Abstract
Objectives
To gather preliminary data concerning the feasibility of using 7 salivary mRNAs--IL-8; IL-1β; dual specificity phosphatase 1 (DUSP1); H3 histone family 3A (H3F3A); ornithin decarboxylase antizyme 1 (OAZ1); S100 calcium-binding protein P (S100P); and spermidine/spermine N1-acetyltransferase 1 (SAT1)—for detecting development of oral squamous cell carcinoma (OSCC) in oral lichen planus (OLP) patients and OSCC patients whose disease was in remission.
Materials and Methods
Saliva samples were collected from five study groups (25 subjects/group): newly-diagnosed OSCC; OSCC-in-remission; disease-active OLP; disease-inactive OLP; and normal controls. The salivary mRNA levels were determined by a pre-amplification RT-qPCR approach with nested gene-specific primers. Mean fold changes between each pair of study groups were analyzed by the Mann-Whitney U test.
Results
Salivary levels of OAZ1, S100P, and DUSP1 mRNAs were significantly higher in newly-diagnosed OSCC patients, compared to: 1) normal controls (p=0.003; p=0.003; and p<0.001, respectively); 2) OSCC-in-remission (p<0.001; p=0.001; and p<0.001, respectively); 3) disease-active OLP (p<0.001; p=0.016; and p<0.001, respectively); and 4) disease-inactive OLP (p=0.043; p<0.001; and p<0.001, respectively). No significant differences were found in the levels of salivary IL-8, IL-1β, H3F3A and SAT1 mRNAs between newly-diagnosed OSCC patients and the normal controls (p=0.093, 0.327, 0.764 and 0.560, respectively).
Conclusion
Salivary OAZ1, S100P and DUSP1 mRNAs are candidate biomarkers for detecting OSCC development in OSCC patients in remission and in OLP patients.
Clinical Relevance
The results of this study serve as the basis for a further large-scale study which may lead to a non-invasive screening method for early detection of OSCC.
Keywords: Saliva, mRNA, Oral Cancer, Oral Lichen Planus, Biomarkers
Introduction
Using salivary biomarkers is a promising non-invasive approach for early detection of oral squamous cell carcinoma (OSCC) and has become an area of strong research interest in recent years. The use of salivary biomarkers for OSCC detection would be especially important for known high-risk groups--such as patients with oral lichen planus (OLP) and patients with a previous history of OSCC who remain at risk although they have been treated and currently show no visible sign of the disease. It is the non-invasive aspect of salivary biomarker screening that is so important for these high-risk patients, because invasive biopsies are currently the only way to detect OSCC in its early stages. Thus, for these patients, monitoring for OSCC involves a lifelong ordeal of periodic biopsies. Therefore, using any non-invasive, reliable salivary biomarker specifically for these two groups would be of great clinical significance, and the search for such biomarkers has been a focus of our ongoing research for several years.
More than 40 potential salivary biomarkers for early detection of OSCC have been identified in the literature so far [1]. However, of the more than 40 previously reported potential OSCC salivary biomarkers, only 4 of them, proteins of IL-6, IL-8, basic fibroblastic growth factor (b-FGF), and endothelin-1, have been investigated in the above mentioned two high-risk groups.[2-5] So the feasibility of using any of the more than 40 previously reported OSCC salivary biomarkers for detecting OSCC development in these two high-risk groups is mostly undetermined. To investigate this feasibility, we decided to measure the levels of each one of the previously reported potential OSCC salivary biomarkers specifically in the two high-risk groups, comparing those levels with those of normal controls and with diagnosed OSCC groups. We began that process by first measuring the levels of the potential OSCC salivary biomarkers that had been reported by 2007. Our findings showed that the levels of b-FGF and endothelin-1 were significantly elevated in OLP patients and OSCC patients-in-remission who showed no visible sign of OSCC, respectively, to a degree that there were no significant differences in their levels from the levels found in newly-diagnosed OSCC patients.[5,4] These results indicate that the previously reported potential OSCC salivary biomarkers, based on comparing their levels in the newly-diagnosed OSCC patients with the levels in non-OSCC controls, may or may not be suitable for detecting OSCC in the two above-mentioned high-risk groups.
Seven salivary mRNAs: IL-8, IL-1β, dual specificity phosphatase 1 (DUSP1), H3 histone family 3A (H3F3A), ornithin decarboxylase antizyme 1 (OAZ1), S100 calcium-binding protein P (S100P), and spermidine/spermine N1-acetyltransferase 1 (SAT1) had been found to show significantly higher levels in some groups of OSCC patients, when they were compared to the levels found in normal controls; and therefore they have been suggested to be potential OSCC salivary biomarkers. [6-8] However, their levels had never been measured specifically in the two high-risk groups described above (OLP patients, and OSCC patients who have been treated and are in remission) and compared with levels measured in normal controls. Since such measurement and comparison is considered an essential first step to establish any candidate salivary biomarker for detecting OSCC in these two groups, that testing was a logical focus for this pilot study.
Therefore, the objective of this pilot study was to gather preliminary data concerning the feasibility of using these seven salivary mRNAs for oral cancer detection in OLP patients and in OSCC patients in remission, by measuring the levels of the seven salivary mRNAs in those two high-risk patient groups and comparing them with the levels found in a group of newly-diagnosed OSCC patients and with a group of normal controls. If the levels of any of these 7 salivary constituents were found to be significantly different in newly-diagnosed OSCC patients when compared to the levels found in these two high-risk groups (in individuals not [yet] diagnosed (or re-diagnosed) with active OSCC), then we would conclude that those salivary constituents would be strong candidate biomarkers for early detection of OSCC, specifically for these two high-risk groups. If no significant differences were found between the newly-diagnosed OSCC patients and the OLP patients, or in OSCC patients in remission, for a given salivary constituent, then this salivary constituent would not be likely to signal early development of OSCC in OLP patients or in OSCC patients in remission.
Material and Methods
1. Patient groups and recruitment
Twenty-five /human subjects were recruited for each of five groups, during the period from September 1, 2009 to August 15, 2011, from the Stomatology Center at Texas A&M Health Science Center (TAMHSC)-Baylor College of Dentistry in Dallas, and from referrals by local dentists and surgeons who had diagnosed or treated OSCC and/or OLP patients. The recruitment protocol used was approved by the Institutional Review Board of TAMHSC-Baylor College of Dentistry, and informed consent was obtained from each patient prior to saliva collection. The five groups of human subjects from which saliva samples and clinical information were collected were as follows:
Group A: OSCC patients, defined as newly-diagnosed OSCC patients before treatment started.
Group B: OSCC patients-in-remission, defined as OSCC post-treatment patients, having gone for at least two years without any evidence of recurrence or second primary OSCC.
Group C: Disease-active OLP patients, defined as OLP patients who were newly diagnosed, had symptomatic lesions and the treatment of OLP had not been started.
Group D: Disease-inactive OLP patients, defined as OLP patients who had been treated previously and showed either no clinically visible lesions of OLP or the asymptomatic reticular type of OLP at the time of saliva collection (See below for explanation).
Group E: Normal controls, defined as persons who had never been diagnosed with either OSCC or OLP, and showed no symptoms of either disease.
Most of the OLP patients in our Stomatology Center, at which we had a database of 800 OLP patients by 2009, had their diseases well-controlled by therapies. These patients were often in the state of remission (asymptomatic), and came to our clinic only once or twice a year, solely for the purpose of monitoring for possible malignant transformation. Clinical presentation of these patients in terms of symptoms and/or signs was distinctively different from those patients who had newly-onset multiple symptomatic lesions, and whether the different degrees of OLP disease activity might affect salivary mRNAs levels was unknown. Therefore, we divided the OLP patients into “disease-active” and “disease-inactive” subgroups, and compared the amount of each mRNA biomarker in each group separately. For analysis purposes, we also combined these two groups as Group F, OLP patients as a whole, and compared the result with Group E, the normal controls (see Results).
The detailed inclusion and exclusion criteria for each group are listed in Table 1. The rationale for these criteria has been delineated in our previous work [4-5].
Table 1. The Inclusion and Exclusion Criteria for the Study Groups (OSCC= Oral Squamous Cell Carcinoma; OLP= Oral Lichen Planus).
| GROUPS | INCLUSION CRITERIA | EXCLUSION CRITERIA |
|---|---|---|
|
A: Newly-
Diagnosed OSCC Patients |
Patients who…
|
|
|
B: OSCC
Patients-in- Remission |
Patients who…
|
|
|
C: Disease-
Active OLP Patients |
Patients who …
|
|
|
D: Disease-
Inactive OLP Patients |
Patients who…
|
|
|
E: Normal
Controls |
Subjects who… |
“Non-smoker” was defined as a person who had smoked fewer than 100 cigarettes in his/her lifetime and had not smoked at all within one calendar year prior to beginning participation in the study, and who had not smoked pipe or cigar or used smokeless tobacco, for any more than a total of 6 months in their lifetime, and not at all within the calendar year prior to beginning participation in the study.
“Ex-smoker” was defined as a person whose last smoking or other use of tobacco products was at least 20 years prior to beginning participation in the study.
“One alcoholic drink” was defined as approximately 150 ml of wine, 330 ml of beer, or 30 ml of hard liquor.
2. Saliva collection
Whole saliva was collected from the participants between 6am and 12pm, using a previously published protocol [9,4]. Participants were asked to refrain from eating, drinking, or using oral hygiene procedures on the day of saliva collection. A water mouth rinse was administered prior to saliva sample collection. Five minutes after the oral rinse, the participant was asked to sit upright and spit into a 50ml Falcon tube kept on ice. A maximum of 8 ml of saliva were collected within 30 minutes.
3. Sample processing
Saliva samples were processed immediately after collection according to a previously published method [4,8,7,10]. The saliva samples were centrifuged at 2,600g for 15 minutes, at 4°C. The supernatant was separated from the pellet, and the RNase inhibitor (Superase-In, Ambion Inc., Austin, TX) was added to the supernatant-- 5ul Superase-In/ml of supernatant. All samples were stored at −80 °C in aliquots until further use.
4. RT- pre-amplification and qPCR
A pre-amplification RT-qPCR approach with nested gene-specific primers was used, according to the previously described methods [8,7,10]. After thawing, the total RNA of the saliva sample was extracted, using the RNeasy mini kit (Qiagen, Valencia, CA), and all samples were then treated with TURBO DNA-free (Ambion, Grand Island, NY) to remove genomic DNA.
Nested PCR assay, a RT-PCR pre-amplification followed by qPCR, was used according to Hu et al [10]. The outer and inner primer sequences for six of the seven mRNA biomarkers (IL-8, IL-1β, DUSP1, OAZ1, S100P, and SAT), along with two reference genes (RPS9 and β-actin), were adopted from the study by Brinkmann et al [7]. The H3F3A primer sequences are as follows:
Outer forward primer for pre-amplification: AGCGTCTGGTGCGAGAAATT
Outer reverse primer for pre-amplification: GCACACAGGTTGGTGTCTTCAA
Inner forward primer for qPCR: CGCTTCCAGAGCGCAGCTAT
Inner reverse primer for qPCR: TCTTCAAAAAGGCCAACCAGAT
The qPCR was performed with Bio-Rad ITAQ Fast SYBR ROX kit (Bio-Rad, Hercules, CA) on Bio-Rad CFX96 Real-Time System (Bio-Rad, Hercules, CA). Each saliva sample was tested in triplicate. An inter-run calibrator of 0.125 ug/ul human RNA (Strategene qPCR Human Reference Total RNA, Agilent Technologies, Santa Clara, CA) was added in each experiment with the saliva samples. The amount of the seven mRNAs was normalized by β–actin and RPS9 [7]. The results were recorded by the Bio-Rad CFX Manager Software version 2.0. The Kruskal-Wallace test was used to analyze the differences in the normalized quantity (ΔCq) of each salivary mRNA biomarker among the study groups. Normalized quantity (ΔCq) between each pair of study groups was analyzed by the Mann-Whitney U test. The mean and median fold changes between each pair of patient vs. control groups were calculated by the Pfaffl method [11].
Results
The demographic data and clinical information concerning the OSCC and OLP patient groups are listed in Table 2.
Table 2. Study Groups - Demographic Data, OSCC Clinical Stage, OLP Clinical Types, and Locations of the Lesions in the Patients.
| Groups |
Number
of Subjects |
Mean
Age (Range) |
Gender | OSCC Stage* | OLP Clinical Types** | Location of the Lesion(s)*** | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | I | II | III | IV | R/P | E/A |
Com-
bination |
B/L | G/A/R | T | V | F | P | |||
|
A: Newly-Diagnosed
OSCC Patients |
25 | 55.96 (26-81) |
18 | 7 | 11 | 4 | 2 | 8 | - | 1 | 4 | 17 | 0 | 3 | 1 | ||
|
B: OSCC Patients-in-
Remission |
25 | 65.04 (35-95) |
16 | 9 | 15 | 3 | 3 | 4 | - | 1 | 3 | 18 | 0 | 3 | 0 | ||
|
C: Disease-Active OLP
Patients |
25 | 63.80 (45-83) |
4 | 21 | - | 3 | 4 | 18 | 17 | 18 | 8 | 6 | 0 | 1 | |||
|
D: Disease-Inactive OLP
Patients |
25 | 61.64 (47-80) |
10 | 15 | - | 4 | 5 | 16 | 17 | 15 | 9 | 4 | 0 | 0 | |||
| E: Normal Controls | 25 | 60.88 (32-85) |
13 | 12 | - | - | - | ||||||||||
|
F: Combined OLP
Patients (C+D) |
50 | 62.72 (45-83) |
14 | 36 | - | 7 | 9 | 34 | 34 | 33 | 17 | 10 | 0 | 1 | |||
The OSCC stage is based on the TNM classification.
R/P= reticular and/or plaque; E/A= erosive and/or atrophic; Combination= some combination of reticular, plaque, erosive and atrophic forms. The OLP clinical types listed under Group D were the presenting types when the patient was diagnosed (biopsied). At the time of saliva collection, all Group D patients showed either no OLP lesions or showed asymptomatic reticular type lesions, as specified in the inclusion criteria.
B/L= bucal mucosa or labial mucosa; G/A/R= gingival, alveolar ridge or retromolar pad; T= tongue; V= vestibule; F= floor of mouth; P= palate.
There was no significant difference in the mean age among the six study groups (p=0.104). Most OSCC patients (60%) in Group B, and 44% of patients in Group A, had been diagnosed at Stage I, and the most commonly affected site was the tongue. The most common initial clinical presentation in our OLP patients (both Groups C and D) was a combination of reticular, erosive and atrophic types; and the most commonly affected sites were bucal/labial mucosa and gingiva (Table 2).
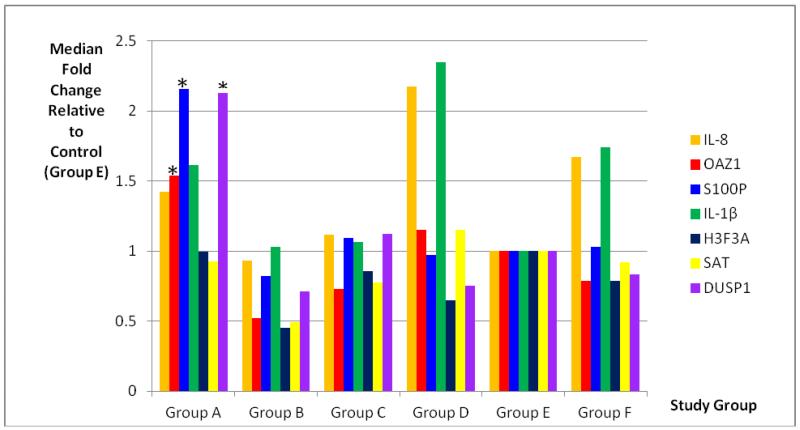
The mean and standard deviation of the threshold temperature (Cq) and normalized quantity (ΔCq) of each mRNA in the study groups are listed in Table 3. There were significant differences in the normalized quantity (ΔCq) among the study groups in the mRNAs of IL-8, OAZ1, S100P, IL-1β, H3F3A, SAT and DUSP1 (p=0.008, p<0.001, p=0.002, p=0.006, p=0.017, p=0.004, and p<0.001, respectively). The mean and median fold changes of each pair of the study groups and the statistical results for each of the mRNA biomarkers are listed in Table 4. The median fold changes of the seven salivary mRNAs in each patient group, compared to the levels found in the normal control group, are also illustrated in Figure 1.
Table 3. Mean and Standard Deviation of the Threshold Temperature (Cq) and Normalized Cq (ΔCq) of Each mRNA in Each Study Group.
|
Salivary
mRNAs |
Group A
(Newly- Diagnosed OSCC Patients) |
Group B
(OSCC Patients- in- Remission) |
Group C
(Disease-Active OLP) |
Group D
(Disease- Inactive OLP) |
Group E
(Normal Controls) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cq | ΔCq | Cq | ΔCq | Cq | ΔCq | Cq | ΔCq | Cq | ΔCq | |
| IL-8 | 16.39 ± 1.99 |
−3.17 ± 1.83 |
16.85 ± 3.09 |
−2.46 ± 2.55 |
17.18 ± 2.96 |
−2.74 ± 0.89 |
16.94 ± 2.92 |
−3.49 ± 1.06 |
17.31 ± 2.58 |
−2.27±1.18 |
| OAZ1 | 18.12 ± 1.68 |
−1.44 ± 0.96 |
19.61 ± 2.80 |
0.30 ± 0.78 |
19.55 ± 2.60 |
−0.37 ± 0.60 |
19.48 ± 2.63 |
−0.96 ± 1.97 |
18.87 ± 2.37 |
−0.71 ± 0.77 |
| S100P | 18.78 ± 2.06 |
−0.78 ± 1.42 |
19.79 ± 2.63 |
0.49 ± 0.96 |
19.68 ± 2.52 |
−0.23 ± 1.68 |
20.97 ± 2.77 |
0.54 ± 0.88 |
20.00 ± 2.95 |
0.42 ± 1.39 |
| IL-1β | 17.13 ± 2.03 |
−2.43 ± 1.30 |
17.12 ± 3.06 |
−2.19 ± 2.35 |
17.82 ± 2.44 |
−2.09 ± 2.51 |
16.99 ± 2.46 |
−3.44 ± 1.18 |
17.42 ± 2.56 |
−2.16 ± 0.96 |
| H3F3A | 18.58 ± 1.80 |
−0.98 ± 1.13 |
19.42 ± 3.46 |
0.12 ± 2.77 |
18.90 ± 2.97 |
−1.02 ± 0.83 |
19.88 ± 2.87 |
−0.55 ± 0.80 |
18.53 ± 2.67 |
−1.05 ± 1.01 |
| SAT | 17.02 ± 1.71 |
−2.54 ± 1.39 |
17.34 ± 2.44 |
−1.97 ± 2.13 |
17.54 ± 2.42 |
−2.38 ± 0.92 |
17.45 ± 2.22 |
−2.99 ± 0.70 |
16.76 ± 2.21 |
−2.82 ± 0.87 |
| DUSP | 18.56 ± 1.56 |
−1.00 ± 0.68 |
19.95 ± 2.85 |
0.64 ± 0.80 |
19.97 ± 2.89 |
0.06 ± 0.84 |
20.74 ± 2.91 |
0.31 ± 0.79 |
19.55 ± 2.67 |
−0.02 ± 1.05 |
Table 4. Mean and Median Fold Changes of Each mRNA Biomarker in Pairs of the Study Groups, and the Statistical Results.
| mRNAs | Study Groups | Compared With… |
Mean Fold
Change |
Median Fold
Change |
p-value |
|---|---|---|---|---|---|
| IL-8 | Group A (Newly-Diagnosed OSCC) |
Group B (OSCC, Patients-in- Remission) |
1.47 | 1.53 | 0.240 |
| Group C (Disease-Active OLP) |
1.35 | 1.27 | 0.479 | ||
| Group D (Disease-Inactive OLP) |
0.83 | 0.65 | 0.410 | ||
| Group E (Normal Controls) | 1.86 | 1.42 | 0.093 | ||
| Group F (Combined OLP) | 1.06 | 0.85 | 0.946 | ||
| Group C (Disease- Active OLP) |
Group D (Disease-Inactive OLP) |
0.62 | 0.51 | 0.016* | |
| Group F (Combined OLP, C+D) |
Group E (Normal Controls) | 1.76 | 1.67 | 0.006* | |
| OAZ1 | Group A (Newly-Diagnosed OSCC) |
Group B (OSCC, Patients-in- Remission) |
3.16 | 2.95 | <0.001* |
| Group C (Disease-Active OLP) |
2.10 | 2.10 | <0.001* | ||
| Group D (Disease-Inactive OLP) |
1.42 | 1.34 | 0.043* | ||
| Group E (Normal Controls) | 1.66 | 1.54 | 0.003* | ||
| Group F (Combined OLP) | 1.73 | 1.96 | <0.001* | ||
| Group C (Disease- Active OLP) |
Group D (Disease-Inactive OLP) |
0.67 | 0.64 | 0.082 | |
| Group F (Combined OLP, C+D) |
Group E (Normal Controls) | 0.96 | 0.79 | 0.357 | |
| S100P | Group A (Newly-Diagnosed OSCC) |
Group B (OSCC, Patients-in- Remission) |
2.37 | 2.62 | 0.001* |
| Group C (Disease-Active OLP) |
1.46 | 1.97 | 0.016* | ||
| Group D (Disease-Inactive OLP) |
2.45 | 2.22 | <0.001* | ||
| Group E (Normal Controls) | 2.29 | 2.16 | 0.003* | ||
| Group F (Combined OLP) | 1.89 | 2.10 | 0.001* | ||
| Group C (Disease- Active OLP) |
Group D (Disease-Inactive OLP) |
1.68 | 1.13 | 0.097 | |
| Group F (Combined OLP, C+D) |
Group E (Normal Controls) | 1.21 | 1.03 | 0.969 | |
| IL-1β | Group A (Newly-Diagnosed OSCC) |
Group B (OSCC, Patients-in- Remission) |
1.08 | 1.57 | 0.421 |
| Group C (Disease-Active OLP) |
1.26 | 1.52 | 0.357 | ||
| Group D (Disease-Inactive OLP) |
0.60 | 0.75 | 0.02* | ||
| Group E (Normal Controls) | 1.2 | 1.61 | 0.327 | ||
| Group F (Combined OLP) | 0.79 | 0.96 | 0.544 | ||
| Group C (Disease- Active OLP) |
Group D (Disease-Inactive OLP) |
0.47 | 0.50 | 0.04* | |
| Group F (Combined OLP, C+D) |
Group E (Normal Controls) | 1.39 | 1.68 | 0.018* | |
| H3F3A | Group A (Newly-Diagnosed OSCC) |
Group B (OSCC, Patients-in- Remission) |
1.90 | 2.19 | 0.026* |
| Group C (Disease-Active OLP) |
0.98 | 1.16 | 0.954 | ||
| Group D (Disease-Inactive OLP) |
1.37 | 1.47 | 0.077 | ||
| Group E (Normal Controls) | 0.95 | 0.99 | 0.764 | ||
| Group F (Combined OLP) | 1.16 | 1.26 | 0.323 | ||
| Group C (Disease- Active OLP) |
Group D (Disease-Inactive OLP) |
1.40 | 1.27 | 0.024* | |
| Group F (Combined OLP, C+D) |
Group E (Normal Controls) | 0.83 | 0.79 | 0.120 | |
| SAT | Group A (Newly-Diagnosed OSCC) |
Group B (OSCC, Patients-in- Remission) |
1.43 | 1.88 | 0.048* |
| Group C (Disease-Active OLP) |
1.12 | 1.20 | 0.318 | ||
| Group D (Disease-Inactive OLP) |
0.75 | 0.81 | 0.367 | ||
| Group E (Normal Controls) | 0.82 | 0.93 | 0.560 | ||
| Group F (Combined OLP) | 0.92 | 1.01 | 0.955 | ||
| Group C (Disease- Active OLP) |
Group D (Disease-Inactive OLP) |
0.67 | 0.68 | 0.03* | |
| Group F (Combined OLP, C+D) |
Group E (Normal Controls) | 0.90 | 0.92 | 0.529 | |
| DUSP1 | Group A (Newly-Diagnosed OSCC) |
Group B (OSCC, Patients-in- Remission) |
3.00 | 2.99 | <0.001* |
| Group C (Disease-Active OLP) |
2.08 | 1.89 | <0.001* | ||
| Group D (Disease-Inactive OLP) |
2.46 | 2.83 | <0.001* | ||
| Group E (Normal Controls) | 1.96 | 2.13 | <0.001* | ||
| Group F (Combined OLP) | 2.26 | 2.60 | <0.001* | ||
| Group C (Disease- Active OLP) |
Group D (Disease-Inactive OLP) |
1.18 | 1.49 | 0.181 | |
| Group F (Combined OLP, C+D) |
Group E (Normal Controls) | 0.83 | 0.83 | 0.261 |
indicates p<0.05
Figure 1.
Median Fold Changes of the Salivary mRNAs in Each Patient Group Compared to the Levels Found in Normal Controls, analyzed by Mann Whitney U test. Significantly higher levels of OAZ1, S100P and DUSP1 were found in Group A (indicated by *) when compared to all other study groups.
Three of the seven salivary mRNAs, OAZ1, S100P, and DUSP1, showed significantly higher levels in the newly-diagnosed OSCC patients, compared to levels found in 1) normal controls (p=0.003; p=0.003; and p<0.001, respectively); 2) OSCC in-remission (p<0.001; p=0.001; and p<0.001, respectively); 3) OLP disease-active (p<0.001; p=0.016; and p<0.001, respectively); 4) OLP disease-inactive (p=0.043; p<0.001; and p<0.001, respectively); and 5) the combined two OLP groups, Group F (p<0.001; p=0.001; and p<0.001, respectively).
There were no significant differences in the levels of salivary IL-8 and IL-1β mRNAs in the newly-diagnosed OSCC patients compared to the levels found in OSCC patients-in-remission (p=0.24 and 0.421, respectively) or in normal controls (p=0.093 and 0.327, respectively). Although significantly elevated levels of H3F3A and SAT1 mRNAs were found in OSCC patients compared to the levels found in OSCC patients-in-remission (p=0.026 and 0.048, respectively), there were no significant differences in the levels of these two mRNAs in OSCC patients compared to the levels found in normal controls (p=0.764 and 0.560, respectively).
There were no significant differences in the levels of salivary IL-8, IL-1β, H3F3A and SAT1 in newly-diagnosed OSCC patients compared to the levels found in Group F, the OLP patients as a whole (p=0.946, 0.544, 0.323 and 0.955, respectively). On the other hand, significantly different levels of these four salivary mRNAs were found in OLP disease-active patients compared to the levels found in OLP disease-inactive patients (p=0.016; 0.04, 0.024 and 0.03, respectively), while there were no significant differences in the levels of OAZ1, S100P and DUSP1 mRNAs in the OLP disease-active patients compared to the levels found in disease-inactive OLP patients (p=0.082; 0.097 and 0.181, respectively). Salivary IL-8 and IL-1 β mRNAs also showed significantly higher levels in Group F, the OLP patients as a whole, when compared to the levels found in normal controls (p=0.006 and 0.018, respectively).
Discussion
Our results suggested that salivary OAZ1, S100P and DUSP1 mRNAs are good candidate biomarkers for OSCC detection in OSCC patients-in-remission and in OLP patients, regardless of OLP disease activity. All three of these genes/proteins have been found to be involved in various human cancers (see below); however, the mRNAs of these three genes have not been reported to be found in the saliva of patients with any types of cancers other than OSCC [6-8]. Therefore, the levels of these 3 salivary mRNAs in patients with other types of cancers are unknown, and whether significantly elevated levels of these 3 salivary mRNAs would indicate specifically OSCC but not other types of cancer development remains to be investigated. The current knowledge about the involvement of these three genes/proteins in OSCC is also very limited (see below), and no study has been reported previously regarding their involvement in OLP.
Ornithine decarboxylase antizyme (OAZ) inhibits ornithine decarboxylase (ODC), the rating-limiting enzyme in polyamine synthesis [12]. As increased ODC activity has been found in most human cancers [13-14], it has been suggested that OAZ functions as a tumor suppressor [15-17]. OAZ1 has been found to be important in DNA repair and in the metastatic potential of the human OSCC cell line [18]. Because OAZ expression is induced by a polyamine-dependent mechanism [19], our finding of elevated salivary OAZ1 mRNAs in the OSCC patients also suggests an increased polyamine level in OSCC.
S100P is a member of the S100 calcium-binding protein family [20]. It has been found to be over-expressed in pancreatic, breast, colon, prostate and lung cancers, and contributes to malignant features via increased cell proliferation, survival, motility and invasion [21]. S100P mRNA expression was found to be significantly increased in anoikis (detachment-induced apoptosis)-resistant OSCC cells, compared to anoikis-sensitive OSCC cells, indicating S100P involvement in the metastatic process in OSCC [22]. However, Sapkota et al.[23] investigated the mRNA expression profile of 16 S100 gene-family members, including S100P, and did not find significantly increased levels of S100P mRNAs in the OSCC tissue specimens. At this time, whether salivary mRNAs reflects the mRNA changes in the OSCC tissue is still unclear. Further investigation on S100P alterations in OSCC is needed.
DUSP1 is a subtype of type I cysteine-based protein tyrosine phosphatase, which is involved in various signaling pathways [24]. DUSP1 expression is increased in pancreatic cancer and in early-stage colon, prostate and bladder cancers [24]. Salivary DUSP1 mRNA has been found to be significantly elevated in OSCC patients compared to normal controls in two of three previously published studies of salivary mRNA biomarkers for OSCC [6,8]. However, the role and/or changes of DUSP1 in OSCC development and progression are still unclear.
In our study, salivary IL-8, IL-1β, H3F3A and SAT1 mRNAs did not appear to be good candidate biomarkers for OSCC detection in OLP patients, as the levels of these salivary mRNAs in our OLP patients (who had no evidence of OSCC development) showed no significant difference from the levels found in our OSCC patients. Although all these four gene products have been found to be involved in carcinogenesis, changes of IL-8 and IL-1β also have been found in OLP patients. Salivary IL-8 has been reported in previously published studies to be significantly elevated in OLP patients [25-26]. Significantly increased levels of IL-1β, via secretion by the oral keratinocytes and the tissue-infiltrated mononuclear cells, have also been reported in OLP when compared to the levels found in non-inflamed oral mucosa [27]. Our findings of significantly elevated salivary IL-8 and IL-1β mRNAs in OLP patients (Group F) when compared to the levels found in normal controls appear to correlate with those previously reported findings for these two cytokines at the protein level. Interestingly, salivary IL-8, IL-1β, H3F3A and SAT1 mRNAs also showed significantly different levels between OLP disease-active and disease-inactive patients (Table 3). These results indicate that these four salivary mRNAs may be influenced by the degree of oral inflammation.
Based on this preliminary data, we concluded that salivary OAZ1, S100P and DUSP1 mRNAs are good candidate biomarkers for OSCC detection in patients who have a history of OSCC but have had no evidence of it recurring after treatment, and in OLP patients, regardless of current level of OLP disease activity. Further large-scale studies that include OSCC patients who have had recurrence, as well as OLP patients with OSCC development, will be required to confirm the utility of using these three salivary mRNA biomarkers for OSCC detection in these two high-risk groups, and to determine the sensitivity and specificity for these three candidate biomarkers.
Acknowledgements
This study was sponsored by the National Institute of Dental and Craniofacial Research (1R21DE018757-01A2). We thank Dr. John O’Brien, Baylor University Medical Center, Dallas, Texas; Dr. Jacqueline Plemons, Dr. John Wright and Dr. Harvey Kessler, Texas A&M University-Baylor College of Dentistry, and other oral surgery and oral medicine colleagues who referred patients to this study. We also want to thank Mr. Mark Lawson at Bio-Rad Laboratories, for his technical assistance with Bio-Rad CFX software for gene analysis.
Footnotes
Conflict of Interest Statement
The authors declare that they have no conflict of interest involved with their work in this study.
References
- 1.Wu J, Yi C, Chung H, Wang D, Chang W, Lee S, Lin C, Yang Y, Yang W. Potential biomarkers in saliva for oral squamous cell carcinoma. Oral Oncology. 2010;46:226–231. doi: 10.1016/j.oraloncology.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Rhodus NL, Cheng B, Myers S, Miller L, Ho V, Ondrey F. The feasibility of monitoring NF-kappaB associated cytokines: TNF-alpha, IL-1alpha, IL-6, and IL-8 in whole saliva for the malignant transformation of oral lichen planus. Mol Carcinog. 2005;44(2):77–82. doi: 10.1002/mc.20113. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann RR, Yurgel LS, Campos MM. Evaluation of salivary endothelin-1 levels in oral squamous cell carcinoma and oral leukoplakia. Regul Pept. 2011;166(1-3):55–58. doi: 10.1016/j.regpep.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Cheng YS, Rees T, Jordan L, Oxford L, O’Brien J, Chen HS, Wong D. Salivary endothelin-1 potential for detecting oral cancer in patients with oral lichen planus or oral cancer in remission. Oral Oncol. 2011;47(12):1122–1126. doi: 10.1016/j.oraloncology.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorugantula L, Rees T, Plemons J, Chen H, Cheng YS. Salivary basic fibroblast growth factor in patients with oral squamous cell carcinoma or oral lichen planus. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114(2):215–222. doi: 10.1016/j.oooo.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH, Wong DT. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10(24):8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 7.Brinkmann O, Kastratovic DA, Dimitrijevic MV, Konstantinovic VS, Jelovac DB, Antic J, Nesic VS, Markovic SZ, Martinovic ZR, Akin D, Spielmann N, Zhou H, Wong DT. Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol. 2011;47(1):51–55. doi: 10.1016/j.oraloncology.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elashoff D, Zhou H, Reiss J, Wang J, Xiao H, Henson B, Hu S, Arellano M, Sinha U, Le A, Messadi D, Wang M, Nabili V, Lingen M, Morris D, Randolph T, Feng Z, Akin D, Kastratovic DA, Chia D, Abemayor E, Wong DT. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev. 2012;21(4):664–672. doi: 10.1158/1055-9965.EPI-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navazesh M. Methods for collecting saliva. Ann NY Acad Sci. 1993;694:72–77. doi: 10.1111/j.1749-6632.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z, Zimmermann BG, Zhou H, Wang J, Henson BS, Yu W, Elashoff D, Krupp G, Wong DT. Exon-level expression profiling: a comprehensive transcriptome analysis of oral fluids. Clin Chem. 2008;54(5):824–832. doi: 10.1373/clinchem.2007.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nuclei Acids Research. 2001;29(9):2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller J, Fong W, Canellakis E. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci USA. 1976;73:1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pegg A. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48:759–774. [PubMed] [Google Scholar]
- 14.Shantz L, Levin V. Regulation of ornithine decarboxylase during oncogenic transformation: mechanisms and therapeutic potential. Amino Acids. 2007;33:213–223. doi: 10.1007/s00726-007-0531-2. [DOI] [PubMed] [Google Scholar]
- 15.Koike C, Chan D, Zetter B. Sensitivity to polyamine-induced growth arrest correlates with antizyme induction in prostate carcinoma cells. Cancer Res. 1999;59:6109–6112. [PubMed] [Google Scholar]
- 16.Iwata S, Sato Y, Asada M, Takagi M, Tsujimoto A, Inaba T, Yamada T, Sakamoto S, Yata J, Shimogori T, Igarashi k, Mizutani S. Anti-tumor activity of antizyme which targets the ornithine carboxylase (ODC) required for cell growth and transformation. Oncogene. 1999;18:165–172. doi: 10.1038/sj.onc.1202275. [DOI] [PubMed] [Google Scholar]
- 17.Tsuji T, Usui S, Aida T, Tachikawa T, Hu G, Sasaki A, Matusumura T, Todd R, Wong D. Induction of epithelial differentiation and DNA demethylation in hamster malignant oral keratinocyte by ornithine decarboxylase antizyme. Oncogene. 2001;20:24–33. doi: 10.1038/sj.onc.1204051. [DOI] [PubMed] [Google Scholar]
- 18.Tsuji T, Katasurano M, Ibaragi S, Shima K, Sasaki A, Hu G. Ornithine decarboxylase antizyme upregulates DNA-dependent protein kinase and enhances the nonhomologous end-joining repair of DNA double-strand breaks in human oral cancer cells. Biochemistry. 2007;46(31):8920–8932. doi: 10.1021/bi7000328. [DOI] [PubMed] [Google Scholar]
- 19.Rom E, Kahana C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc Natl Acad Sci USA. 1994;91:3959–3963. doi: 10.1073/pnas.91.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salama I, Malone P, Mihaimeed F, Jones J. A review of the S100 proteins in cancer. EJSO. 2008;34(2008):357–364. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Gibadulinova A, Tothova V, Pastorek J, Pastorekova S. Transcriptional regulation and functional implication of S100P in cancer. Amino Acids. 2011;41:885–892. doi: 10.1007/s00726-010-0495-5. [DOI] [PubMed] [Google Scholar]
- 22.Kupferman M, Patel V, Sriuranpong V, Amornphimoltham P, Jasser S, Mandal M, Zhou G, Wang J, Coombes K, Multani A, Pathak S, Gutkind J, Myers J. Molecular analysis of anoikis resistance in oral cavity squamous cell carcinoma. Oral Oncol. 2007;43(5):440–454. doi: 10.1016/j.oraloncology.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Sapkota D, Bruland O, Bue O, Bakcer H, Elgindi O, Vasstrand E, Ibruhim S. Expression profile of the S100 gene family members in oral squamous cell carcinomas. J Oral Pathol Med. 2008;37:607–615. doi: 10.1111/j.1600-0714.2008.00683.x. [DOI] [PubMed] [Google Scholar]
- 24.Patterson K, Brummer T, O’Brien P, Daly R. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Lin M, Zhang S, Wang Z, Jiang L, Shen J, Bai J, Gao F, Zhou M, Chen Q. NF-kappaB-dependent cytokines in saliva and serum from patients with oral lichen planus: a study in an ethnic Chinese population. Cytokine. 2008;41(2):144–149. doi: 10.1016/j.cyto.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Rhodus NL, Cheng B, Myers S, Bowles W, Ho V, Ondrey F. A comparison of the pro-inflammatory, NF-kappaB-dependent cytokines: TNF-alpha, IL-1-alpha, IL-6, and IL-8 in different oral fluids from oral lichen planus patients. Clin Immunol. 2005;114(3):278–283. doi: 10.1016/j.clim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto T, Osaki T. Characteristic cytokines generated by keratinocytes and mononuclear infiltrates in oral lichen planus. J Invest Dermatol. 1995;104:784–788. doi: 10.1111/1523-1747.ep12606990. [DOI] [PubMed] [Google Scholar]