Abstract
There is growing interest in understanding the effects of host-microbial interactions on host physiologic processes. Much of the work in this arena is logically focused on the interaction at mucosal surfaces as this is a primary site of interaction. However, there is ample evidence to suggest that the effects of the microbiota have a much farther reach including the systemic immune system. While there are some similarities to effects at mucosal surfaces (i.e. reduced numbers of adaptive immune cells, diminished innate responses), there are some important differences that we highlight such as the response to immunogens and bacterial antigens. We propose that understanding the details of how specific components of the microbiota influence the systemic immune system likely will have significant impact on our understanding the pathophysiology of a variety of autoimmune diseases.
Keywords: Microbiome, systemic immunity, commensal bacteria, host microbial interactions
1. Introduction
Intestinal contents contain bacteria that increase in number along the cephalocaudal axis (~103/gram in the duodenum to ~1012/gram in the distal colon) (1, 2). Colonization of the intestine with bacteria occurs shortly after birth and is influenced by the route of delivery (3, 4). Human infants and mouse pups delivered vaginally are initially colonized with predominantly Lactobacillus and Prevotella species, prominent vaginal commensals. In contrast, infants delivered by Cesarean section are predominantly colonized with Staphylococcus species, prominent skin commensals. While on a milk-based diet, the intestinal diversity of mouse pups and human infants narrows to harbor mostly lactate producers. After weaning, the diversity increases to resemble that of the mother’s colon, reflecting dietary change to solid food (4, 5). While there are differences at the species level the most prominent genera in adult human intestine are similar to that in adult mice and include Bacteroidetes, Lachnococci, Rhuminococci, Lactobacilli, and Prevotellae (6, 7).
Since 1989 when the hygiene hypothesis was first published (8), much attention has been paid to how exposures to microbes influence immune activity. While some studies suggest the benefits of exposure to environmental microbial products by reducing the incidence of atopy (reviewed by Finlay in this issue (ref), (9)), other microbial exposures, particularly EBV infection, are associated with autoimmune disease (reviewed in (10)). Certainly, genetic variations also influence immune reactivity, and thus, host microbe interactions in these contexts.
Typically, commensals and pathogens are largely kept at bay through mucosal barriers and its immune mechanisms (reviewed by Eberl in this issue (ref)) creating systemic immune ignorance except under circumstances of innate deficiencies in the mucosal immune system (11, 12) or breaches in mucosal barrier functions. Nevertheless, numerous studies have demonstrated a substantial effect by the presence of gut commensals on the development of the systemic immune system and its function, which will be the focus of this review.
2. Role of commensals in development of the systemic immune system
Analysis of the germ-free mouse has greatly aided our understanding of the role of microbes in immune development. Like mucosal immunity, the systemic immune system is profoundly affected by the absence of commensal bacteria. Not only is the anatomy affected, but also the function of the innate and adaptive immune responses.
2.1 Immune organs
Studies in germ-free mice demonstrated the effect of bacterial colonization on the development of secondary lymph organs. Spleens and peripheral lymph nodes (LNs)1 of germ-free mice are hypoplastic, and mesenteric lymph nodes (MLNs) are often absent. Medullary cords are thinner, and germinal centers are reduced in number and size. The primary immune organs, thymus and bone marrow have normal appearing architecture (13, 14).
2.2 Cellular populations
Commensal microbes affect the numbers and function of B cells, T cells, and innate immune cells.
2.2.1 B cells
Bone marrow and splenic B cell numbers are greatly reduced in germ-free mice. The lack of commensal organisms greatly impairs the basal production of IgA (reviewed by MacPherson in this issue (ref)) as well as IgG and IgM. The effects of the microbiota are not just on B cell development in the local mucosa and regional lymph nodes. The effect is systemic as in the bone marrow of 8–12 week old germ-free mice fed an antigen-free diet, compared to conventionally housed2 mice, demonstrate 2-, 5-, and 17-fold reductions in IgM+, IgG+, and IgA+ B cells, respectively, in the bone marrow (despite no obvious alterations in architecture). The spleen of germ free mice contained significant reductions (50–75%) in the number of IgM+ and IgA+ B cells (but not IgG+ B cells) versus conventional mice. By 52 weeks of age, IgM+ B cells numbers in both the bone marrow and spleen are similar in germ free and conventionally housed mice, while the defects in IgG+ B cells in the bone marrow and IgA+ B cells in the bone marrow and spleen persist (14, 15). When splenocytes from germ-free mice are cultured ex vivo, IgM is secreted but IgG is undetectable (16). These studies suggest that commensal microorganisms influence B cell maturation and class switching, although the effect may be indirect due to microbial influences on T cell populations. Future studies will need to address the specific cellular and molecular mechanisms that direct these effects.
2.2.2 T cells
A recent study indicates that exposure to the microbiota does not affect T cell populations in the thymus (15). In one study using transgenic mice designed to limit the repertoire of TCR genes, thymically derived Treg cells had TCR gene usage that was found to be distinct from that used by Tregs in the colon. Furthermore, Foxp3+ thymocytes could not be created by retroviral transduction of colonic Treg TCR genes into Rag1−/− mice (17). These data suggest commensal microbiota do not influence thymically derived TCR usage. However, one recent study suggests that Treg cells with TCRs directed against the members of the commensal microbiota develop in the thymus (18). This is clearly an area where additional studies will be useful to better understand these discrepant results.
Peripheral development of T cells is greatly affected by microbiota. Multiple studies have demonstrated that the absolute number of splenic T cells is diminished up to 50% in germ-free mice, but in contrast to mucosal sites, the ratio of naïve and memory T cells is unaffected (15, 19–21). Specifically, the microbiota exerts considerable effects on the CD4+ T helper cell population. The profile of splenic and cord blood T cells is skewed towards IL-4 producing Th2 cells in both germ-free mice and human neonates (21, 22). Specific bacteria can affect the development of Th subsets. Three of the best-studied examples are the effect of Bacteroides fragilis on Th1 cells, segmented filamentous bacteria (SFB) on the development of Th17 cells, and Clostridia species on regulatory T cells.
Bacteroides fragilis is a gram-negative anaerobe that colonizes the lower gastrointestinal tract of humans and mice whose immunomodulatory properties have been well characterized. Its zwitterionic capsular polysaccharide A is immunodominant (23–25). As noted above, germ-free mice have hypoplastic follicles and reduced numbers of CD+ T cells in the spleen. Upon colonization with B. fragilis, and specifically exposure of the host to PSA, CD4+ T cell numbers in the spleen increase to that of conventionally housed mice and lymphocyte follicles form. The mechanism for this increase in CD4+ T cell repopulation was shown to be due to DC internalization of PSA and activation in the intestine and migration to mesenteric lymph nodes. Additionally, the skewed Th2 profile of germ-free mice was corrected by colonization with B. fragilis in a PSA-dependent manner. B. fragilis and PSA alone were sufficient to induce an expansion of CD4+ IFN-γ+ Th1 cells in germ-free mice, restoring the Th1/Th2 balance to that of conventionally housed mice (21). In contrast, in the intestinal mucosa PSA induces an expansion of CD4+ Foxp3+ IL-10-producing T cells in a TLR2-dependent manner, resulting in protection from experimental colitis (26, 27). Therefore, mucosa-microbial interactions can be distinct from the effects of the microbiota on systemic immunity even for the same microbial antigen.
Th17 cells develop under the control of the transcription factor RORγt, which is preferentially expressed in the small intestine lamina propria and thymus. Signals from IL-6 in the presence of TGF-β stimulate expression of RORγt in immature T cells and result in their differentiation so that they produce IL-17 (28). The majority of Th17 cells appear to develop within the small intestine lamina propria. Shortly after birth, Th17 cells are undetectable in mice, but increase in number as the mouse ages in a conventional environment. At weaning, they represent the majority of IL-17 producing cells in the lamina propria. This finding correlates with bacterial colonization of the mouse. In support of this hypothesis, Th17 cells are substantially diminished in germ-free mice. After reintroduction of fecal contents from conventional mice, Th17 cells begin to appear in conventionalized mice about two weeks after colonization, and plateau in numbers by six weeks after colonization (29). Using antibiotics and mice from two vendor sources, specific colonization of mice with SFB, an as yet unculturable gram-positive bacteria with a minimal genome belonging to Clostridiales (30), were sufficient to stimulate differentiation of Th17 cells within the lamina propria (31). The relationship of additional commensal bacterial species and Th17 development is an area that needs further refinement as SFB do not colonize humans.
Furthermore, commensal bacteria, and specifically SFB, are required for the development of systemic Th17 responses. In the K/BxN mouse model of spontaneous inflammatory arthritis, autoantibodies to glucose-6-phosphate isomerase (GPI) and autoreactive T cell responses are required to initiate disease. Arthritis does not occur in mice housed under germ-free conditions as anti-GPI antibodies, B cells, and germinal center formation are significantly decreased. Additionally, germ-free mice have decreased follicular T helper cells. While the number of activated T cells in the spleens of the germ-free and K/BxN mice is similar, T cells have diminished responsiveness to antigen challenge with GPI. In this model, 30–50% of Th17 cells circulate from the small intestine under conventional housing conditions, but are absent in germ-free mice. Colonizing germfree K/BxN mice with SFB re-establishes arthritis, and using antibiotics to eliminate SFB in conventionally housed mice prevents disease (32). Similar results were obtained in the experimental autoimmune encephalomyelitis mouse model of multiple sclerosis (33).
Regulatory T cells are also affected by the presence of specific members of the intestinal microbiota. Studies have shown that the majority of Tregs reside within the gut. In germ-free animals, Treg cells are present in normal to increased numbers in the lamina propria of the small intestine but are reduced in the colon. Colonization of germ-free mice with a cocktail of 46 species of Clostridia resulted in increased numbers of colonic Tregs to that of conventionally housed mice while small intestinal levels of Tregs remained unchanged. Interestingly, three weeks after colonization of germ-free mice with this Clostridia mixture, the number of Treg cells found in the lung, liver, and spleen significantly increased to levels of conventionally housed mice (34), suggesting a systemic effect of this commensal genus on Treg development. Additional studies using a human fecal sample treated with chloroform and gavaged into germ-free mice confirmed that 17 Clostridia strains could expand the Treg population in mice (35).
2.2.3 Innate immunity
Initial studies suggested that macrophages, DCs, neutrophils, and NK cells in the spleen and LNs are unaffected in germ-free mice (13, 36, 37), but later studies demonstrated that while the total numbers of cells and their subsets were unaffected, function is altered (38, 39). These studies reveal that commensal organisms are required to prime the innate immune system.
Neutrophil function is altered by in the presence of microbial products. Neutrophils isolated from the bone marrow of germ-free mice or mice treated with broad-spectrum antibiotics have reduced ex vivo killing of the pathogens Staphlyococcus aureus and Streptococcus pneumonia. The mechanism was linked to microbial peptidoglycan from commensal microbiota that circulates systemically. After interaction with Nod1, peptidoglycan stimulates neutrophil killing of bacteria. Priming neutrophils from germ-free mice with peptidoglycan restored their capacity to opsonize bacteria (38).
Priming of the innate immune system also affects the activity of DCs and NK cells. Microbial stimulation of splenic DCs from germ-free mice fails to produce IL-15, type I IFNs, IL-6, TNF-α, IL-12, and IL-18. However, DCs are not globally unresponsive, as they can increase expression of CD86, Ccl5, and Fpr1. The lack of type I IFNs and IL-15 production by DCs further impairs NK cell function, demonstrated by a lack of IFN-γ after poly(I:C) injection and uncontained systemic murine cytomegalovirus infection in germ-free mice. Transfer of NK cells from germ free-mice into conventionally housed mice demonstrated that the defect of NK cell function in germ-free mice was cell extrinsic as injection of poly(I:C) resulted in normal levels of IFN- γ production in the transferred cells from germ-free mice (39).
2.3 Microbial exposure early versus late in life
The hygiene hypothesis suggests that microbial exposures early in life influence immune development and alter risk for disease later in life (8). Yet, very few studies address the role of early colonization on systemic immune development. Mucosal transcriptome analysis demonstrates “normalization” of the mucosal system in germ-free mice 8–16 days after colonization (40). Colonization of germ-free mice by co-housing with conventionally raised mice at one or three weeks of age continue to have elevated IL-10 and TGF-β in the sera and unaffected splenic cellular populations, similar to germ-free controls (41). This provocative result suggests that very early colonization with commensal organisms has lasting effects on systemic immunity. More studies will be required to better investigate how the timing of specific microbial colonization affects systemic immunity.
3. Effect of Commensals on Systemic Responses to Antigen
Germ-free mice can mount a robust immune response following immunization with purified antigen (42–44), but mice with a depleted microbiota resulting from treatment with broad-spectrum antibiotics are unable to mount a sufficient immune response to pathogen due to a lack of innate cell priming (45–48). These data point to the crucial role for APCs to be persistently activated at mucosal surfaces in order to mount appropriate immune responses to pathogens. Once the adaptive immune system has been educated by antigen, it can react independently of the presence of commensal microorganisms.
3.1 Immunization
Though immunization of germ free mice with T-dependent and T-independent antigens initially leads to a delayed IgM response, this is followed by greater numbers of antigen-specific IgG producing B cells in the spleen (42, 44). Immunization of antigen-free mice compared to germ-free mice and then conventional mice with dinitrophenyl-keyhole limpet hemocyanin results in greater T cell responses when T cells and APCs from the spleens of the immunized mice are stimulated ex vivo. This study demonstrates that the lower the antigenic load in the mouse, the greater the ability of the APCs to generate more T cell proliferative responses. The source of T cells did not have an impact (43).
Why is there an apparent hyper-responsiveness of the B and T cells to immunogen in the absence of microbiota? One possibility is that without the diversification of the T and B cell repertoire by the presence of commensal microorganisms during development, and without bystander activation of immune cells to microbial products in the environment (e.g. skin flora), immunization with a single antigen leads to a hyper-focused immune response. Studies using monocolonized germ-free mice and/or conventionally housed mice treated with broad-spectrum antibiotics would address this hypothesis and aid in our understanding of how microbes in our environment shape immunization.
3.2 Infection
One major purpose of the intestinal microbiota is to provide local resistance to colonization by pathogens (46, 48). In addition it also provides systemic resistance to infections by mechanisms that are now only begun to be understood. This later concept was nicely demonstrated by multiple studies with influenza A. Broad-spectrum antibiotic treatment of mice prior to influenza A infection results in greater mortality and morbidity. In the first study, TLR activation in the gut was associated with improved innate responses that activated T cells for clearing the influenza infection (47). A second study demonstrated that innate type I IFN responses by macrophages required the presence of commensal organisms in order to effectively activate CD8+ T cells for clearance of influenza A and lymphocytic choriomeningitis virus infections (45). These data are in accordance with previous studies (38, 39) demonstrating the need for innate cell priming by commensal organisms for optimal immune function.
4. Role of commensal bacteria in autoimmunity
Numerous recent studies show associations between the composition of the microbiota and risk for autoimmune disease. Many studies of large human cohorts have identified shifts in bacterial phyla and species diversity when comparing healthy controls to diseased individuals (49–51). However, direct links between microbes and human disease have not been made. Therefore, animal models have been utilized in an attempt to study the interaction between microbes and their host. In this setting, the hypothesis that microbial alterations lead to autoimmune disease can be tested.
4.1 Inflammatory bowel disease
Dysbiosis (defined as altered composition of the microbiota) occurs in patients with inflammatory bowel disease (IBD). Specifically there is loss of members of the Clostridiales and enrichment for Enterobacteriaceae (49). However, no single species has been identified to associate with disease.
Animal models of colitis suggest a role of gut microbes in modulating inflammation. In colitis induced by dextran sodium sulfate (DSS), germ-free mice show a worse response to this toxin in part due to a lack of anti-inflammatory actions of microbe-derived short-chain fatty acids (SCFAs) on neutrophils (52). This effect, though, is likely complex and includes effects of microbes on non-immune cell types such as the maintenance of epithelial stem cell proliferation (53). Additionally, some bacterial species or their antigens can help protect mice from experimental colitis even when housed in conventional settings. As mentioned previously, PSA from B. fragilis can expand local IL-10+ regulatory T cells. This results in protection of mice from colitis induced by either trinitrobenzene sulphonic acid (TNBS) or the transfer of CD4+CD45Rbhigh T cells in the presence of Helicobacter hepaticus (26, 27). Clostridia species that induce systemic expansion of Treg cells also protect mice from TNBS colitis through the production of IL-10 and TGB-β (35).
In contrast, another commensal organism, Bacteroides thetaiotamicron, causes colitis in a genetically susceptible mouse model. Mice that are deficient in Il10rb and transgenic for a dominant-negative Tgfbr2 on CD4 T cells (dnKO mice) develop spontaneous colitis with 100% incidence by four weeks of age. Disease is characterized by diffuse colonic inflammation with neutrophil and macrophage recruitment, crypt abscesses and dropout, and epithelial hyperproliferation. Th1 and Th17 cells are expanded in the lamina propria and mesenteric lymph nodes. Systemically mice have elevated cytokines IL-6, TNF-α, and IFN-γ. Colitis can be treated with the antibiotics ciprofloxacin and metronidazole. Through a series of microbial re-colonization of antibiotic-treated dnKO mice, the commensal bacteria B. theta was demonstrated to be a causative organism for the initiation of colitis (54, 55).
Dietary changes can also impact the microbiota and modulate the immune responses leading to colitis in animal models. Mice fed a Western-style diet high in saturated fat results in a shift of the microbiota leading to increased abundance of Bacteroidetes, decreased abundance of Firmicutes, and the appearance of Bilophila wadsworthia. Germ-free mice monocolonized with B. wadsworthia have increased Th1 cytokines in the colon, expanded Th1 cells in the MLNs and spleen, and DCs from the MLNs and spleen that produce increased IL-12p40. Finally, colitis in Il10−/− mice and DSS-treated mice is worse in the presence of this bacterium (56).
Intestinal commensal Clostridiales are known to generate SCFAs by digestion of insoluble fiber in the host’s diet. SCFAs increase the number and function of colonic regulatory T cells. In the T cell transfer model of colitis in which naïve T cells are injected into Rag1−/− recipients, co-transfer of Treg cells decreases the severity of colitis. When the Rag1−/− recipients were given SCFAs in their drinking water, they demonstrated even less severe colitis by histology and reduced inflammatory cytokines (57).
4.2 Rheumatoid arthritis
Like IBD, rheumatoid arthritis (RA) likely has multiple pathways that may lead to disease. One pathway implicates interactions between microbiota and systemic autoimmune disease. Up to 70% of patients with RA have antibodies to citrullinated peptides termed CCP (58, 59). Citrullination is a natural post-translational modification of the amino acid arginine by the enzyme peptidylarginine deiminase (PAD). Anti-CCP antibodies have been shown to be pathologic in an animal model of RA (60), and citrullinated protein is bound with higher affinity by MHC class II HLA-DR4 alleles that associate with RA (61). Porphymonas gingivalis is the only known microbe that expresses PAD and citrullinates host proteins (62); it is a common cause of periodontal disease despite the fact that it is a minor component of the oral microbiome both in health and disease (63). Interestingly, studies have shown a correlation between RA, anti-CCP antibodies, and periodontal disease, specifically colonization of the oral cavity with P. gingivalis (50, 64, 65).
In mouse models of inflammatory arthritis, the presence of gut commensals or their antigens is important to initiate disease. For example germ-free rats develop arthritis at 100% incidence when immunized intradermally with adjuvant containing heat-killed Mycobacterium bovis BCG or peptidoglycan from Streptococcus epidermidis, but only at 20% incidence when rats are housed conventionally, suggesting that microbial flora helps establish systemic tolerance to bacterial antigens (66). Conversely, in spontaneous models of arthritis due to genetic manipulations, mice housed in germ-free conditions fail to develop arthritis (32, 67). K/BxN mice develop arthritis due to autoimmune Th17 responses to GPI only after exposure to SFB (32). SKG mice, which have a mutation in the T cell ZAP-70 signal transduction protein, rely upon fungal β-glucan stimulation of DCs in order to develop spontaneous arthritis (67).
4.3 Type I diabetes
In a small study of children with type 1 diabetes, compared to age-, sex-, and HLA-matched controls, an expansion of Bacteroidetes and reduced abundance of lactate- and butyrate-producing species associated with antibodies to β-islet cells (51), again suggesting an anti-inflammatory and protective role for microbially produced SCFAs.
The incidence of diabetes in the non-obese diabetic (NOD) mouse model of type I diabetes relies upon interaction with gut microbiota. In some colonies, diabetes reaches 100% in germ-free housing conditions (68), and protection from diabetes in the NOD mouse is associated with the presence of SFB (69). NOD mice deficient in MyD88 have an altered gut flora compared to MyD88-sufficient NOD mice with an overrepresentation in the families of Lactobacillaceae (Firmicutes), Rikenellaceae (Bacteroidetes), and Porphyromonadaceae (Bacteroidetes). The lack of MyD88 in NOD mice in a conventional facility did not associate with increased incidence of diabetes. When placed in germ-free conditions, however, MyD88−/− NOD mice had increased diabetes incidence (70). These data suggest that interactions of innate signaling with commensal organisms are linked to autoimmune susceptibility.
5. Conclusions
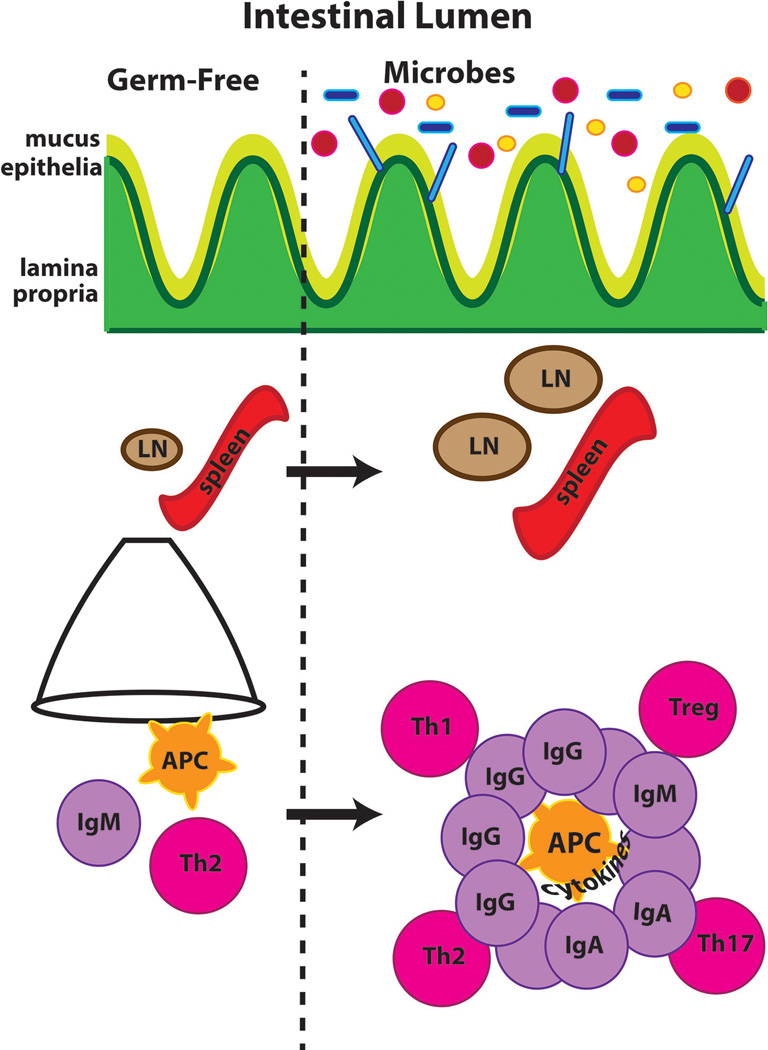
Studies in germ-free mice have demonstrated that our microbiome significantly impacts systemic immune function. A model based on the published data discussed above is presented in Figure 1. Population studies of microbial shifts in individuals with disease lend support to the animal data.
Figure 1. Model of microbial influences on systemic immune development.
Germ-free mice have hypoplastic secondary immune organs with little B cell differentiation and Th2 skewed responses. Upon colonization, the architecture of secondary immune organs is restored. T and B cell responses normalize to represent a more diverse repertoire, which is dependent upon microbial interactions with innate antigen presenting cells (APC).
As the human microbiome project expands our understanding of the microbes that live with us, we will gain a better appreciation of their role in shaping our systemic immune system. Many studies have demonstrated how single organisms can modulate systemic immune responses, but these have been limited to investigations in the germ-free setting or after broad-spectrum antibiotic treatment. How specific microbes interact with the host in the natural microbial background will be the challenge of future studies. Additionally, our knowledge of bacterial metabolites beyond just SCFAs is expanding and these products will be critical new tools to test the effect of these microbial products on systemic immunity.
Additionally, there are limitations in our understanding of the plasticity of early microbial influences on immune development. The window of time for systemic immune education and the length in time of the immune effects from microbial exposure during this window remains to be addressed.
Highlights.
Germ-free mice demonstrate hypoplastic secondary immune organs, reduced T and B cells, and decreased innate responses.
Germ free mice demonstrate exaggerated responses to immunogens.
Specific commensal bacteria can stimulate Th1, Th17, and Treg responses.
Systemic and mucosal responses to bacteria and bacterial antigens are not necessarily the same.
Associations have been made between altered intestinal microbiota and risk of autoimmune diseases.
Acknowledgements
This work was supported by the NIH (DK09707901) and fellowships from W.M. Keck foundation for research in molecular medicine and the American College of Rheumatology Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: APC, antigen presenting cell; CCP, cyclic citrullinated peptide; DC, dendritic cell; GPI, glucose-6-phosphate isomerase; IBD, inflammatory bowel disease; MLN, mesenteric lymph node; LN, lymph node; PAD, peptidylarginine deiminase; PSA, polysaccharide A; RA, rheumatoid arthritis; SCFA, short-chain fatty acid; SFB, segmented filamentous bacteria; SPF, specific-pathogen free; TNBS trinitrobenzene sulphonic acid.
For the purpose of this review, the terms conventionally housed or SPF housed will be used interchangeably to reference mice that have intact commensal microbiota.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences of the United States of America. 1998 Jun 9;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006 Feb 24;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010 Jun 29;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Biphasic assembly of the murine intestinal microbiota during early development. The ISME journal. 2013 Jun;7(6):1112–1115. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011 Mar 15;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut microbes. 2011 Mar-Apr;2(2):99–104. doi: 10.4161/gmic.2.2.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012 Jun 14;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strachan DP. Hay fever, hygiene, and household size. Bmj. 1989 Nov 18;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, Heederik D, Piarroux R, von Mutius E Group GTS. Exposure to environmental microorganisms and childhood asthma. The New England journal of medicine. 2011 Feb 24;364(8):701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 10.Lossius A, Johansen JN, Torkildsen O, Vartdal F, Holmoy T. Epstein-Barr virus in systemic lupus erythematosus, rheumatoid arthritis and multiple sclerosis-association and causation. Viruses. 2012 Dec;4(12):3701–3730. doi: 10.3390/v4123701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature reviews Immunology. 2004 Jun;4(6):478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 12.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder TF, Hardt WD, Bercik P, Verdu EF, McCoy KD, Macpherson AJ. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009 Jul 31;325(5940):617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. The American journal of pathology. 1963 Apr;42:471–483. [PMC free article] [PubMed] [Google Scholar]
- 14.Hooijkaas H, Benner R, Pleasants JR, Wostmann BS. Isotypes and specificities of immunoglobulins produced by germ-free mice fed chemically defined ultrafiltered "antigen-free" diet. European journal of immunology. 1984 Dec;14(12):1127–1130. doi: 10.1002/eji.1830141212. [DOI] [PubMed] [Google Scholar]
- 15.Pereira P, Forni L, Larsson EL, Cooper M, Heusser C, Coutinho A. Autonomous activation of B and T cells in antigen-free mice. European journal of immunology. 1986 Jun;16(6):685–688. doi: 10.1002/eji.1830160616. [DOI] [PubMed] [Google Scholar]
- 16.Vitetta ES, Grundke-Iqbal I, Holmes KV, Uhr JW. Cell surface immunoglobulin. VII. Synthesis, shedding, and secretion of immunoglobulin by lymphoid cells of germ-free mice. The Journal of experimental medicine. 1974 Apr 1;139(4):862–876. doi: 10.1084/jem.139.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011 Oct 13;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, Bry L, Kraj P, Kisielow P, Ignatowicz L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013 May 9;497(7448):258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobber R, Hertogh-Huijbregts A, Rozing J, Bottomly K, Nagelkerken L. The involvement of the intestinal microflora in the expansion of CD4+ T cells with a naive phenotype in the periphery. Developmental immunology. 1992;2(2):141–150. doi: 10.1155/1992/57057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee WT, Yin XM, Vitetta ES. Functional and ontogenetic analysis of murine CD45Rhi and CD45Rlo CD4+ T cells. Journal of immunology. 1990 May 1;144(9):3288–3295. [PubMed] [Google Scholar]
- 21.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005 Jul 15;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Prescott SL, Macaubas C, Holt BJ, Smallacombe TB, Loh R, Sly PD, Holt PG. Transplacental priming of the human immune system to environmental allergens: universal skewing of initial T cell responses toward the Th2 cytokine profile. Journal of immunology. 1998 May 15;160(10):4730–4737. [PubMed] [Google Scholar]
- 23.Tzianabos AO, Finberg RW, Wang Y, Chan M, Onderdonk AB, Jennings HJ, Kasper DL. T cells activated by zwitterionic molecules prevent abscesses induced by pathogenic bacteria. The Journal of biological chemistry. 2000 Mar 10;275(10):6733–6740. doi: 10.1074/jbc.275.10.6733. [DOI] [PubMed] [Google Scholar]
- 24.Tzianabos AO, Russell PR, Onderdonk AB, Gibson FC, 3rd, Cywes C, Chan M, Finberg RW, Kasper DL. IL-2 mediates protection against abscess formation in an experimental model of sepsis. Journal of immunology. 1999 Jul 15;163(2):893–897. [PubMed] [Google Scholar]
- 25.Cobb BA, Wang Q, Tzianabos AO, Kasper DL. Polysaccharide processing and presentation by the MHCII pathway. Cell. 2004 May 28;117(5):677–687. doi: 10.016/j.cell.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008 May 29;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 27.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2010 Jul 6;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006 Sep 22;126(6):1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008 Oct 16;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell host & microbe. 2011 Sep 15;10(3):260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009 Oct 30;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010 Jun 25;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proceedings of the National Academy of Sciences of the United States of America. 2011 Mar 15;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011 Jan 21;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013 Aug 8;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, McLoughlin RM, Cobb BA, Charrel-Dennis M, Zaleski KJ, Golenbock D, Tzianabos AO, Kasper DL. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. The Journal of experimental medicine. 2006 Dec 25;203(13):2853–2863. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson NS, Young LJ, Kupresanin F, Naik SH, Vremec D, Heath WR, Akira S, Shortman K, Boyle J, Maraskovsky E, Belz GT, Villadangos JA. Normal proportion and expression of maturation markers in migratory dendritic cells in the absence of germs or Toll-like receptor signaling. Immunology and cell biology. 2008 Feb;86(2):200–205. doi: 10.1038/sj.icb.7100125. [DOI] [PubMed] [Google Scholar]
- 38.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature medicine. 2010 Feb;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012 Jul 27;37(1):171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 40.El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, Dore J, Dekker J, Samsom JN, Nieuwenhuis EE, Kleerebezem M. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal immunology. 2012 Sep;5(5):567–579. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 41.Hansen CH, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, Tlaskalova-Hogenova H, Hansen AK. Patterns of early gut colonization shape future immune responses of the host. PloS one. 2012;7(3):e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bos NA, Ploplis VA. Humoral immune response to 2,4-dinitrophenyl--keyhole limpet hemocyanin in antigen-free, germ-free and conventional BALB/c mice. European journal of immunology. 1994 Jan;24(1):59–65. doi: 10.1002/eji.1830240110. [DOI] [PubMed] [Google Scholar]
- 43.Hooper DC, Molowitz EH, Bos NA, Ploplis VA, Cebra JJ. Spleen cells from antigen-minimized mice are superior to spleen cells from germ-free and conventional mice in the stimulation of primary in vitro proliferative responses to nominal antigens. European journal of immunology. 1995 Jan;25(1):212–217. doi: 10.1002/eji.1830250135. [DOI] [PubMed] [Google Scholar]
- 44.Schuler W, Lehle G, Weiler E, Kolsch E. Immune response against the T-independent antigen alpha (1 leads to 3) dextran. I. Demonstration of an unexpected IgG response of athymic and germ-free-raised euthymic BALB/c mice. European journal of immunology. 1982 Feb;12(2):120–125. doi: 10.1002/eji.1830120205. [DOI] [PubMed] [Google Scholar]
- 45.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012 Jul 27;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012 Jun 22;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011 Mar 29;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infection and immunity. 2012 Nov;80(11):3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A, Lipuma L, Attur M, Pillinger MH, Weissmann G, Littman DR, Pamer EG, Bretz WA, Abramson SB. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis and rheumatism. 2012 Oct;64(10):3083–3094. doi: 10.1002/art.34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, Orivuori L, Hakala S, Welling GW, Harmsen HJ, Vaarala O. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013 Apr;62(4):1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009 Oct 29;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005 Jan 4;102(1):99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell host & microbe. 2011 May 19;9(5):390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, Allen PM. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008 Mar 4;5(3):e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012 Jul 5;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013 Aug 2;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Jaarsveld CH, ter Borg EJ, Jacobs JW, Schellekens GA, Gmelig-Meyling FH, van Booma-Frankfort C, de Jong BA, van Venrooij WJ, Bijlsma JW. The prognostic value of the antiperinuclear factor, anti-citrullinated peptide antibodies and rheumatoid factor in early rheumatoid arthritis. Clinical and experimental rheumatology. 1999 Nov-Dec;17(6):689–697. [PubMed] [Google Scholar]
- 59.Rantapaa-Dahlqvist S, de Jong BA, Berglin E, Hallmans G, Wadell G, Stenlund H, Sundin U, van Venrooij WJ. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis and rheumatism. 2003 Oct;48(10):2741–2749. doi: 10.1002/art.11223. [DOI] [PubMed] [Google Scholar]
- 60.Kuhn KA, Kulik L, Tomooka B, Braschler KJ, Arend WP, Robinson WH, Holers VM. Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. The Journal of clinical investigation. 2006 Apr;116(4):961–973. doi: 10.1172/JCI25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. Journal of immunology. 2003 Jul 15;171(2):538–541. doi: 10.4049/jimmunol.171.2.538. [DOI] [PubMed] [Google Scholar]
- 62.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infection and immunity. 1999 Jul;67(7):3248–3256. doi: 10.1128/iai.67.7.3248-3256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011 Nov 17;10(5):497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC, Holers VM, Kuhn KA, O'Dell JR. Antibody responses to Porphyromonas gingivalis (P. gingivalis) in subjects with rheumatoid arthritis and periodontitis. International immunopharmacology. 2009 Jan;9(1):38–42. doi: 10.1016/j.intimp.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikuls TR, Thiele GM, Deane KD, Payne JB, O'Dell JR, Yu F, Sayles H, Weisman MH, Gregersen PK, Buckner JH, Keating RM, Derber LA, Robinson WH, Holers VM, Norris JM. Porphyromonas gingivalis and disease-related autoantibodies in individuals at increased risk of rheumatoid arthritis. Arthritis and rheumatism. 2012 Nov;64(11):3522–3530. doi: 10.1002/art.34595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohashi O, Kuwata J, Umehara K, Uemura F, Takahashi T, Ozawa A. Susceptibility to adjuvant-induced arthritis among germfree, specific-pathogen-free, and conventional rats. Infection and immunity. 1979 Dec;26(3):791–794. doi: 10.1128/iai.26.3.791-794.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshitomi H, Sakaguchi N, Kobayashi K, Brown GD, Tagami T, Sakihama T, Hirota K, Tanaka S, Nomura T, Miki I, Gordon S, Akira S, Nakamura T, Sakaguchi S. A role for fungal {beta}-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. The Journal of experimental medicine. 2005 Mar 21;201(6):949–960. doi: 10.1084/jem.20041758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world--recent facts and figures. Immunology today. 1993 May;14(5):193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 69.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2011 Jul 12;108(28):11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008 Oct 23;455(7216):1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]