acad Sci USA. 1997;94:7082–7087. doi: 10.1073/pnas.94.13.7082. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Partial list of P. aeruginosa PAO1 genes induced more than 2.5-fold after 48 hours of poplar root contact versus contact with glass wool.
Partial list of P. aeruginosa PAO1 genes repressed more than 3-fold with after 48 hours of poplar root contact versus contact with glass wool.
Strains and plasmids used.
Partial list of poplar tree root genes induced more than 15-fold after 12 hours of P. aeruginosa PAO1 contact versus no bacteria.
Partial list of poplar tree root genes repressed more than 20-fold after 12 hours of P. aeruginosa PAO1 contact versus no bacteria.
Primers used for verifying the P. aeruginosa knockout mutations.


