Abstract
The hepatitis C virus (HCV) internal ribosome entry site (IRES) drives non-canonical initiation of protein synthesis necessary for viral replication. HCV IRES functional studies have focused on 80S ribosome formation, but have not explored roles after the 80S ribosome is poised at the start codon. Here, we report that mutations of an IRES domain that docks in the 40S subunit’s decoding groove and cause only a local perturbation in IRES structure result in conformational changes in the IRES-rabbit 40S subunit complex. Functionally, we find the mutation decreases IRES activity by inhibiting the first ribosome translocation event, and modeling suggests that this effect is through an interaction with a single ribosomal protein. The HCV IRES’ ability to manipulate the ribosome provides insight into how the ribosome’s structure and function can be altered by bound RNAs, including those derived from cellular invaders.
Keywords: hepatitis C virus, internal ribosome entry site, cryo-electron microscopy, ribosome structure, ribosome translocation, ribosomal protein S5, viral RNA
INTRODUCTION
Hepatitis C virus (HCV) is the most common blood-borne virus in the United States and is estimated to infect ~3% of the world’s population. HCV’s genome is a single-stranded, positive-sense RNA molecule and thus after release of the genome into the cytoplasm, the first step of viral replication is translation of a single open reading frame to yield the viral proteins. The HCV genomic RNA is therefore similar to a cellular messenger RNA (mRNA), serving as the template for protein synthesis by the translation machinery. However, unlike cellular mRNAs that originate in the nucleus and are capped on their 5′ end by 7-methylguanosine and polyadenylated on the 3′ end (both important translation initiation signals), the HCV genomic RNA is delivered directly to the cytoplasm lacking both a cap and a poly(A) tail (Fig. 1a). As a result, translation of the HCV RNA is initiated by a mechanism that differs substantially from the mechanism used by the cell to translate its own mRNAs. Specifically, HCV uses an internal ribosome entry site (IRES) RNA at the 5′ end of the viral genome 1 to hijack the cellular translation machinery. The HCV IRES RNA sequence is highly conserved among HCV isolates and genotypes 2,3. This conservation underscores the HCV IRES RNA’s importance to the viral replication cycle and reflects the specificity of the interactions between the IRES RNA and the translation machinery.
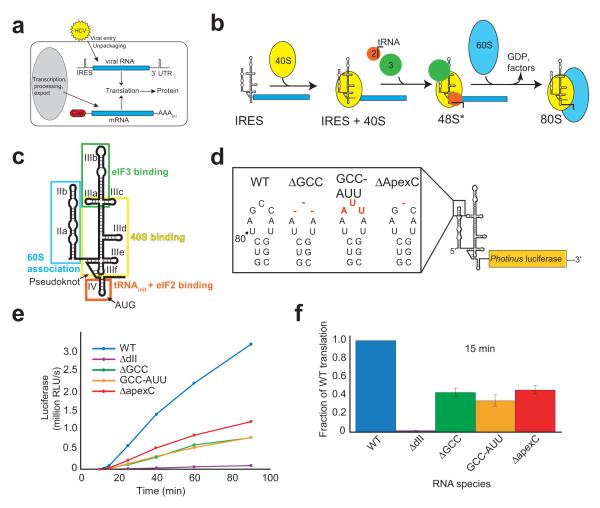
Figure 1. In vitro translation analysis of dIIb mutations.
(a) HCV viral RNA and cellular mRNA differ in their origin and features. HCV viral RNA is delivered directly to the cytoplasm lacking a cap and poly-(A) tail, while cellular mRNA is produced and processed in the nucleus before exportation to the cytoplasm with a cap and tail. However, both are translated by the same cellular machinery, mandating different mechanisms of initiation. (b) Simplified diagram HCV IRES 80S ribosome assembly mechanism. The IRES first binds the 40S subunit (yellow), then eukaryotic initiation factor (eIF) 3 (green) and the ternary complex (eIF2-GTP-Met-tRNAi, red and line), and finally after GTP hydrolysis and eIF release the 60S subunit (blue) joins to form an 80S ribosome. Asterisk denotes a difference from canonical 48S complexes. (c) Cartoon representation of the secondary structure of the HCV IRES. The location of the start AUG is shown. Boxes areas indicate the parts of the IRES involved in different steps of the mechanism shown in panel b. (d) Schematic of mutations (red) made to domain IIb in the context of the uncapped and unpolyadenylated monocistronic Photinus luciferase reporter. Wild-type (WT) RNA is shown to the left. (e) Time course of a translation assay from 0 to 90 min as measured by produced luciferase relative light units (RLUs). (f) Fifteen minute translation assay with RLUs calculated as a fraction of the wild type IRES. Error bars represent one s.e.m for three independent triplicate experiments.
Studies of the HCV IRES’ mechanism have focused on how the IRES assembles an 80S ribosome directly at the AUG start codon, revealing a mechanism different from canonical translation initiation 4. In the canonical pathway the 5′ cap is recognized by the eukaryotic initiation factor (eIF) 4F complex, followed by binding of the 43S particle (which includes the 40S subunit, the eIF2-GTP-met-tRNAimet ternary complex, eIF3, and other factors). The subunit then scans the mRNA to locate the start codon, at which time factor release and 60S subunit association yield an 80S ribosome 5. In contrast, the HCV IRES first directly binds the 40S subunit 6-9 followed by binding of eIF3 and the ternary complex 10-12 (Fig. 1b). Subsequent GTP hydrolysis, eIF release, and binding of a 60S subunit yield an 80S ribosome placed directly at the start codon 13,14. In addition, the HCV IRES can use eIF2-independent pathways under conditions of cellular stress to generate 80S ribosomes 15,16.
The HCV IRES’ function is conferred by its structure. The IRES adopts an extended global architecture 17, within which specific RNA structural domains drive different steps in 80S ribosome formation (Fig. 1c) 18: domain III binds the 40S subunit and eIF3 6,7,19; domain IV provides the AUG initiation codon for interaction with the ternary complex 11,12,20; the pseudoknot is important for placement of the AUG in the 40S subunit decoding groove 21; and domain II is involved in eIF2-catalyzed GTP hydrolysis 14, removal of eIF3j 13, 60S subunit joining 11 and the configuration of RNA in the decoding groove 22. These findings, combined with the observation that the HCV IRES alters the conformation of the 40S subunit when it binds 23, indicate that the IRES RNA is an active manipulator of the translation machinery, not just a binding site for the ribosome and factors.
Among the aforementioned IRES domains, domain II (dII) is particularly intriguing: it induces changes in the conformation of the 40S subunit 23 and it is docked within the ribosome’s decoding groove to interact with ribosomal protein (rp) S5 24,25, a protein known to contact E-site tRNA 26-28. Hence, we set out to study the role of subdomain IIb (dIIb), the part of the IRES that penetrates deep into the ribosome’s decoding groove. We have discovered that mutations made to dIIb alter the conformation of the IRES-40S subunit complex and inhibit the first round of ribosome translocation. The modeled position of this domain adjacent to rpS5 suggests this effect may be due to alteration of a contact with this protein. This is the first evidence that the HCV IRES directly influences a step after assembly of the 80S ribosome, and may have implications for our understanding of translation initiation in general.
RESULTS
IRES dIIb affects the rate of protein synthesis
We previously reported that mutating the dIIb apical loop (Fig. 1d) changes the configuration of the HCV RNA in the decoding groove of 40S subunit-HCV IRES RNA complexes and reduces IRES-driven protein synthesis 22. This is consistent with other studies that show dIIb is important for HCV IRES-mediated translation initiation 11,29,30. To monitor protein production as a function of time, we measured the ability of three dIIb mutants to translate a downstream luciferase (LUC) reporter sequence (uncapped and not polyadenylated) in rabbit reticulocyte lysate (RRL) over a 90 minute time course (Fig. 1d & e). All three dIIb mutants produced LUC at a decreased rate compared to wild-type (WT), but at a greater rate than an IRES with dII deleted (ΔdII). We observed differences in LUC production after only 15 minutes (Fig. 1f) and the mutants continued to make protein at a roughly constant rate but slower than WT. Hence, the activity of the IRES is inhibited, but not abolished. Assuming identical elongation and ribosome termination rates on each RNA, this result shows that the dIIb mutations slow but do not halt translation initiation.
Conformation of IRES-40S subunit complex influenced by dIIb
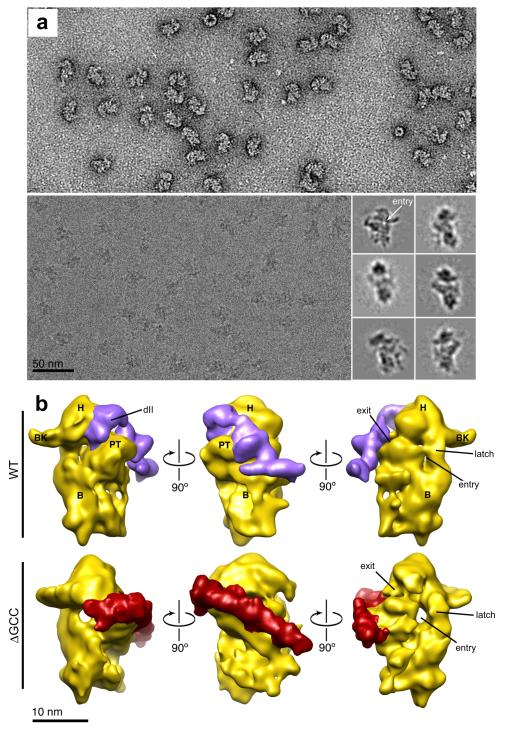
Cryogenic electron microscopy (cryo-EM) reconstructions of HCV IRES-40S subunit complexes show that binding of full-length WT HCV IRES induces structural changes in the subunit, but the ΔdII deletion mutant does not cause this conformational change 23. This raised the question: what is the conformation of the 40S subunit when bound to our dIIb mutant RNAs? To answer this, we visualized ΔGCC IRES RNA-40S subunit complexes by electron microscopy using both negative staining (to assess sample purity, homogeneity, and concentration) and cryo-EM (to generate a three-dimensional structure) (Fig. 2a). We obtained the reconstruction of the complex at a resolution comparable to previous HCV IRES-40S subunit reconstructions (17-20 Å). The structure of the ΔGCC IRES-40S subunit complex compared to the WT-40S subunit complex revealed surprising differences in the position and orientation of IRES domains as well as the conformation of the 40S subunit (Fig. 2b). Specifically, in the mutant, dII does not loop away from the subunit’s surface to contact the side of the head and enter the decoding groove; rather it lies across the platform. This change in the position of dII is accompanied by a rotation of the overall IRES orientation relative to the body of the 40S subunit. This cryo-EM structure cannot eliminate the possibility that the IRES’ location and conformation is an average of several similar structures. In other words, the IRES’ position on the 40S subunit and conformation could be more dynamic due to the ΔGCC deletion, potentially explaining why structural features visible in the WT IRES are not seen in the mutant IRES and why the position of the IRES appears rotated relative to the subunit. However, this possibility does not alter the conclusion that the relatively small deletion mutation at the tip of dIIb is altering the IRES-40S subunit interaction and is completely consistent with our observation that this mutation lowers, but does not eliminate, IRES activity. The conformation of the 40S subunit when bound to the mutant IRES has some features similar to the WT IRES-bound state, but also marked differences. In both, the latch formed between rRNA helices 18 and 34 is closed (this latch is open in the apo40S) 23,31. However, the entry site (where mRNA enters the decoding groove) is much more open in the mutant IRES complex; this feature is clearly visible in class averages assembled from individual particle images (Fig. 2a). This result is also consistent with a previous study showing that mutation of dIIb alters the configuration of the mRNA in the decoding groove 22. In summary, the dIIb mutant IRES binds the 40S subunit using the same side of the subunit that is used by the WT IRES. However, the structures are considerably different and thus deleting the apical nucleotides of dIIb substantially affects the global conformation of the complex. These changes are different from those caused by ΔdII 23, suggesting that mutation of dIIb perturbs translation initiation differently than does ΔdII.
Figure 2. Electron microscopy of HCV IRES mutant ΔGCC in complex with the 40S subunit.
(a) Representative electron micrographs of ΔGCCIRES-40S complexes stained using negative stain (top) and complexes embedded in vitrified ice (bottom). Inset, six classes of projection averages of particles in vitrified ice showing different views of the complex calculated using MSA and MRA programs in IMAGIC 54. The readily-observed open entry tunnel is shown (arrow). (b) Top: previously determined reconstruction of wild-type HCV IRES bound to the 40S ribosomal subunit 23 from left to right: solvent-accessible, exit-channel and solvent-inaccessible sides. 40S subunit is shown in yellow and key structural features are labeled: H=head; PT=platform; BK=beak; B=body. The entry and exit sites for mRNA are indicated. We calculated the density corresponding to the IRES (purple) from a difference map between the wild-type (WT) IRES-40S and apo40S 23. Location of dII is indicated. Bottom: reconstruction of the ΔGCC mutant IRES bound to the 40S ribosomal subunit (yellow) showing the three views as in the top panel. The mutant IRES is shown in red, calculated from a difference map between the ΔGCC IRES-40S reconstruction and apo40S 23.
Subdomain IIb influences a step after 80S ribosome formation
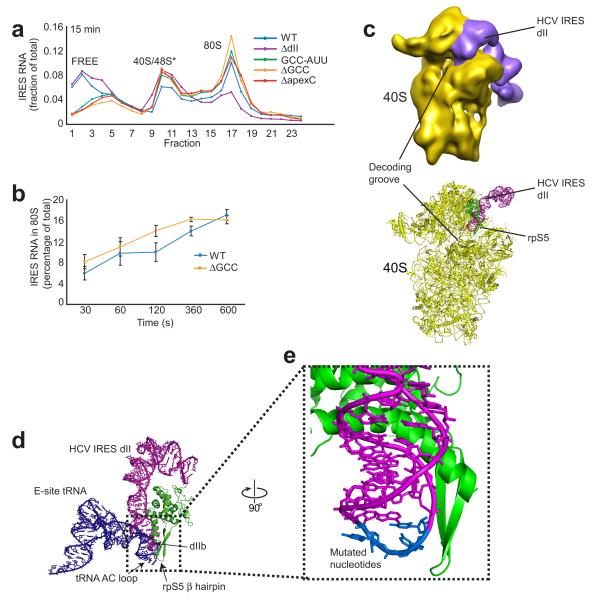
The structure of the mutant IRES–40S complex and translation initiation efficiencies of all three dIIb mutants raises the question of which step in initiation is affected by these mutations. Removal of domain II (ΔdII), or replacing the entire dIIb apical loop with an ultrastable UUCG tetraloop has been reported to inhibit 80S ribosome formation 11,12,14, suggesting our targeted dIIb mutations would do the same. To test this, we assayed ribosome assembly in RRL, separating the resultant ribosomal complexes by sucrose density gradient ultracentrifugation. Consistent with previous reports 11, after 15 minutes we observed robust 80S formation with WT IRES and the ΔdII mutant showed depressed levels (Fig. 3a). However, contrary to expectation, all of the targeted dIIb mutants formed both IRES-40S and IRES-48S* complexes and IRES-80S ribosomes as well as the WT IRES RNA (Fig. 3a). This result indicated that these mutations do not inhibit the formation of 80S ribosomes on the HCV IRES RNA. Furthermore, a 10 minute time course showed no difference in the rate of 80S formation between the dIIb mutants and WT (Fig. 3b). We can confirm from these data, combined with the translation initiation data (Fig. 1e), that mutation of dIIb does not inhibit 80S ribosome formation on the IRES. Thus, the altered position of the ΔGCC mutant IRES RNA on the 40S subunit and different conformation of the 40S subunit revealed by cryo-EM (relative to that with WT IRES) can support 80S ribosome formation, but some subsequent event is slowed by mutation of dIIb.
Figure 3. Wild-type (WT) and mutant IRES ribosome assembly assays and position of domain IIb.
(a) Graph of measured radiolabeled IRES RNA migration through a sucrose gradient after 15 minute incubation in rabbit reticulocyte lysate (RRL) followed by ultracentrifugation. 40S and 48S* are indistinguishable in our sucrose gradient. (b) Amount of 80S complex formed at time points from 0.5 to 10 min. Error bars represent one s.e.m of three independent experiments. (c) Top: cryo-EM reconstruction of the full-length HCV IRES RNA (purple) bound to mammalian 40S subunit (yellow) 23. Bottom: crystal structure of 40S subunit from Tetrahymena thermophila (yellow) 32 and the NMR structure of HCV IRES domain II (dII, purple) 33 placed into the cryo-EM reconstruction (not shown). RpS5 is green and structural features are labeled. (d) Comparison of the orientation of E-site bound tRNA (blue) and HCV IRES dII (purple) within the decoding groove. Position of dII is based on the model shown in panel d and previously published 22,24, while the E-site tRNA is from a crystal structure of the T. thermophilus 70S ribosome 55. RpS5 (S7 in bacteria) is green, its β hairpin and the tRNA anticodon (AC) loop are indicated. (e) Close-up view of the position of domain IIb (dIIb, purple) near the β-hairpin of rpS5 (green). The location of the nucleotides that were mutated in this study are blue and labeled.
Modeled local interaction between dIIb and rpS5
To determine which interactions of dIIb with the ribosome are disrupted by mutation, we modeled the placement of dII on the ribosome by docking a 40S subunit structure 32 and the structure of dII 33 into the cryo-EM density of the complete WT HCV IRES-40S subunit complex 22,23 (Fig. 3c). Our placement of dII is consistent with the model of Boehringer et al. based on an IRES•80S complex 24, and contains the added detail provided by the crystal structure of a 40S subunit. We compared this model with the crystal structure of a bacterial ribosome bound to tRNAs to determine to what degree dII’s modeled contacts to the 40S subunit are similar to a bound E-site tRNA’s (Fig. 3d). The two RNAs overlap in one location: the anticodon (AC) loop of the tRNA overlaps with the modeled position of the HCV IRES dIIb, and both are positioned directly against the β-hairpin structure of rpS5 (S7 in bacteria) (Fig. 3c). Specifically, we observed that the mutated nucleotides in dIIb are directly adjacent to the rpS5 β-hairpin (Fig. 3e). The modeled position of dIIb is consistent with observations of dII crosslinking to rpS5 25, with the aforementioned IRES-80S ribosome model 24, and with the role of rpS5’s β-hairpin in contacting tRNA.
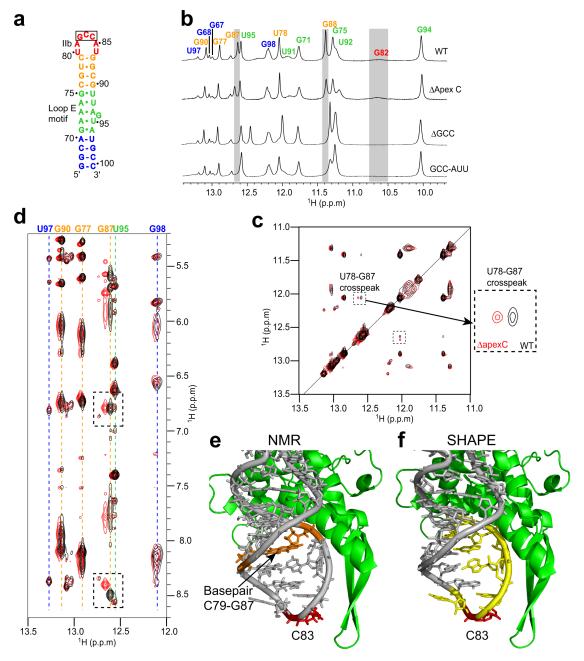
The putative location of our dIIb mutations adjacent to the β-hairpin of rpS5 suggests they disrupt a specific contact between dIIb and rpS5. Previous chemical probing of these mutant RNAs show no global change in IRES secondary structure, but it remains possible that the mutations change the overall structure of dII. To assess this, we characterized the structures of the dIIb mutant RNAs using nuclear magnetic resonance (NMR). We generated WT and mutant samples comprising nt 76-100 of the HCV IRES (Fig. 4a), which contains dIIb and is identical to RNAs used to solve the structure of dII 33. Comparison of the 1-dimensional 1H spectra obtained in water shows very little change in the chemical shifts or relative intensity of the peaks when WT is compared to the three mutant RNAs (Fig. 4b). The largest chemical shift is seen in the imino protons of G87 and G88, which are adjacent to the apical loop of dIIb. The visibility of these imino proton peaks in all spectra shows base-pairing is unaltered by dIIb mutation.
Figure 4. Characterization of the structural changes induced by dIIb mutation.
(a) Secondary structure of the RNA sequence (previously solved 33) used to characterize the structural changes induced by mutating domain IIb (dIIb). Elements color-coded to match other panels. (b) 1-D 1H-NMR spectra (in water) of the wild-type (WT) and dIIb mutant RNAs. This part of the spectrum contains resonances from the base imino protons with assignments for WT shown at the top. The gray boxes indicate the most shifted resonances in all three mutant spectra. (c) Overlaid WT and ΔapexC 2-D 1H-NOESY NMR spectra (in water). The portion of the spectra that contains the cross-peaks between imino protons is shown with the G87 and U78 imino protons cross-peak indicated. WT spectrum is black, mutant is red. (d) Same overlaid spectra and color scheme as in panel c, showing the cross-peaks between imino protons and other protons. The location of the cross-peaks between the G87 imino proton and the C79 amino protons are boxed, assignments of imino proton resonances are above the spectrum matching the colors of panels a and b. (e) Close-up view of the tip of dIIb against the β hairpin of rpS5 (green). The C79-G87 base-pair (orange) and location of the single base deletion (C83) in ΔapexC (red) are indicated. (f) Same view as panel e, but showing the location of previously-reported increases in chemical modification in the ΔapexC mutant (yellow) 22.
The ΔapexC mutant’s spectrum is most similar to WT’s; therefore this RNA elicits the post-80S ribosome functional effect with the smallest change in loop structure. In addition, we predict the single deleted nucleotide (C83) contacts the β-hairpin of rpS5 (Fig. 3e). Therefore, we subjected this mutant to additional NMR experiments. The portion of the 2-dimensional 1H-NOESY spectra in water that contains the imino-imino crosspeaks almost perfectly overlaps when the WT and ΔapexC data are overlaid; the largest shift occurs to the G87 imino (Fig. 4c). Likewise, we observed that spectra showing the crosspeaks between the imino protons and other protons shows only small chemical shift changes (Fig. 4d). For example, crosspeaks between the imino of G87 and C79 amino protons are shifted but intense (Fig. 4d), again confirming that this base-pair at the base of dIIb still forms in the mutant RNA (Fig. 4e). Changes to the spectra are limited to nucleotides adjacent to the deletion, showing that structural changes are localized to the dIIb apical loop. This result is consistent with published chemical probing data of the ΔapexC (and other mutant) RNAs using selective 2′-OH acylation analyzed by primer extension (SHAPE) in that changes in the chemical probing pattern are limited to the apical loop in the IRES-40S subunit complex (Fig. 4f) 22. Taken together, our data show that deletion of C83 (and likely the other mutations) induces a local structural perturbation that, we propose, disrupts dIIb’s interaction with the β-hairpin of rpS5; this is accompanied by a global change in the structure of the IRES-40S complex and with the inhibition of a step after 80S ribosome formation.
AUG start codon placement unaffected by dIIb mutations
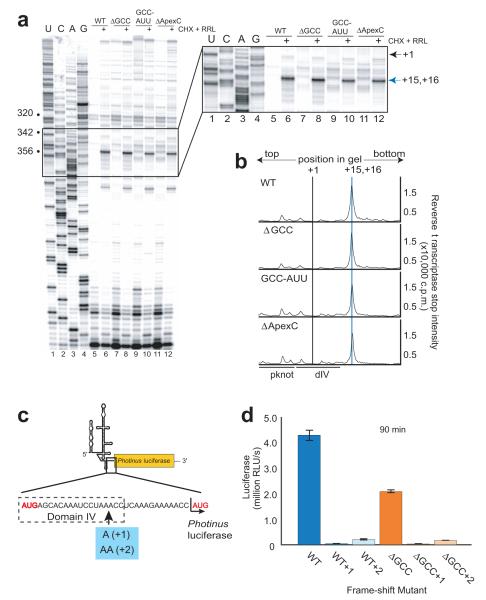
To identify the post-80S ribosome formation step affected by dIIb mutations, we first explored the possibility that they cause incorrect AUG start codon placement. To test this, we used primer extension inhibition (toeprinting) analysis 10,34-36 on WT and the dIIb mutants in the unbound and 80S ribosome-bound states (the latter is formed in RRL with cycloheximide; CHX) (Fig. 5a, compare lanes 5,7,9,11 with 6,8,10,12). As expected, we observe no toeprint on unbound RNAs However, when bound to the 80S ribosome, all mutants produced similar toeprints at the +15, +16 nts (where A of the initiation codon AUG is +1) downstream of the initiation codon (Fig. 5a & b). This indicates that the AUG is properly positioned in all of these complexes and the mutations likely affect a step after 80S ribosome assembly on the IRES.
Figure 5. Biochemical analysis of AUG docking and potential frame-shifting.
(a) Denaturing sequencing gel of the reverse transcription and toeprinting of wild-type (WT) and mutant IRES RNAs with the relevant part of the gel boxed and expanded to the right. Dideoxy sequencing reaction in lanes 1-4, free IRES in lanes 5, 7, 9 and 11 and IRES-80S complexes (formed by incubation in rabbit reticulocyte lysate, RRL with cycloheximide, CHX) in lanes 6, 8, 10 and 12. Nucleotide numbers are bulleted on the left, the A of the AUG is indicated by the grey arrow (+1) and the toeprint is indicated by the blue arrow (+15, +16) to the right of the expanded gel. (b) Graph of quantitated, normalized and background-corrected IRES-80S toeprints from panel a. +1 and +15, +16 are indicated by grey and blue lines, respectively (pseudoknot, pknot; domain IV, dIV)The location of IRES secondary structural domains are indicated beneath the graphs. (c) Cartoon of the uncapped, unpolyadenylated monocistronic Photinus reporter. The region of the RNA between the viral AUG and luciferase AUG (both highlighted in red) is expanded below. One or two adenosines (blue box) were added for frameshift analysis. (d) Graph of 90 minute translation assay for WT and ΔGCC reporters without any mutations or with the addition of one or two adenosine residues. Y-axis represents luciferase activity in relative light units (RLUs) detected and error bars represent one s.e.m of three independent triplicate experiments.
Subdomain IIb mutants initiate in the correct reading frame
Mutations to the β-hairpin of the bacterial homolog of rpS5 (rpS7) have been shown to increase the rate of frameshifting 37. We therefore considered the possibility that the dIIb mutants, by disrupting contact with rpS5, could cause frameshifting during or after the first translocation event, and thus an apparent decrease in translation. To test this, we conducted translation assays using reporters with one or two nucleotides after the IRES AUG start codon but before the AUG of the LUC open reading frame. If the ribosome sometimes slips out of frame on the dIIb mutant RNAs, the addition of these nucleotides would rescue a 1 or 2 nt frameshift (Fig. 5c). We did not observe a partial rescue (Fig. 5d); in fact, introduction of these nucleotides decrease translation efficiency even more than the dIIb mutants. Hence, the decreased translation initiation efficiency of the dIIb mutants is not due to frameshifting.
Domain IIb mutation does not affect peptide bond formation
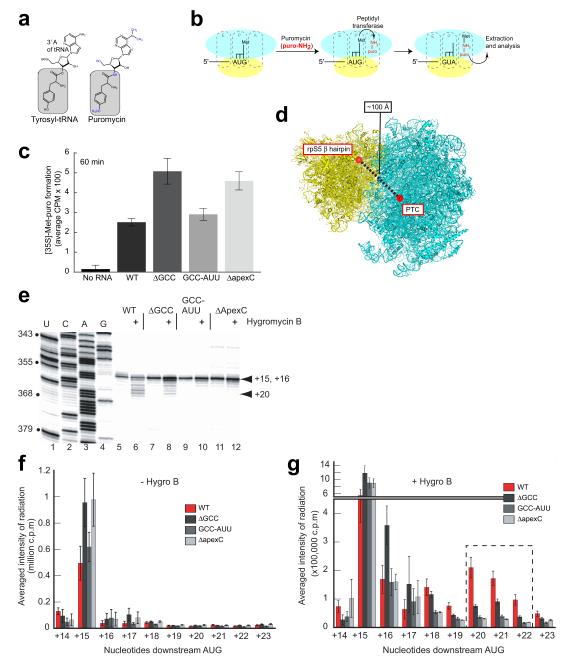
Slowed initial peptide bond formation after 80S ribosome assembly would decrease the overall rate of protein production and explain a decrease in protein synthesis (Fig. 1e), we hypothesized dIIb mutations affect peptide bond formation. We tested the ability of 80S ribosomes formed on dIIb mutants to catalyze peptide bond formation using the aminoglycoside puromycin to accept an amide linkage from the amino acid on the P-site tRNA when the A site of the ribosome is vacant (Fig. 6a & b) 10,38. We generated 80S ribosome complexes in RRL supplemented with [35S]-methionine using WT and dIIb mutant IRES RNAs truncated to end after the AUG start codon. This results in 80S ribosome-IRES complexes with [35S]-met-tRNAimet in the P site and a vacant A site. The amount of [35S] methionine transferred to added puromycin indicated the ability of the 80S ribosomes to catalyze peptide bond formation. We found that the dIIb mutant IRES-80S complexes were as competent to form a peptide bond as were complexes formed on WT IRES (Fig. 6c). In fact, two of the mutants exhibited a reproducible increase in the production of the puromycin-methionine product compared to WT, which is surprising since the location of dIIb and its putative interaction with rpS5 is ~100 Å from the peptidyl transferase center (PTC) of the large subunit (Fig. 6d). This result suggests that although the dIIb mutations are not decreasing peptide bond formation, their effects are felt in distal parts of the ribosome.
Figure 6. Puromycin and toeprinting assays with antiobiotic.
(a) Structures of tyrosyl-tRNA (left) and the puromycin (right), differences in grey and blue. (b) Cartoon of the puromycin assay (40S subunit yellow, 60S blue) moving from left to right: IRES-40S formation, then Met-tRNAi and puromycin (puro-NH2) binding in the 60S subunit P and A sites, respectively. Peptidyl transferase results in methionine bound to puromycin via a noncanonical amide linkage (met-puro), then extraction. (c) Quantitated and background-corrected graph of met-puro formation after 60 minutes on wild-type (WT) and mutant IRES RNAs. Error bars: one s.e.m of three independent duplicate experiments. (d) Yeast 80S ribosome crystal structure 56 (40S subunit yellow, 60S cyan) with approximate locations and distance between the IRES domain IIb (dIIb)-rpS5 interaction and the peptidyl transferase center (PTC) shown. (e) Relevant part of the toeprint gel with the dideoxy sequencing reactions in lanes 1-4, free IRES-80S complexes in rabbit reticulocyte lysate (RRL) without any antibiotic in lanes 5, 7, 9 and 11 as well as initiating and elongating IRES-80S complexes formed in RRL with hygromycin B in lanes 6, 8, 10 and 12. Black arrowhead represents initiating complexes (+15, +16) and blue arrowhead represents elongating complexes (+20) on the right, nucleotide numbers are bulleted, on the left. (f) Graph of quantitated, normalized, and background-corrected toeprints without antibiotic from panel e (WT IRES red, dIIb mutants grey). (g) Same as in panel f except graph represents toeprints with antibiotic. Error bars represent one s.d. of three independent experiments.
The first translocation event is promoted by subdomain IIb
We hypothesized that mutation of dIIb, while allowing 80S ribosome formation and peptide bond formation, inhibits the first round of translocation in which the P site-bound initiator tRNA and start codon AUG move to the E site. To test this, we used toeprinting analysis to directly detect ribosome movement on the WT and mutant IRES RNAs. We conducted these assays in RRL both in the absence and presence of the antibiotic hygromycin B, which inhibits translocation 39. In the absence of antibiotic, all IRESs produced a toeprint consistent with the start codon AUG placed in the P site of 80S ribosomes (Fig. 6e, lanes 5, 7, 9 and 11). We did not see stops due to elongating ribosomes, likely because these ribosomes move to a position that prevents primer or reverse transcriptase binding. Quantitation of several replicates of this experiment corrected for slight differences in loading or labeling intensity and showed slightly increased toeprint intensity for the dIIb mutants compared to WT, indicating more 80S ribosomes are paused at this initial assembly location (Fig. 6f). When antibiotic was included, the drug captured ribosomes that had undergone one or two rounds of translocation, indicated by the presence of toeprint stops at the +20 to +22 position (Fig. 6e, lane 6). In contrast, we did not observe these strong downstream toeprint stops on the mutant RNAs (Fig. 6e, lanes 8, 10, and 12). Again, we quantified multiple replicates to correct for loading differences and found that 80S ribosomes formed on a dIIb mutants were slow to move from their initial position (Fig. 6g). Specifically, 33.3% of the WT RNA was left in an untranslocated state, while 54.9%, 58%, and 62% of ΔGCC, ΔapexC, and GCC-AUU did not translocate, respectively. Thus, mutations in dIIb decreased translocation by ~50%, which is consistent with the activity measurements of figure 1. Mutation of HCV IRES dIIb therefore results in 80S ribosomes with reduced ability to undergo the first round of translocation. We conclude that an intact dIIb is needed for the first translocation step, when initiation transitions to elongation.
DISCUSSION
We have discovered that mutations in the apical loop of dII of the HCV IRES inhibit the first translocation event after the formation of the 80S ribosome. HCV IRES-driven 80S ribosome formation and the first step of translocation can therefore be decoupled, a function of dII distinct from previously defined roles of HCV IRES domains. The involvement of dIIb in the first round of translocation could be explained by two broad mechanisms. First, dII’s position in the E site mandates that it move to make way for the P-site tRNA to translocate, evident in cryo-EM reconstructions of the HCV IRES bound to a 40S subunit and 80S ribosome 23,24. Based on the aforementioned HCV IRES-80S ribosome cryo-EM reconstruction, it has been suggested that contacts between a different portion of dII and the L1 stalk of the large subunit could facilitate domain II displacement from the E site, but this IRES-ribosome contact and potential function has not been tested 24. Also, a recent crystal structure of HCV IRES domain IIa (not studied) bound to an inhibitor suggest that conformational changes in parts of domain II not studied here may effect movement of the domain from the E site 40, but again this has not been demonstrated functionally. In the portion of dII studied here, dIIb mutations could potentially inhibit dII displacement, and this would slow translocation by sterically hindering the movement of tRNA. This “failure-to-move” phenotype then could be ascribed to the loss of a specific IRES interaction with rpS5 necessary for dII ejection from the E site, although an analogous role for rpS5 in tRNA ejection has not been reported. This idea does not eliminate the possibility that the L1 stalk and dII conformational changes also help move dII. The second potential explanation is that dIIb actively promotes an event within the ribosome that is important for the first round of translocation, again likely through a specific interaction of dIIb with rpS5 and subsequent conformational changes in the IRES-ribosome complex. Mutation of dIIb would then either interfere with this event, or the IRES would be unable to actively promote this event. These two broad mechanisms are both consistent with our structural and functional data; they are not mutually exclusive.
Given the fact HCV IRES dIIb is positioned to interact directly with the β-hairpin of rpS5, examining known functions of rpS5 (and from bacterial ortholog rpS7) could give insight into how dIIb influences translocation. During elongation, rpS5 (S7 in bacteria) plays a role in maintaining the reading frame and in overall fidelity 37,41,42. More specifically to the β-hairpin, in bacteria truncation of this structure results in destabilization of the E-site tRNA and an increase in frameshifting and reverse translocation 37. However, our data shows no evidence for frameshifting induced by the dIIb mutants. Although not explicitly shown to be dependent on rpS5 or S7, it was reported that during elongation the presence of tRNA in the E site influences the fidelity of aminoacyl-tRNA (ac-tRNA) selection in the A site 43, but other studies find little or no evidence for this 44-46. Hence, it seems unlikely that the presence of dIIb directly influences entry of A-site tRNA. Overall, we find no known role for rpS5 that readily explains the effect we observed by mutating HCV IRES dIIb, consistent with the notion that the IRES is co-opting this feature of the ribosome to manipulate the complex in a noncanonical way or that rpS5 plays an undiscovered role during translation initiation.
Mutation of dIIb disrupts a putative interaction with rpS5, changes the structure of the IRES-40S subunit complex compared to that of a WT IRES, and inhibits the first translocation step. These observations suggest that a specific rpS5-dIIb interaction induces allosteric changes that propagate through the ribosome. Indeed, there is evidence for a network of interactions within the ribosome that could cause this and also for similar conformational changes induced by bound initiation factors eIF1 and 1A. Specifically, rpS5 interacts with rRNA helices 29, 30, and 42 (refs. 47,48), and these helices interact with eIF1A (ref. 49). Another network links rpS5 to the eIF1 binding site through rpS14 and rRNA helix 23 (refs. 41,50,51). This is noteworthy because binding of eIF1 and 1A induce a conformational change in the 40S that strongly resembles that induced by the WT HCV IRES 23,31, and eIF1A is known to act with eIF5B after 80S formation to commit the ribosome to elongation 52,53. This last point raises the interesting possibility that HCV IRES dIIb may induce the same effect as eIF1, 1A, and 5B, to promote a late step during initiation. This notion, speculative at this point, is appealing because a minimal reconstitution of HCV IRES-driven translation initiation does not require eIF1 or 1A 10, and thus dIIb could substitute for these absent factors. Although not a part of this study, higher resolution structures of mutant HCV IRESs in complex with 80S ribosomes and chemical probing of the rRNA in these complexes before and after translocation could provide insight into the putative allosteric changes associated with this translocation-slowing phenotype.
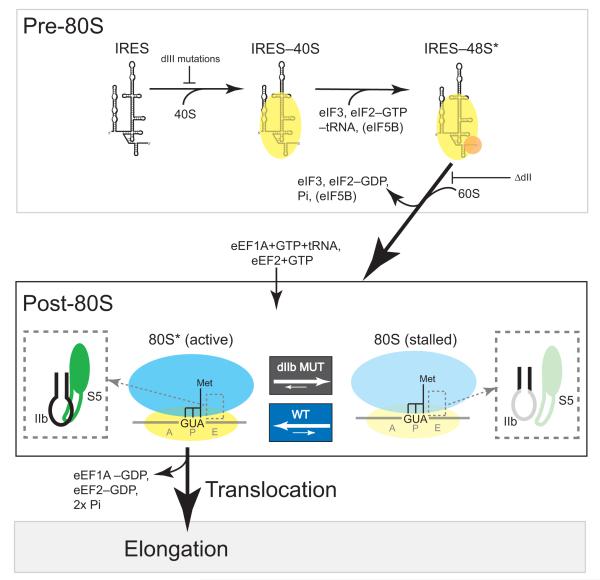
We would like to propose the following model to explain the role of HCV IRES dIIb in events that occur within the IRES•ribosome complex prior to and during the first translocation step (Fig. 7). First, the IRES assembles an 80S ribosome such that the ribosome is poised at the start codon with an initiator tRNA in the P site. We propose that within this ribosome, dIIb contacts the β-hairpin of rpS5 thereby stabilizing a conformation of the ribosome that is conducive to translocation. Delivery of ac-tRNA to the ribosome by eukaryotic elongation factor (eEF) 1A and subsequent peptide bond formation is then followed by rapid and efficient eEF2-catalyzed translocation. In the case of the dIIb mutants, the mutation induces a local change in structure in the apical loop and this perturbs the interaction with rpS5, affecting 40S subunit conformation. This IRES-40S complex is still capable of progressing to 80S ribosome, but the resultant ribosome’s conformational equilibrium is shifted towards a state with an inhibited ability to translocate. Although ac-tRNA may still be delivered to the A site and a peptide bond formed, the mutant-bound ribosome stalls at the start site. However, because the ribosome samples conformations, these ribosomes are not permanently stalled but occasionally sample a productive state where they are able to translocate. In summary, our data supports a model in which dIIb selects a productive state from the conformational ensemble, while ribosomes bound to IRES with mutated dIIb spend more time in an unproductive state and transition to elongation less efficiently.
Figure 7. Model of domain IIb (dIIb)’s role in HCV IRES translation initiation.
Top box: simplified pathway for HCV IRES-driven translation initiation up to the point of 80S ribosome formation. This process includes 40S subunit binding, ternary complex (eIF2-met-tRNAi-GTP) binding, eIF3 binding, and then subunit joining and factor release. Below this are the post-80S events that we hypothesize occur within the newly formed 80S ribosome. Briefly, dIIb interacts with the β hairpin of rpS5 (dashed box: dIIb in black, rpS5 in green) and this favors a conformation that is fully competent to accept a tRNA into the A site (delivery catalyzed by eukaryotic elongation factor 1A, eIF1A) and subsequent translocation (catalyzed by eEF2). We term this fully competent conformation “80S*”. When dIIb is mutated (right), the local structure of the dIIb apical loop shifts towards an inactive conformation and the productive interaction with rpS5 is lost (dashed box: dIIb in gray, rpS5 in light green). This favors an 80S ribosome state that stalls prior to translocation. In our model, the active (80S*) and inactive (80S) states are in dynamic equilibrium and the presence of an intact dIIb shifts the equilibrium towards 80S*, thus promoting progression to elongation.
Our data open another door to understanding the intricacies of translation initiation and ribosome function. Fundamentally, the ribosome is a Brownian machine sampling many conformations; protein factors and tRNA binding shift the conformational equilibrium, providing efficiency and directionality. Thus, the ribosome is programmed to be manipulated by its binding partners. This inherent characteristic of the ribosome is critical for canonical translation processes and allows subtle and robust regulation of ribosome function. Our results reveal these principles are exploited by a single loop of the HCV IRES, supporting the view of the HCV IRES as a dynamic manipulator of the translation machinery and lending insight into how the translation machinery works in cap-dependent and cap-independent pathways.
ONLINE METHODS
Plasmid construction and cloning
We constructed pUC19-based plasmids containing the HCV genotype Ib wild type (nucleotides 40-372) and ΔdII mutant (nucleotides 119-372) sequences flanked by a 5′ hammerhead and 3′hepatitis delta ribozyme as previously described 6,17,22. Plasmids with the WT or mutant HCV IRES between two luciferase genes were made by PCR amplification of the desired sequence and ligation into the EcoRI and NcoI sites of plasmid pRL (gift of A. Willis, Medical Research Council Toxicology Unit, Leicester, UK) 57. The plasmid used to generate RNA for toeprinting analysis (contains 85 additional 3′ nucleotides on the wild type HCV genotype 1a as well as the primer binding site) was a kind gift of P. Lukavsky (Central European Institute of Technology, Masaryk University, Brno, Czech Republic), 14. We generated the genotype 1b ΔdII mutant used in toeprinting by PCR amplification of the desired sequence and ligation into the Hind III and XbaI sites of this plasmid. All mutants were made using the QuickChange mutagenesis kit (Stratagene).
RNA Preparation
We made RNAs for assembly assays and puromycin experiments using DNA generated by PCR using M13 -41 forward and reverse primers (5′-GGTTTTCCCAGTCACGAC-3′ and 5′-GGAAACAGCTATGACCATG-3′, respectively) and the relevant plasmid template. The PCR products were used for in vitro transcription reactions as described 58. We purified and concentrated RNA as described 22. Monocistronic Photinus luciferase RNAs were made from PCR templates using forward T7-HCV and reverse photinus primers (5′-TAATACGACTCACTATAGGGCTCCCCTGTGAGGAACTACTGTCTT-3′ and 5′-TTACACGGCGATCTTTCCGCCCTTCTT-3′, respectively) using the T7 MegaScript kit (Ambion). RNAs were DNase treated, then purified with TRI®Reagent (Sigma) and chloroform followed by isopropanol, 100% and 75% ethanol precipitations, respectively. We made RNAs for toeprinting from EcoR1-linearized plasmids in the same manner as the luciferase RNAs.
Radiolabeling RNA and Primers
For assembly assays, we 5′-radiolabeled RNA as described 22 and diluted it to approximately 1,000 cpm μL−1. DNA primers were 5′-radiolabeled in a reaction containing 800 pmol primer in the same conditions as the RNA, then mixed with 20 μL 9M urea loading buffer, loaded directly onto a 10% urea denaturing gel, purified, and diluted to approximately 25,000 cpm μL−1.
Ribosome Assembly Assays
We diluted 5′-radiolabeled HCV IRES RNAs to ~1000 cpm μL−1, heated to 85 °C for 30 s, then cooled on the desktop. 1 μL of this RNA was then added to a mixture containing 30 μL rabbit reticulocyte lysate (RRL), 0.5 μL amino acid mixture minus leucine, 0.5 μL amino acid mixture minus methionine (all provided in RRL translation kit, micrococcal nuclease treated, Promega), and 18 μL RNase-free water. Reactions then were incubated at 30 °C for the desired time, then halted by the addition of ribosome association dilution buffer (50 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgCl2, and 1 mM DTT) and placed on ice. The reactions were analyzed with 10-35% sucrose gradients in ribosome association dilution buffer by ultracentrifugation in a SW41 Ti swinging bucket rotor at 36,000 rpm (~222,000 ×g) for 3 hours. We fractionated the gradients into ~0.5 mL fractions using a BIOCOMP Gradient Stationip and Gilson FC203B fraction collector. 200 μL of each fraction was blotted onto membranes, air dried, and analyzed using a phosphorimager. We quantified the spots using ImageQuant software and reported each as a fraction of the total radiation.
Luciferase Assays
We conducted translation assays as described 22 with the following exceptions: for the time-point experiment, reactions were brought up to 125 μL volume so that 25 μL could be removed at each time point, which were then halted with 200 μL cold 1X Passive Lysis Buffer (Promega) and placed at −80 °C to ensure no further activity.
Toeprinting Assays
We completed toeprinting assays essentially as described 22. For 48S-bound IRES, 10.75 μL RRL was mixed with 0.5 μL RNasin® Plus and 0.5 mM GMPPNP, incubated at 30 °C for 5 min followed by addition of 0.5 μg toeprint RNA in a final volume of 15 μL. For 80S-bound IRES, 10.8 μL RRL we mixed 0.5 μL RNasin® Plus (Promega) and one of the following: no antibiotic, 3 mg mL−1 cycloheximide or 2 mg mL−1 hygromycin b, incubated at 30 °C for 5 min. This incubation was followed by addition of 0.5 μg toeprint RNA in a final volume of 15 μL. We made the ladder used for analysis with wild type toeprint RNA reverse transcribed with SuperScript® III Reverse Transcriptase (Life Technologies) with annealing and extension temperatures at 45 °C.
Puromycin Assays
We first biotinylated 3′ truncated IRES RNAs (nts 40-344 which stop after the AUG codon) using the 5′ EndTag™ Nucleic Acid Labeling System (Vector Laboratories). Briefly, 65 μg of RNA was phosphorylated with ATPγS using T4 polynucleotide kinase in a 20 μL reaction, at 37 °C for one hour. Biotinylation then was carried out upon the addition of ~385 μg biotin (long arm) maleimide for one hour at 65 °C. Reactions then were extracted with equal volume phenol-chloroform-isoamyl alcohol pH 6.7 (25:24:1) (Fisher), precipitated with 1/10th volume 3 M NaOAc pH 5.2 and 3 volumes 100% cold ethanol overnight. RNA was pelleted and washed with 70% ethanol, dried and resuspended in 10 μL RNase-free water. Concentration was determined by absorbance at UV (260 nm). To conduct the assay, we mixed 30 μL RRL with 16.5 μL RNase-free water and 1 μL L-[35S]-methionine (>1000 Ci mmol−1) and incubated at 30 °C for 15 minutes to allow aminoacylation of initiator methionine tRNA. 2.25 μg biotinylated RNA was added to the reaction and incubated at 30 °C for 25 minutes for 80S ribosome formation. Reactions were mixed with one tube (0.6 mL) streptavidin paramagnetic beads (MagneSphere®, Promega, prewashed three times with 0.5X SSC buffer [0.0187 mM trisodium citrate dehydrate, pH 7.2 and 0.187 NaCl], and once with 300 μL ribosome association dilution buffer), resuspended in 50 μL ribosome association dilution buffer, and incubated at 30°C for 10 minutes. Complexes were then washed six times with 500 μL ribosome association dilution, and resuspended such that reactions were split into duplicates with and without 1 mM puromycin. The assay was then carried out at 35 °C for 60 minutes. Puromycin was extracted with 500 μL 200 mM potassium phosphate buffer pH 8 and 500 μL ethyl acetate with continuous shaking for 10 minutes at 35 °C. The upper ethyl acetate layer was removed and mixed with 7 mL ScintiSafe (Fisher) liquid scintillation fluid and counts per minute were averaged between two 10 minute count times. This method of immobilizing the IRES-80S ribosomes greatly reduced background levels of puromycin-[35S]-methionine formation compared to results for 80S ribosomes purified by ultracentrifugation in sucrose gradients.
Nuclear Magnetic Resonance
We collected NMR spectra at 25 °C on a Varian 900 MHz spectrometer, using Standard Varian Biopack pulse sequences for all experiments. This included both 1-dimensional spectra as well as all homonuclear 2-dimensional spectra employing a 3919 watergate for water suppression. NOESY (Biopack pulse sequence, WBNOESY) spectrum was collected with 256 indirect points. Two-dimensional data were processed using a Gaussian weighting function in the direct dimension and a sinebell weighting function in the indirect dimension.
Negative stain Electron Microscopy
We prepared the ΔGCC mutant IRES RNA as described above, and the 40S ribosomal subunits from RRL as previously described 6. We assembled IRES-40S subunit complex as previously described 23. We applied the complex to freshly glow discharged carbon coated 400 mesh copper grids and stained with 0.75% uranyl formate as described 59. Samples were viewed on a 120 kV transmission electron microscope (FEI, Hillsboro, OR). Images were recorded at a nominal magnification of 40,000x using a bottom mount 4k × 4k Gatan slow scan charge coupled device (CCD) camera.
Preparation of complexes for Electron Cryomicroscopy
Purified ΔGCC mutant IRES RNA in RNase-free water was heated to 70 °C for 2 minutes, then cooled to room temperature. Buffer solution was added to a final concentration of 20 mM Tris-HCl pH 7.4, 100 mM KOAc, 2.5 mM MgCl2, 1 mM DTT, 40 mM KCl. Purified 40S subunits were added at a 1:1 molar ratio with the IRES RNA to a final concentration of 500 nM complex. Complex was stored on ice until diluted (generally to 100 nM) and used in microscopy.
Electron Cryomicroscopy Data Collection & Single Particle Reconstruction
We prepared vitrified samples of the ΔGCC mutant IRES-40S subunit complexes at 100 nM using an FEI Vitrobot. Briefly, 3.5 μL was applied to a Quantifoil holey carbon grid (Vitrobot chamber was at 4 °C and 100% humidity). After a 20 s pause, the grid was blotted with filter paper (force = 0, blot time = 2 s) and plunged into liquid ethane. Frozen samples were loaded onto a Gatan cryo-holder and inserted into a FEI Tecnai F20 operating at 200 kV equipped with a field emission gun. Images were collected at a nominal magnification of 62,000x using a 4k × 4k Tietz CMOS detector. Images were binned two times yielding a pixel size of 2.66 Å per pixel. Approximately 29,000 particles were selected from 1,790 images using Electron Micrographe Utility (cryoem.ucsf.edu). Class averages were determined using five consecutive rounds of MSA (multivariate statistical analysis) and MRA (multireference alignment) in IMAGIC 54. CTF parameters for each image were determined using CTFFIND3 (ref. 60). An initial model for refinement and three-dimensional reconstruction was generated by filtering a previously published apo40S reconstruction (EMD-1346) model to 40 Å 31. Initial parameters were generated during cycles of randomized search and refinement using FREALIGN v 8.08 (ref. 61). After initial parameters were determined, consecutive cycles of local refinement and reconstruction were carried out until there was no apparent improvement in the alignment. Resolution of the three-dimensional model was calculated with the program RMEASURE 62 and determined to be ~17.5 Å. The density was normalized using MAPMAN 63 and filtered to 20 Å using BFACTOR. Difference maps presented in the figures were calculated using MAPMAN 63. Reconstructions and difference maps were assembled as displayed in Figure 2B using UCSF Chimera 64.
ACKNOWLEDGMENTS
The authors thank the members of the Kieft Lab and R. Davis, D. Bentley, D. Barton, and T. Evans for useful suggestions and discussions, and M. Ruehle, T. Blumenthal, T. Cech, and M. Johnston for critical reading of this manuscript. We also thank C. Spahn (Institut für Medizinische Physik und Biophysik, Charite – Universitätsmedizin Berlin, Berlin, Germany) for data files and advice with structural modeling, P. Lukavsky (Central European Institute of Technology, Masaryk University, Brno, Czech Republic) for NMR resonance assignments and a plasmid, A.Willis (Medical Research Council Toxicology Unit, Leicester, UK) for a plasmid, and G. Armstrong and E. Eisenmesser for assistance in NMR data collection and processing. This work was supported by NIH grant GM08134603 to JSK. MEF was supported as an American Heart Association pre-doctoral fellow (#0815655G). JSK is an Early Career Scientist of the Howard Hughes Medical Institute. The Gonen laboratory is supported by the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
M.E.F. conducted all biochemical experiments. J.S.K. and M.E.F. conducted and analyzed the NMR experiments. B.S.V, D.S., T.G. and J.S.K conducted the cryo-EM experiments, with structure calculation by B.S.V. Results were interpreted by M.E.F., B.S.V., J.S.K and T.G. M.E.F. and J.S.K designed the study overall, and wrote the manuscript. All authors contributed to figure construction.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
ACCESSION NUMBERS
The cryo-EM map of the mutant HCV IRES-rabbit 40S subunit complex has been deposited with accession number EMDB-5527.
REFERENCES
- 1.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–83. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukh J, Purcell RH, Miller RH. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc Natl Acad Sci U S A. 1992;89:4942–6. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds P, et al. Sequence variability in the 5′ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74(Pt 4):661–8. doi: 10.1099/0022-1317-74-4-661. [DOI] [PubMed] [Google Scholar]
- 4.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 5.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–50. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lytle JR, Wu L, Robertson HD. The ribosome binding site of hepatitis C virus mRNA. J Virol. 2001;75:7629–36. doi: 10.1128/JVI.75.16.7629-7636.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–55. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–80. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci U S A. 2004;101:16990–5. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–41. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–64. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieft JS, et al. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–29. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 18.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–71. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–82. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–68. [PMC free article] [PubMed] [Google Scholar]
- 21.Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–69. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA. 2011;17:1258–73. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–62. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 24.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Fukushi S, et al. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem. 2001;276:20824–6. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 26.Wower J, Scheffer P, Sylvers LA, Wintermeyer W, Zimmermann RA. Topography of the E site on the Escherichia coli ribosome. EMBO J. 1993;12:617–23. doi: 10.1002/j.1460-2075.1993.tb05694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 28.Doring T, Mitchell P, Osswald M, Bochkariov D, Brimacombe R. The decoding region of 16S RNA; a cross-linking study of the ribosomal A, P and E sites using tRNA derivatized at position 32 in the anticodon loop. EMBO J. 1994;13:2677–85. doi: 10.1002/j.1460-2075.1994.tb06558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odreman-Macchioli F, Baralle FE, Buratti E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J Biol Chem. 2001;276:41648–55. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- 30.Kalliampakou KI, Psaridi-Linardaki L, Mavromara P. Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett. 2002;511:79–84. doi: 10.1016/s0014-5793(01)03300-2. [DOI] [PubMed] [Google Scholar]
- 31.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–6. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 33.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–8. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 34.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–69. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–20. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 36.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–25. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 37.Devaraj A, Shoji S, Holbrook ED, Fredrick K. A role for the 30S subunit E site in maintenance of the translational reading frame. RNA. 2009;15:255–65. doi: 10.1261/rna.1320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monro RE, Marcker KA. Ribosome-catalysed reaction of puromycin with a formylmethionine-containing oligonucleotide. J Mol Biol. 1967;25:347–50. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- 39.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–94. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 40.Dibrov SM, et al. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc Natl Acad Sci U S A. 2012;109:5223–8. doi: 10.1073/pnas.1118699109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert F, Brakier-Gingras L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J Biol Chem. 2003;278:44913–20. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- 42.Galkin O, et al. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA. 2007;13:2116–28. doi: 10.1261/rna.688207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geigenmuller U, Nierhaus KH. Significance of the third tRNA binding site, the E site, on E. coli ribosomes for the accuracy of translation: an occupied E site prevents the binding of non-cognate aminoacyl-tRNA to the A site. EMBO J. 1990;9:4527–33. doi: 10.1002/j.1460-2075.1990.tb07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petropoulos AD, Green R. Further in vitro exploration fails to support the allosteric three-site model. J Biol Chem. 2012;287:11642–8. doi: 10.1074/jbc.C111.330068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–7. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, et al. Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc Natl Acad Sci U S A. 2011;108:16980–5. doi: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malygin AA, Yanshina DD, Karpova GG. Interactions of human ribosomal proteins S16 and S5 with an 18S rRNA fragment containing their binding sites. Biochimie. 2009;91:1180–6. doi: 10.1016/j.biochi.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 48.Ian’shina DD, Malygin AA, Karpova GG. Binding of human ribosomal protein S5 with the 18S rRNA fragment 1203-1236/1521-1698. Mol Biol (Mosk) 2006;40:460–7. [PubMed] [Google Scholar]
- 49.Yu Y, et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–82. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antunez de Mayolo P, Woolford JL., Jr Interactions of yeast ribosomal protein rpS14 with RNA. J Mol Biol. 2003;333:697–709. doi: 10.1016/j.jmb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–97. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acker MG, et al. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol. 2009;385:491–506. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol Cell Biol. 2007;27:2384–97. doi: 10.1128/MCB.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 55.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 56.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–9. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
ONLINE METHODS REFERENCES
- 57.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–8. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 58.Keel AY, Easton LE, Lukavsky PJ, Kieft JS. Large-scale native preparation of in vitro transcribed RNA. Methods Enzymol. 2009;469:3–25. doi: 10.1016/S0076-6879(09)69001-7. [DOI] [PubMed] [Google Scholar]
- 59.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 61.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 62.Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J Struct Biol. 2007;157:201–10. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 63.Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr. 1996;52:826–8. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]