Abstract
Transcription factors induced by the LH surge play a vital role in reprogramming the gene expression in periovulatory follicles. The present study investigated the role of RUNX2 transcription factor in regulating the expression of Runx1, Ptgs2, and Tnfaip6 using cultured granulosa cells isolated from PMSG-primed immature rats. hCG or forskolin+PMA induced the transient increase in Runx1, Ptgs2, and Tnfaip6 expression, while the expression of Runx2 continued to increase until 48 h. The knockdown of the agonist-stimulated Runx2 expression increased Runx1, Ptgs2, and Tnfaip6 expression and PGE2 levels in luteinizing granulosa cells. Conversely, the over-expression of RUNX2 inhibited the expression of these genes and PGE2 levels. The mutation of RUNX binding motifs in the Runx1 promoter enhanced transcriptional activity of the Runx1 promoter. The knockdown and overexpression of Runx2 increased and decreased Runx1 promoter activity, respectively. ChlP assays revealed the binding of RUNX2 in the Runx1 and Ptgs2 promoters. Together, these novel findings provide support for the role of RUNX2 in down-regulation of Runx1, Ptgs2, and Tnfaip6 during the late ovulatory period to support proper ovulation and/or luteinization.
Keywords: Runx2, Runx1, Ptgs2, Tnfaip6, Granulosa cells, LH, Ovary
1. INTRODUCTION
The preovulatory gonadotropin (FSH/LH) surge initiates multipath, yet intertwined processes that lead to the expansion of cumulus oocyte complex (COC), ovulation, and corpus luteum (CL) formation. These changes are accomplished by spatio-temporally regulated expression of genes in periovulatory follicles. Specific transcription factors induced by the LH surge directly control the dynamic changes in periovulatory gene expression, thus playing a central role in successful ovulation of COCs and luteal transformation [reviewed in (Richards, 2007; Russell and Robker, 2007; Sirotkin, 2010; Stouffer et al., 2007)].
Previously, we and others have demonstrated that two members of the Runt related transcription factor family, Runx1 and Runx2, were highly induced by the LH surge in periovulatory follicles of rat, mouse, and human ovaries (Hernandez-Gonzalez et al., 2006; Jo and Curry, Jr., 2006; Park et al., 2010). However, their temporal expression pattern was distinct; Runx1 expression was highly induced shortly after the LH surge, and declined rapidly around the time of ovulation (Jo and Curry, Jr., 2006), whereas Runx2 expression progressively increased and reached the highest level in the forming CL (Park et al., 2010). In situ hybridization and immunohistochemical analyses further verified the tissue localization of Runx1 and Runx2 to granulosa and cumulus cells of periovulatory follicles and forming CL (Jo and Curry, Jr., 2006; Park et al., 2010).
RUNX1 and RUNX2 are members of the Core Binding Factor (CBF/PEBP2/RUNX) family. CBF is a heterodimeric transcription factor composed of α and β subunits; the α subunit is encoded by one of three Runx genes (Runx1, Runx2, and Runx3) and the β subunit is encoded by a single gene, CBFβ. CBFβ does not bind to DNA directly, but enhances DNA binding of RUNX proteins (Bae et al., 1993; Ogawa et al., 1993; Speck et al., 1999). Both RUNX1 and RUNX2 have been shown to be essential for the development of various tissues by controlling cell proliferation and differentiation [reviewed in (Coffman, 2003; Otto et al., 2003)]. However, RUNX1 and RUNX2 contribute distinct biological processes by regulating tissue specific gene expression (Geoffroy et al., 2002; Lutterbach and Hiebert, 2000). For instance, RUNX1 plays a vital role in hematopoiesis by regulating specific hematopoietic genes (Lutterbach and Hiebert, 2000). RUNX2 is known to regulate the expression of specific osteogenic genes and is considered as a master regulator for osteogenesis (Komori, 2002; Komori, 2008; Komori, 2010).
Structurally, RUNX proteins contain multiple trans-activation and repression domains in addition to a conserved DNA-binding Runt domain (Durst and Hiebert, 2004; Westendorf, 2006). Therefore, RUNX proteins can increase or inhibit the transcriptional activity of their respective target genes in a context dependent manner [reviewed in (Cameron and Neil, 2004)]. RUNX proteins are commonly thought of as a potent transcriptional activators because early studies identified them as positive regulators for numerous genes in diverse cell types (Komori, 2010; Lutterbach and Hiebert, 2000; Otto et al., 2003; Pratap et al., 2006). More recently, their role as a transcriptional repressor has also been appreciated in a number of cell types (Otto et al., 2003; Shimizu et al., 2010; Sun et al., 2009; Vitolo et al., 2007). Intriguingly, among the list of the genes that are negatively regulated by RUNX proteins include Runx genes themselves (Brady et al., 2009; Brady and Farrell, 2009; Drissi et al., 2000). All three Runx genes contain multiple RUNX binding sites in their promoter regions (Rini and Calabi, 2001; Wong et al., 2011; Xiao et al., 2001). Indeed, experimental evidence has been accumulating in support of auto- or cross-regulatory mechanisms for their own expression (Brady et al., 2009; Brady and Farrell, 2009; Drissi et al., 2000; Ghozi et al., 1996; Spender et al., 2005; Wong et al., 2011). For instance, RUNX2 was shown to suppress its own expression by direct binding to its own promoter and 5’ UTR region in osteoblastic cell lines (Drissi et al., 2000). In T-cell receptor activated cells, RUNX1 self-represses its own expression by acting on its distal promoter region (Wong et al., 2011). Moreover, a recent study by Spencer et al. (Spender et al., 2005) has reported cross-regulation between different RUNX family members; RUNX3 represses Runx1 expression in human B cells.
Interestingly, our previous microarray data showed that the knockdown of Runx2 by siRNA resulted in increased levels of mRNA for Runx1 in cultured granulosa cells, suggesting intra-familiar regulation between these two transcription factors. The microarray data also revealed that Ptgs2 and Tnfaip6 expression were higher in Runx2 siRNA-treated cells. Importantly, Ptgs2 and Tnfaip6 expression are transiently induced in periovulatory follicles and were found to be crucial for proper ovulation and/or COC expansion (Davis et al., 1999; Fulop et al., 1997; Fulop et al., 2003; Joyce et al., 2001; Ochsner et al., 2003; Sirois et al., 2004; Yoshioka et al., 2000).
In the present study, we hypothesize that RUNX2 plays a role in down-regulation of Runx1, Ptgs2, and Tnfaip6 expression in luteinizing granulosa cells. To test this hypothesis, the temporal expression of these genes was first assessed in luteinizing granulosa cells in culture. Then, using the experimental approaches of silencing and over-expression of Runx2, we determined whether modulation of Runx2 expression affects Runx1, Ptgs2, and Tnfaip6 expression. In addition, we investigated the molecular mechanisms of RUNX2 regulation on Runx1 expression at the promoter level in luteinizing granulosa cells.
2. MATERIALS AND METHODS
2.1. Materials
Unless otherwise noted, all chemicals and reagents were purchased from Sigma Chemical Co. (St. Louis, MO). Molecular biological enzymes, molecular size markers, oligonucleotide primers, and culture media were purchased from Invitrogen by Life Technologies, Inc. (Carlsbad, CA).
2.2. Animals
Sprague Dawley rats were obtained from Harlan, Inc. (Indianapolis, IN) and maintained at the University of Kentucky Laboratory Animal Resources. Animal protocols for the rat study were approved by University of Kentucky Animal Care and Use Committees. Immature rats (24–25 days old) were injected with pregnant mare serum gonadotropin (PMSG, 10 IU) to stimulate follicular development. Forty eight hours later, the animals were injected with human chorionic gonadotropin (hCG, 10 IU) to induce ovulation and formation of CL. Animals were killed at the time of hCG administration and defined times after hCG.
2.3. Isolation and culture of rat granulosa cells
Ovaries were collected from immature rats at 48 h post-PMSG and punctured to isolate granulosa cells as described previously (Park et al., 2008). The granulosa cells were pooled, filtered, and pelleted by centrifugation. The cells were cultured in Opti-MEM (Invitrogen) media supplemented with 0.05 mg/ml of gentamycin, and 1x ITS (insulin, transferin, and selenium). The cells were cultured in the absence or presence of various reagents at 37 °C in a humidified atmosphere of 5% CO2.
2.4. Quantification of Runx1, Runx2, Ptgs2, and Tnfaip6 mRNA
Total RNA was isolated from rat ovaries and cultured granulosa cells using a Trizol™ reagent (Invitrogen) or RNeasy mini kit (QIAGEN, Inc., Valencia, CA), respectively. To measure levels of rat Runx1, Runx2, Ptgs2, and Tnfaip6 mRNA, we used Real-time PCR as described previously (Park et al., 2008). Oligonucleotide primers corresponding to each gene were designed using PRIMER3 software and listed in Table 1. The specificity for each primer set was confirmed by both running the PCR products on a 2 % agarose gel and analyzing the melting (dissociation) curve using the MxPro Real-time PCR analysis program (Stratagene, La Jolla, CA) after each Real-time PCR reaction. All samples were measured in duplicate or triplicate and the amplification efficiency of each transcript primer set was determined by running a standard curve. The relative abundance of the target transcripts was normalized to the endogenous reference gene L32 for rats and calculated according to the Pfaffl method (Pfaffl, 2001).
Table 1.
List of primers used for Real-time PCR
| Gene Name |
Reference (Accession no./Primer sets, 5'-3') |
|---|---|
| L32 |
BC061562 GAAGCCCAAGATCGTCAAAA AGGATCTGGCCCTTGAATCT |
| Runx2 |
NM_009820 GTTATGAAAAACCAAGTAGCCAGGT GTAATCTGACTCTGTCCTTGTGGAT |
| Runx1 |
NM_017325.1 AACCCTCAGCCTCAAAGTCA GGGTGCACAGAAGAGGTGAT |
| Ptgs2 |
U03389.1 TACCCGGACTGGATTCTACG AAGTTGGTGGGCTGTCAATC |
| Tnfaip6 |
BC158666.1 CGTCTTGCAACCTACAAGCA TTCGGGTTGTAGCAATAGGC |
2.5. Western blot analysis
Nuclear extracts and cytoplasmic fractions were isolated from ovaries or cultured granulosa cells using a Nuclear Extraction Kit (Active Motif, Carlsbad, CA) as described previously (Jo et al., 2004). All lysates were denatured by boiling for 5 min and separated by SDS-PAGE on a 9 % polyacrylamide gel and then transferred onto a nitrocellulose membrane. The membrane was incubated overnight at 4°C in 1% casein solution containing primary antibody against RUNX2 (PC287, Calbiochem, La Jolla, CA), RUNX1 (Ab-1,Calbiochem) or PTGS2 (Cayman Chemical, Ann Arbor, MI). Primary antibody against TATA binding protein (TBP, Abcam) and β-ACTIN (Cell Signaling Technology, Danvers, MA) was used as a loading control for nuclear and cytoplasmic fractions, respectively. The blots were incubated with the respective secondary HRP-conjugated antibody (Santa Cruz Biotechnology) for 1 h. Peroxidase activity was visualized using the SuperSignal® West Pico Chemiluminescent Substrate (Pierce Chemical Co., Rockford, IL).
2.6. Cloning of the rat Runx1 promoter and generation of Runx1 promoter-reporter plasmid constructs
Genomic DNA was isolated from ear samples from rats using an easy-DNA kit (Invitrogen). A 369 bp (−283/+86), 861 bp (−775/+86), and 1349 bp (−1263/+86) fragments of the Runx1 gene were amplified using the primers attached with restriction enzyme sites (Kpn I and Hind III) and cloned into the pGL3 basic vector (Promega, Madison, WI). Site-directed point mutations of the Runx1 promoter were generated using a QuickChange II site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene). The sequences of the oligonucleotide primers used to generate respective Runx1 promoters containing mutations (shown in lowercase) are following: mutant A (5’- AGG CCC GCT CCA CCT GCg cta gcT ACA CGG TGC TCG CTC G −3’), mutant B (5’- AAC TGC GGC TCA ACT CCC ACC AAg cag gcG GAT CAG CCA CAA ACT TAA CC −3’). All constructs cloned into the expression vector were sequenced commercially to verify their authenticity (MWG Biotech, Inc., High Point, NC).
2.7. Knockdown of Runx2 mRNA by siRNA in granulosa cell cultures
Granulosa cells were isolated from ovaries collected at 48 h post-PMSG and transfected with siRNA as described previously (Park et al., 2010). Briefly, granulosa cells were cultured overnight to acclimatize the cells and the next day, cells were transfected with siRNA specific for Runx2 (sense, GCA CGC UAU UAA AUC CAA Att; antisense, UUU GGA UUU AAU AGC GUG Ctg; Ambion, Inc.) or negative control siRNA (Stealth™ RNAi Negative Control Med GC; Invitrogen) using Lipofectamine ™ RNAiMAX (Invitrogen) according to the manufacturer’s instruction. Transfected cells were incubated for 3 h before stimulating with forskolin (FSK; 10 µM) + phorbol 12-myristate 13-acetate (PMA; 20 nM) and further cultured for 24 or 48 h. At the end of culture, the cells were used to isolate total RNA or extract proteins.
2.8. Transient transfection and luciferase reporter assay
Granulosa cells isolated from immature rats (48 h post-PMSG) were transfected with respective firefly luciferase reporter plasmids (pGL3-basic vector or pGL3-Runx1 promoter constructs) and Renilla luciferase vector (pRL-TK vector) using a Lipofectamine 2000 reagent (Invitrogen) as described previously (Liu et al., 2010). The next day, the cells were treated with FSK+PMA. Six hours later, the cells were harvested to measure Firefly and Renilla luciferase activities using a Dual-luciferase reporter assay system (Promega) and each reaction was monitored by Luminescence System in the Tecan Infinite 200 microplate reader (Tecan US, Durham, NC). Firefly luciferase activities were normalized to Renilla luciferase activities and each experiment was performed in quadruplicate at least 3 times.
2.9. Adenoviral mediated over-expression of RUNX2 in vitro
Granulosa cells were isolated from immature rats (48 h post-PMSG) as described above. The cells were incubated with adenoviruses expressing RUNX2 (Ad-RUNX2) (Ge et al., 2009) or containing null control vector (Ad-ψ) at 5 infectious units (IFU). The next day, cells were treated with FSK+PMA for 4 h and then collected for isolation of total RNA for Real-time PCR or proteins for Western blot analysis.
2.10. Measurements of PGE2 level
Granulosa cells isolated from immature rats (48 h post-PMSG) were plated in a 96 well plate and transfected with Runx2 siRNA or NC siRNA or infected with Ad-RUNX2 or Ad-ψ as described above. Upon completion of culture, PGE2 levels were determined using the Biotrak Prostaglandin E2 Enzyme Immunoassay system (Amersham Pharmacia Biotech, USA) according to the manufacturer's protocol at 620 nm wavelength.
2.11. Chromatin immunoprecipitation (ChIP) analysis
ChIP assay was performed on RUNX binding sites in rat Runx1 and Ptgs2 promoter regions using a ChIP kit (Upstate Biotechnology) as described previously (Park et al., 2008). Briefly, chromatins isolated from cultured granulosa cells were immunoprecipitated overnight at 4 °C with anti-RUNX2 antibody (5 µg/reaction; Santa Cruz Biotechnology) or rabbit IgG (5 µg/reaction). The immunoprecipitated chromatin and 1:10 dilution of input chromatin were analyzed by PCR using the primers designed to amplify fragments spanning the RUNX motif in the Runx1 gene [see Fig.4E, RUNX−728 (forward 5’-GTG GGA GTG AGC GTG TGT AA-3’, reverse 5’-TCG GAA GTC AGC CAC TGT C-3’) and RUNX−526 (forward 5’-GAG TGC ATG TCT GCC TGT GT-3’, reverse 5’-ACT CTG GTT TGG GAA CGA TG-3’)] and in the Ptgs2 gene [see Fig.4F, RUNX−260 (forward 5′-GGG GAA GCT GTG ACA TTC TC T-3′, proximal reverse, 5′-CCA TAG GGG CAG GCT TTA CT-3′ ’)]. After 25–30 amplification cycles, PCR products were run on a 2% agarose gel, stained with ethidium bromide, and visualized under UV light.
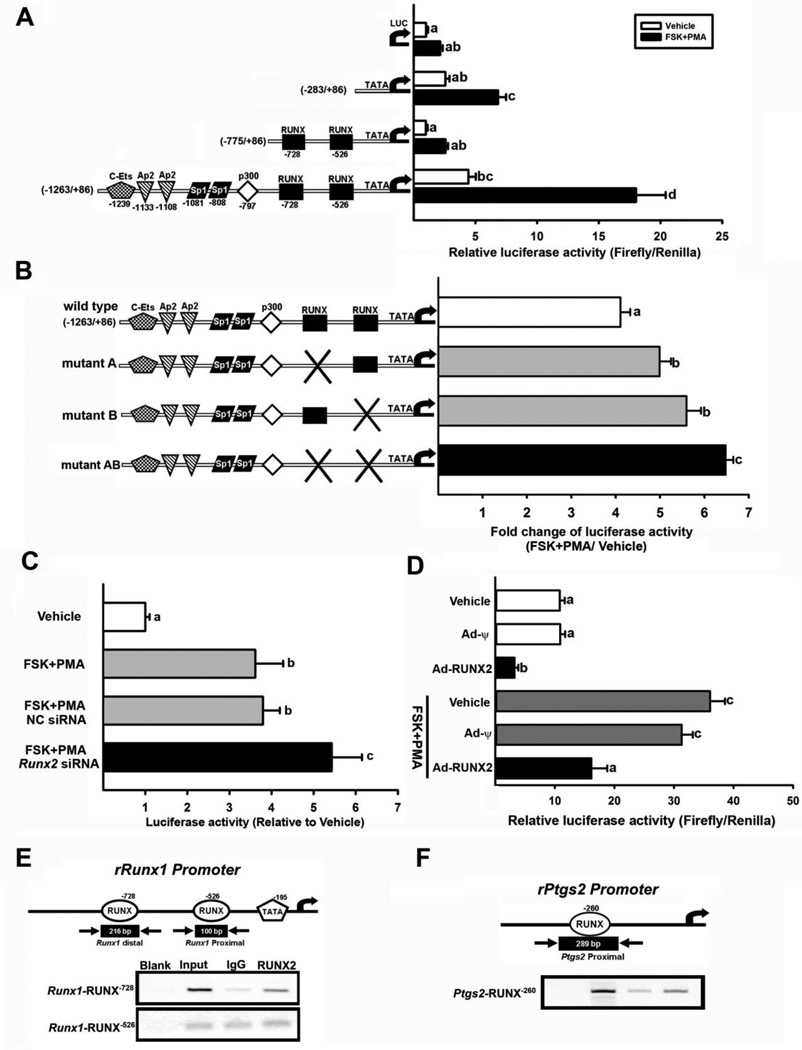
Figure 4. Regulation of transcriptional activity of Runx1 promoter reporter constructs in cultured granulosa cells.
Granulosa cells were isolated from gonadotropin-primed immature rats (48 h post-PMSG). A) The cells were transfected with empty luciferase reporter vector (LUC), −283/+86 bp, −775/+86 bp, or −1263/+86 Runx1-luciferase reporter constructs, treated with FSK+PMA and further cultured for 6 h. Firefly luciferase activities were normalized to Renilla luciferase activities. B, The cells were transiently transfected with wild type, mutant A, mutant B, or mutant AB Runx1-luciferase reporter constructs and stimulated with FSK plus PMA for 6 h. Luciferase activity of each constructs was expressed as a fold change of FSK plus PMA treatment to vehicle treatment. C, The cells were transfected with −1263/+86 Runx1-luciferase reporter constructs overnight, and then treated with NC siRNA or Runx2 siRNA and stimulated with FSK plus PMA. Luciferase activity of FSK plus PMA-treated constructs was normalized to Vehicle treated construct. D, The cells were infected with adenoviruses expressing Runx2 (Ad-Runx2) or control null vector (Ad-ψ), and then transfected with −1263/+86 Runx1-luciferase reporter constructs and stimulated with FSK+PMA for 6 h. Firefly luciferase activities were normalized to Renilla luciferase activities. The experiments were repeated at least 3 times (mean ± SEM). Bars with no common superscripts are significantly different (P < 0.05). E & F, ChIP assays were performed using granulosa cells infected with adenoviruses expressing Runx2 (Ad-RUNX2) at 5 IFU and then stimulated with FSK+PMA for 6 h. DNAs were analyzed by PCR using primers indicated in the text and represented as arrows in E & F. Amplified DNA fragments containing RUNX binding motifs are represented as black boxes with the indicated PCR product size. Experiments were repeated at least three times, each with different cultured granulosa cell samples.
2.12. Statistical analyses
Results were expressed as mean ± SEM. Data were tested for homogeneity of variance by Levene test, and square root or log transformations were performed on data set that showed heterogeneous variance. All data were analyzed by Student’s t-test, or ANOVA (one-way or two-way ANOVA) to determine the significant difference across time of culture or among treatments in vitro. Student t-test was used to assess differences of treatment between groups. If ANOVA revealed significant effects, the means were compared by Tukey’s test, with p < 0.05 considered significant.
3. RESULTS
3.1. hCG induced the transient expression of Runx1, Ptgs2, and Tnfaip6 and the sustained expression of Runx2 in rat preovulatory granulosa cell cultures
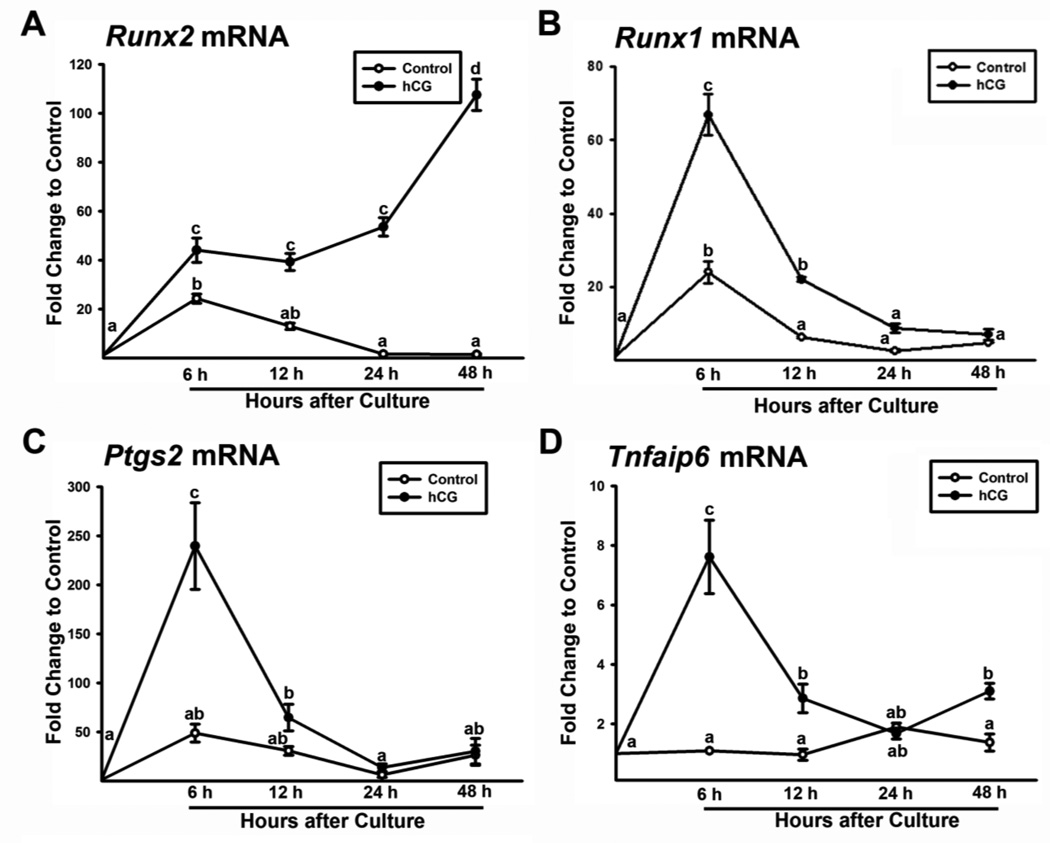
In the previous microarray data, we found that the levels of mRNA for Runx1, Ptgs2, and Tnfaip6 were increased in Runx2 siRNA-treated cells, suggesting negative regulation of these gene expressions by RUNX2. To further study the relationship among these periovulatory genes, we first determined the expression profile of these genes in our granulosa cell culture model. Although the expression pattern of these genes has already been characterized in periovulatory ovaries (Hernandez-Gonzalez et al., 2006; Jo and Curry, Jr., 2006; Joyce et al., 2001; Park et al., 2010; Yoshioka et al., 2000), it is important to verify that LH/hCG induces the unique expression pattern of these genes in our rat granulosa cell cultures. As shown in Fig. 1, hCG increased the expression of Runx1, Runx2, Ptgs2, and Tnfaip6. The levels of Runx2 mRNA increased at 6 h and continued to rise until 48 h (Fig. 1A), while Runx1 expression was transiently increased with the highest level of mRNA at 6 h and declined to basal level at 24 h (Fig. 1B). The up-regulation of Ptgs2 and Tnfaip6 expression was also rapid and transient, which reached highest levels at 6 h (Fig. 1C&D). These results demonstrated that in our granulosa cell cultures, hCG treatment induced transient up-regulation of Runx1, Ptgs2, and Tnfaip6 expression and gradual increases in Runx2 expression similar to those documented in the in vivo system (Jo and Curry, Jr., 2006; Joyce et al., 2001; Park et al., 2010; Yoshioka et al., 2000).
Figure 1. Stimulation of Runx2, Runx1, Ptgs2 and Tnfaip6 expression by hCG in granulosa cells in vitro.
Granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were cultured for 0, 6, 12, 24, or 48 h in medium alone (Control) or with hCG (1 IU/ml). The levels of mRNA for Runx2 (A), Runx1 (B), Ptgs2 (C) and Tnfaip6 (D) were measured by Real-time PCR and normalized to the L32 value in each sample (mean ± SEM; n = 3 independent culture experiments). No common superscripts are significantly different (P < 0.05).
3.2. Alteration of Runx2 expression affected Runx1, Ptgs2, and Tnfaip6 expression in granulosa cell cultures
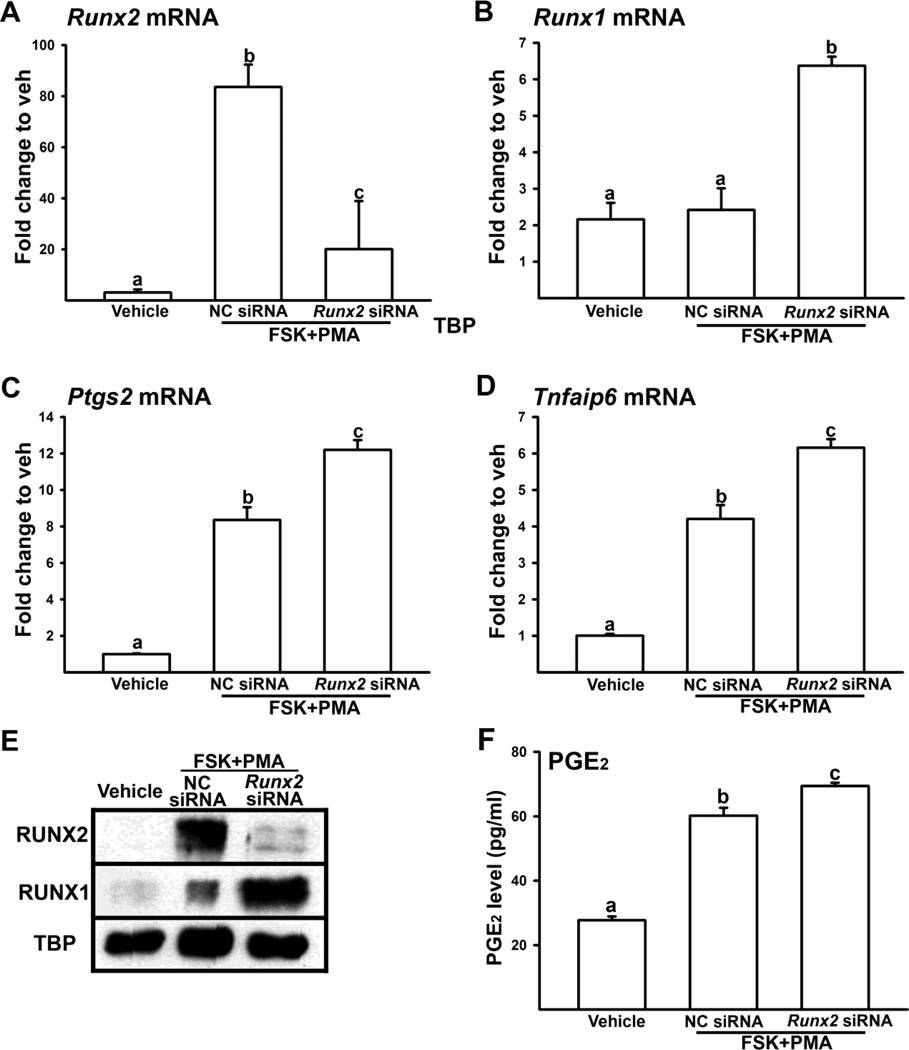
Based on the unique, yet differential expression pattern among Runx1, Runx2, Ptgs2, and Tnfaip6 in periovulatory granulosa cells, we hypothesized that the elevated expression of Runx2 in the late ovulatory period facilitates down-regulation of Runx1, Ptgs2, and Tnfaip6 expression. Therefore, we determined whether the knockdown of Runx2 expression could affect Runx1, Ptgs2, and Tnfaip6 expression in luteinizing granulosa cells. In our granulosa cell culture system, optimal gene silencing was achieved when the cells were acclimatized overnight and then transfected with Runx2 siRNA. However, this overnight acclimation caused LH receptor down-regulation in cultured granulosa cells (Robert et al., 2003; Schwall and Erickson, 1983). Therefore, in many studies from our laboratory (Liu et al., 2009; Liu et al., 2010; Park et al., 2008; Park et al., 2010) and others (Li et al., 2009; Li et al., 2011; Shimada et al., 2007; Sriraman et al., 2003), the combination treatment of forskolin (FSK, activator of adenylate cyclase) and PMA (activator of PKC) was used to mimic the action of LH/hCG in inducing periovulatory genes in granulosa cell cultures. These agonist treatments stimulated the expression of Runx1, Runx2, Ptgs2, and Tnfaip6 with a similar expression pattern compared to those observed in hCG-stimulated granulosa cell cultures (Supplemental Fig. 1). In the present study, we examined the effect of Runx2 silencing at 24 h as well as 48 h after stimulation with these activators. As shown in Fig. 2A & E, Runx2 siRNA effectively suppressed the stimulated expression of Runx2 compared to that in negative control (NC) siRNA-treated cells in both mRNA and protein levels in luteinizing granulosa cells. Moreover, the knockdown of Runx2 expression resulted in increases in levels of mRNA for Runx1, Ptgs2 and Tnfaip6 compared to that in NC siRNA-treated cells (Fig. 2B, C & D). Consistent with the mRNA data, RUNX1 protein level and PGE2 concentrations in Runx2 siRNA-treated cells were higher compared to that in NC siRNA-treated cells (Fig. 2E & F). We also observed similar increases in Runx1, Ptgs2, and Tnfaip6 expression by Runx2 knockdown at 48 h after the agonist treatment (Supplemental fig. 1).
Figure 2. Regulation of Runx1, Ptgs2, and Tnfaip6 expression by Runx2 knockdown in cultured granulosa cells.
Granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were transfected without siRNA (Vehicle) or with negative control siRNA (NC siRNA) or Runx2 siRNA and stimulated with FSK+PMA for 24 h. The levels of mRNA for Runx2 (A), Runx1 (B), Ptgs2 (C), and Tnfaip6 (D) were measured by Real-time PCR and normalized to the L32 value in each sample. E. RUNX1 and RUNX2 proteins in nuclear extracts were assessed by Western blot analyses. Each lane was loaded with 30 µg of nuclear fraction extracted from granulosa cell cultures transfected without or with siRNAs. The membrane was re-probed with a monoclonal antibody against TATA binding protein (TBP) to show the relative loading of nuclear extracts. The blots are representatives of three separate experiments. F. Concentration of PGE2 was measured from granulosa cells transfected without or with siRNAs. Experiments were repeated at least four times, each with different cultured granulosa cell samples (mean ± SEM). Bars with no common superscripts are significantly different (P < 0.05).
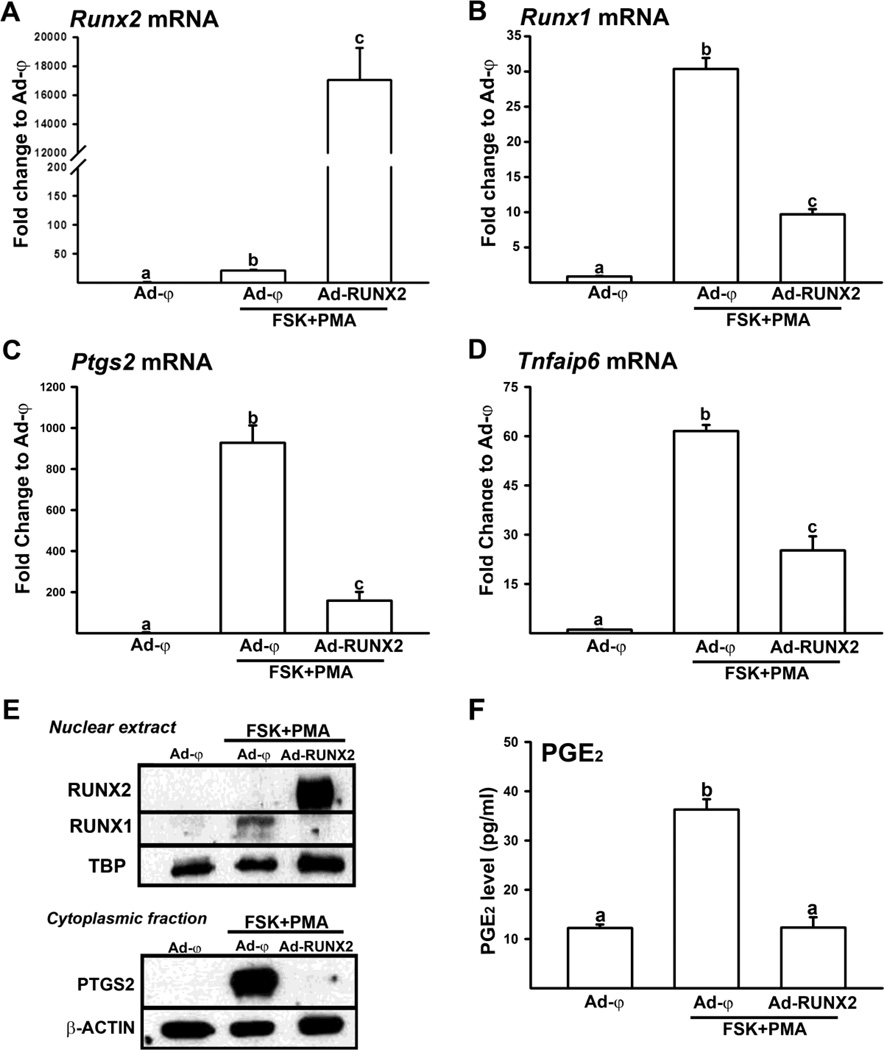
Next, to determine whether the over-expression of Runx2 affects Runx1, Ptgs2, and Tnfaip6 expression, granulosa cells were infected with adenoviruses expressing Runx2 (Ad-RUNX2) or null control vector (Ad-ψ) at 5 infectious units (IFU). The next day, the cells were stimulated with the activators for 4 h, the time period showing the high expression of Runx1, Ptgs2, and Tnfaip6 and relatively low expression of Runx2. The over-expression of Runx2 was confirmed at both mRNA and protein levels. As expected, the agonist treatment increased the expression of Runx1, Ptgs2 and Tnfaip6 in granulosa cells infected with Ad-ψ. But the adenovirus mediated over-expression of Runx2 resulted in dramatic reduction of the agonist-stimulated expression of these genes (Fig. 3B–E). The levels of RUNX1 and PTGS2 protein as well as PGE2 concentrations were also drastically decreased by RUNX2 over-expression (Fig. 3E & F).
Figure 3. Regulation of Runx1, Ptgs2, and Tnfaip6 expression by RUNX2 over-expression in cultured granulosa cells.
Granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were infected with adenoviruses expressing Runx2 (Ad-RUNX2) or containing null control vector (Ad-ψ) at 5 IFU, treated with FSK+PMA and further cultured for 4 h. The levels of mRNA for Runx2 (A) Runx1 (B), Ptgs2 (C), and Tnfaip6 (D) were measured by Real-time PCR and normalized to the L32 value in each sample. E, Western blots show RUNX1 and RUNX2 protein in nuclear extracts and PTGS2 in cytoplasmic fractions isolated from granulosa cells infected with adenoviruses. Each lane was loaded with 30 µg of cellular fractions from granulosa cell infected with adenoviruses. The membrane was re-probed with a monoclonal antibody against TATA binding protein (TBP) or β-ACTIN to show the relative loading. The blots are representatives of three separate experiments. F. Concentration of PGE2 was measured from granulosa cells infected with adenoviruses. Experiments were repeated at least three times, each with different cultured granulosa cell samples (mean ± SEM). Bars with no common superscripts are significantly different (P < 0.05).
3.3. Transcriptional activity of Runx1 promoter reporter constructs was regulated by Runx2 expression in granulosa cell cultures
Data from both silencing and over-expression experiments indicated that RUNX2 negatively affects Runx1 expression. To further investigate the transcriptional regulation of Runx1 expression, we cloned the rat Runx1 promoter fragment (−1263/+86 bp) into the upstream of a firefly luciferase reporter construct. This construct contains 2 consensus RUNX binding motifs as well as binding sites for various transcription activators including Sp1, AP2, and c-Ets. This promoter construct was transfected into preovulatory granulosa cells isolated from PMSG-primed immature rats. The cells were then stimulated with FSK+PMA for 3, 6, 12, or 24 h. We found that the transcriptional activity of the Runx1 promoter construct was highest at 6 h after FSK+PMA treatment and decreased by 24 h, which mimicked the profile of Runx1 mRNA (Data not shown). To further analyze transcriptional contribution of different regions of the Runx1 promoter, three different promoter fragments (−283/+86, −775/+86, and −1263/+86 bp) of the Runx1 gene were cloned and transfected into preovulatory granulosa cells. The cells were stimulated with FSK+PMA for 6 h, a time point showing the highest transcriptional activity. The agonist treatment increased the luciferase activity of −283/+86 and −1263/+86 bp reporter constructs compared to that of the vehicle (Fig. 4A). The Runx1 promoter construct (−1263/+86 bp) containing 2 RUNX motifs as well as binding sites for other transcription activators showed the highest transcriptional activity (Fig. 4A). Meanwhile, the Runx1 promoter construct (−775/+86 bp) containing 2 RUNX binding motifs showed the lowest transcriptional activity compared to two other constructs. This data suggested suppressive attribute of RUNX binding motifs on Runx1 transcriptional activity.
To assess whether the presence of RUNX binding motifs affects the transcriptional activity of the Runx1 promoter, site-directed mutations of the distal binding site (−728/−723 bp, mutant A) and proximal binding site (−260/−255 bp, mutant B) as well as double mutation of these two sites (−728/−723 bp plus −260/−255 bp, mutant AB) were generated. These constructs were transfected into preovulatory granulosa cells and then stimulated with FSK+PMA for 6 h (Fig. 4B). The results showed that mutation of either the distal or proximal RUNX binding site increased Runx1 promoter activities compared to the activities of the wild type construct (Fig. 4B). Moreover, the double mutations of both regions of consensus RUNX binding sites (mutant AB) showed the highest transcriptional activity (Fig. 4B). These findings indicated that RUNX binding sites are involved in the repression of Runx1 promoter activity.
To further determine whether RUNX2 is involved in controlling the Runx1 promoter activity, the expression of Runx2 was altered by Runx2 siRNA and adenovirus expressing Runx2. The cultured granulosa cells were transfected with the −1263/+86 bp Runx1 promoter construct and then stimulated with FSK+PMA. The transcriptional activity of the Runx1 promoter reporter construct was higher in Runx2 siRNA-transfected granulosa cells compared to the cells transfected with NC siRNA, indicating that Runx2 knockdown enhanced the promoter activity of the Runx1 gene (Fig. 4C). In contrast, the over-expression of Runx2 reduced the reporter activity of the Runx1 promoter (Fig. 4D). Together, these data suggest that Runx2 expression negatively affects the promoter activity of rat Runx1 gene in luteinizing granulosa cells.
Next, we determined whether RUNX2 could directly interact with RUNX binding motifs in the Runx1 promoters. ChlP assays were performed in preovulatory granulosa cells infected with Ad-RUNX2. As shown in Fig. 4E, PCR analyses revealed the enrichment of chromatin fragments spanning the distal RUNX binding site (−728), but not proximal RUNX binding site (−526) by immunoprecipitation with RUNX2 antibody. In addition, ChIP assays showed the binding of RUNX2 on the Ptgs2 promoter region in same cultured granulosa cell samples (Fig. 4F)
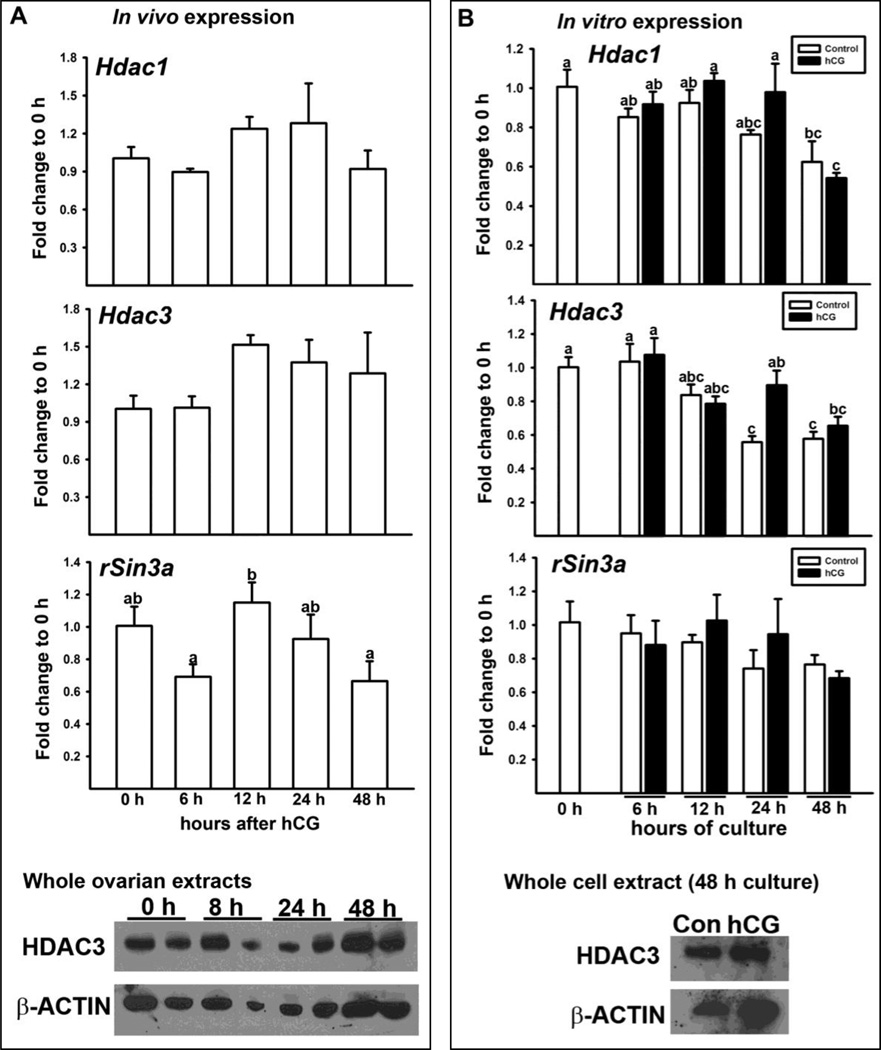
3.4. Expression of HDACs in the ovary in vivo and granulosa cells in vitro
HDACs were found to act as co-repressors of RUNX2 in various cell types (Hug, 2004; Jensen et al., 2007; Jensen et al., 2008; Lamour et al., 2007; Shimizu et al., 2010; Sun et al., 2009; Westendorf, 2006). Therefore, the periovulatory expression of Hdac1, Hdac3 and rSinc3a were assessed in whole ovaries collected at different times after hCG administration. Hdac1, Hdac3 and rSinc3a mRNA were constitutively expressed throughout the periovulatory period (Fig. 5). Similar to the mRNA expression profile, HDAC3 protein expression was also constant in whole ovarian extracts collected at different time intervals after hCG administration (Fig. 5A). Similar to the in vivo data, the expression of these genes was readily detected, but not affected by hCG treatment in cultured granulosa cells (Fig. 5B).
Figure 5. The ovarian expression of Hdacs in vivo and in vitro.
A) Ovaries were collected before or at indicated hours (h) after hCG injection from PMSG-primed immature rats. B) Granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were cultured for 0, 6, 12, 24, or 48 h in medium alone (Control) or with hCG (1 IU/ml). The levels of mRNA for Hdac1, Hdac3, and rSin3a were measured by Real-time PCR and normalized to the L32 value in each sample (mean ± SEM; n = 3 independent culture experiments). HDAC3 protein in whole cell extracts was assessed by Western blot analyses. The membrane was re-probed with β-ACTIN to show the relative loading of whole cell extracts.
4. DISCUSSION
The LH surge causes dynamic changes in gene expression in periovulatory follicle (Espey and Richards, 2002; Richards et al., 1998; Richards, 2007). Interestingly, for a certain cohort of genes, their expression rapidly increases, yet is transient during the preovulatory period [e.g., Ptgs2, Tnfaip6, Pgr, Runx1, C/EBPβ (Fan et al., 2011; Jo and Curry, Jr., 2006; Joyce et al., 2001; Natraj and Richards, 1993; Yoshioka et al., 2000)], while for another group of genes, their expression increases progressively and continues in the forming CL (e.g., Runx2, Cyp11a1, StAR, C/EBPa)(Chen et al., 1999; Hernandez-Gonzalez et al., 2006; Park et al., 2010; Rodgers et al., 1987; Sirois and Richards, 1993). These two distinct patterns of gene expression implicates functional involvement of these genes in the ovulatory process (ovulatory genes) and luteal development (luteal genes), respectively.
As central mediators of LH-initiated cellular events, transcriptional factors induced in periovulatory follicles directly control temporal changes in periovulatory gene expression. So far, studies have primarily focused on identifying periovulatory genes that are up-regulated by LH-induced specific transcription factors (Fan et al., 2011; Jo and Curry, Jr., 2006; Robker et al., 2000). Moreover, little attention was given to the involvement of specific transcription factors in rapid down-regulation of ovulatory gene expression. In the present study, we demonstrated that RUNX2 contributes to rapid down-regulation of a distinctive set of transiently induced preovulatory genes such as Ptgs2, Tnfaip6, and Runx1. Moreover, the negative effect of RUNX2 on Runx1 transcription involves the direct binding of RUNX2, at least on one of two RUNX binding motifs in the promoter of the rat Runx1 gene.
RUNX proteins (RUNX1, 2, 3) are structurally similar and have been shown to exhibit overlapping transcriptional activity on several target genes in different experimental models (Cameron and Neil, 2004; Liu et al., 2010; Park et al., 2008; Zhang et al., 2009). However, in most of the cases the expression of Runx genes is temporally or spatially separated. Recent studies have provided experimental evidence of self- and cross-regulatory mechanisms responsible for the tightly controlled expression of Runx genes (Brady et al., 2009; Brady and Farrell, 2009; Drissi et al., 2000; Spender et al., 2005). As relevant examples to the present study, Wong et al. (Wong et al., 2011) reported that RUNX1 down-regulates its own expression by directly binding to a RUNX consensus site in the promoter upon T-cell receptor activation in CD4+ T cells. Runx1 expression was also suppressed by RUNX3 in human B lymphoid cell lines and this suppression was thought to be important for B cell proliferation (Brady et al., 2009; Brady and Farrell, 2009; Spender et al., 2005). However, there has been, to our knowledge, no report of either positive or negative intra-familial regulation between Runx1 and Runx2. Previously, we have reported that both Runx1 and Runx2 expression are up-regulated by hCG-activated signaling, yet with distinctive temporal expression pattern in periovulatory granulosa cells. Based on the previous microarray data showing the increased expression of Runx1 by siRNA-mediated Runx2 knockdown, we proposed negative regulation of Runx1 by RUNX2 in periovulatory granulosa cells. Indeed, the present data from siRNA and over-expression experiments, along with promoter reporter and ChIP assays collectively pointed out that RUNX2 functions as a transcriptional repressor for the Runx1 gene in luteinizing rat granulosa cells. In addition to Runx1, the present study also identified Ptgs2 and Tnfaip6 as two other potential target genes for RUNX2 in luteinizing granulosa cells. The levels of mRNA for both Ptgs2 and Tnfaip6 are rapidly, yet transiently increased by the LH surge in granulosa and cumulus cells of periovulatory follicles (Fulop et al., 1997; Joyce et al., 2001; Sirois et al., 1992; Yoshioka et al., 2000). This unique temporal expression pattern could be mimicked in vitro by hCG or FSK+PMA treatment, as confirmed in our granulosa cell culture model. Furthermore, the data from silencing and over-expression experiments could be interpreted that RUNX2 is a part of the transcriptional complexes controlling rapid down-regulation of Ptgs2 and Tnfaip6 expression. Both rat Ptgs2 (Liu et al., 2009) and Tnfaip6 harbor multiple consensus RUNX binding motifs in their promoter region, suggesting that RUNX2 could act directly at the promoter level to repress the expression of genes. However, there is another important fact to consider. In the early ovulatory phase, RUNX1, not RUNX2, was found to directly bind to RUNX binding motifs on the Ptgs2 promoter and facilitate the LH-dependent increase in Ptgs2 expression in preovulatory granulosa cell (Liu et al., 2009), suggesting that RUNX1 is involved in the up-regulation of Ptgs2 expression during the early ovulatory period. Unlike Ptgs2 expression, we found that reduction (~45%) of Runx1 expression by Runx1 siRNA had no effect on Tnfaip6 expression in preovulatory granulosa cell cultures (Supplemental Fig. 3). Nonetheless, it is possible that RUNX2 could inhibit the expression of Ptgs2 and Tnfaip6 indirectly via down-regulating the expression of Runx1 during the late ovulatory period. It is well established that the expression of Ptgs2 and Tnfaip6 in periovulatory follicles is vital for successful ovulation and/or cumulus expansion (Davis et al., 1999; Fulop et al., 2003; Lim et al., 1997). Given the relationship between Runx2 and these important ovulatory genes, Ptgs2 and Tnfaip6, the present data suggests a potential role for RUNX2 in fine-tuning the ovulatory process.
We previously demonstrated that RUNX2 is actively involved in the up-regulation of specific luteal genes such as Rgc32, Mmp13, Ptgds, Fabp6 and Abcb1a in luteinized granulosa cells (Park et al., 2010). These studies, together with the present data, revealed the opposing action of RUNX2 in regulating periovulatory gene expression; RUNX2 increases the expression of luteal genes, while simultaneously suppresses the transcription of specific ovulatory genes in luteinizing granulosa cells. But, the present observation may not be surprising, given the fact that RUNX proteins have been often described as a context-dependent transcription factor [reviewed in (Cameron and Neil, 2004)]. For example, RUNX2 was found to act either as an activator or repressor by recruiting and/or interacting with other co-regulators in the promoter region of target genes in non-ovarian cells (Cameron and Neil, 2004; Durst and Hiebert, 2004; Komori, 2010; Otto et al., 2003; Westendorf, 2006). Recent studies using molecular approaches have identified several co-regulators of RUNX2 such as p300 (Westendorf and Hiebert, 1999), SMADs (Zhang et al., 2000), pRB (Thomas et al., 2001) and C/EBPβ (Gutierrez et al., 2002) as a co-activator and histone deacetylase (HDACs) (Hug, 2004; Jensen et al., 2007; Jensen et al., 2008; Shimizu et al., 2010; Sun et al., 2009; Westendorf, 2006), transducin-like enhancer of split (TLE) proteins (Ali et al., 2010), mSin3a (Lutterbach and Hiebert, 2000), and yes associated protein (YAP)(Vitolo et al., 2007) as a co-repressor. Importantly, several of these proteins are also expressed in the periovulatory ovary, including p300/CBP (Ongeri et al., 2005), SMADs (Kaivo-oja et al., 2006), and C/EBPβ (Sterneck et al., 1997). The present data also added HDACs as potential co-repressors of RUNX2 in the periovulatory ovary. Therefore, it is conceivable that the dual actions of RUNX2 observed in our studies are likely mediated by the interaction with these co-activators or co-repressors expressed in granulosa cells of periovulatory follicles.
In conclusion, the expression of Ptgs2, Tnfaip6, and Runx1 dramatically increases in preovulatory granulosa cells in response to the LH surge, yet rapidly declines before ovulation. This rapid down-regulation of Ptgs2, Tnfaip6, and Runx1 during the late ovulatory period is likely orchestrated by multiple factors, including reduction of positive regulators and accumulation of negative regulators. Our findings provide new insight into the complex regulatory mechanisms of Ptgs2, Tnfaip6, and Runx1 expression. RUNX2 acts as a direct transcriptional repressor for the Runx1 gene. The inhibitory effect of RUNX2 on Ptgs2 and Tnfaip6 could be direct and/or indirect through down-regulating Runx1 expression. The identify of and precise mechanisms of interaction between transcriptional regulators expressed in periovulatory follicles needs to be determined to fully appreciate dynamic transcriptional control of genes required for proper ovulation, cumulus expansion and luteal formation.
Supplementary Material
Highlights.
RUNX2 knockdown increases the expression of Runx1, Ptgs2, and Tnfaip6 in the ovary.
RUNX2 over-expression represses Runx1, Ptgs2, and Tnfaip6 expression in the ovary.
RUNX binding motifs are relevant to Runx1 promoter activity
Runx2 knockdown increases transcriptional activity of the Runx1 promoter
RUNX2 represses Runx1 expression by direct binding in the Runx1 promoter.
ACKNOWLEDGEMENT
We thank Drs. Thomas E Curry, Linah Al-Alem and Muraly Puttabyatappa and Ms. Katherine Rosewell for critical reading for the manuscript.
This work was supported by National Institutes of Health Grants by R01HD061617 and R03HD066012 (to MJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have nothing to disclose.
REFERENCES
- Ali SA, Zaidi SK, Dobson JR, Shakoori AR, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4165–4169. doi: 10.1073/pnas.1000620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SC, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- Brady G, Farrell PJ. RUNX3-mediated repression of RUNX1 in B cells. J. Cell Physiol. 2009;221:283–287. doi: 10.1002/jcp.21880. [DOI] [PubMed] [Google Scholar]
- Brady G, Whiteman HJ, Spender LC, Farrell PJ. Downregulation of RUNX1 by RUNX3 requires the RUNX3 VWRPY sequence and is essential for Epstein-Barr virus-driven B-cell proliferation. J. Virol. 2009;83:6909–6916. doi: 10.1128/JVI.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron ER, Neil JC. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23:4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Feng Q, Liu YX. Expression of the steroidogenic acute regulatory protein and luteinizing hormone receptor and their regulation by tumor necrosis factor alpha in rat corpora lutea. Biol. Reprod. 1999;60:419–427. doi: 10.1095/biolreprod60.2.419. [DOI] [PubMed] [Google Scholar]
- Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 2003;27:315–324. doi: 10.1016/s1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- Davis BJ, Lennard DE, Lee CA, Tiano HF, Morham SG, Wetsel WC, Langenbach R. Anovulation in cyclooxygenase-2-deficient mice is restored by prostaglandin E2 and interleukin-1beta. Endocrinology. 1999;140:2685–2695. doi: 10.1210/endo.140.6.6715. [DOI] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva De Sousa LS, Choi JY, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- Espey LL, Richards JS. Temporal and spatial patterns of ovarian gene transcription following an ovulatory dose of gonadotropin in the rat. Biol. Reprod. 2002;67:1662–1670. doi: 10.1095/biolreprod.102.005173. [DOI] [PubMed] [Google Scholar]
- Fan HY, Liu Z, Johnson PF, Richards JS. CCAAT/enhancer-binding proteins (C/EBP)-alpha and -beta are essential for ovulation, luteinization, and the expression of key target genes. Mol. Endocrinol. 2011;25:253–268. doi: 10.1210/me.2010-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop C, Kamath RV, Li Y, Otto JM, Salustri A, Olsen BR, Glant TT, Hascall VC. Coding sequence, exon-intron structure and chromosomal localization of murine TNF-stimulated gene 6 that is specifically expressed by expanding cumulus cell-oocyte complexes. Gene. 1997;202:95–102. doi: 10.1016/s0378-1119(97)00459-9. [DOI] [PubMed] [Google Scholar]
- Fulop C, Szanto S, Mukhopadhyay D, Bardos T, Kamath RV, Rugg MS, Day AJ, Salustri A, Hascall VC, Glant TT, Mikecz K. Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development. 2003;130:2253–2261. doi: 10.1242/dev.00422. [DOI] [PubMed] [Google Scholar]
- Ge C, Xiao G, Jiang D, Yang Q, Hatch NE, Roca H, Franceschi RT. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J. Biol. Chem. 2009;284:32533–32543. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy V, Kneissel M, Fournier B, Boyde A, Matthias P. High bone resorption in adult aging transgenic mice overexpressing cbfa1/runx2 in cells of the osteoblastic lineage. Mol. Cell Biol. 2002;22:6222–6233. doi: 10.1128/MCB.22.17.6222-6233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghozi MC, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez S, Javed A, Tennant DK, van RM, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol. Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Hug BA. HDAC4: a corepressor controlling bone development. Cell. 2004;119:448–449. doi: 10.1016/j.cell.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Jensen ED, Nair AK, Westendorf JJ. Histone deacetylase co-repressor complex control of Runx2 and bone formation. Crit Rev. Eukaryot. Gene Expr. 2007;17:187–196. doi: 10.1615/critreveukargeneexpr.v17.i3.20. [DOI] [PubMed] [Google Scholar]
- Jensen ED, Schroeder TM, Bailey J, Gopalakrishnan R, Westendorf JJ. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J. Bone Miner. Res. 2008;23:361–372. doi: 10.1359/JBMR.071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Curry TE., Jr. Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol. Endocrinol. 2006;20:2156–2172. doi: 10.1210/me.2005-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Gieske MC, Payne CE, Wheeler-Price SE, Gieske JB, Ignatius IV, Curry TE, Jr., Ko C. Development and application of a rat ovarian gene expression database. Endocrinology. 2004;145:5384–5396. doi: 10.1210/en.2004-0407. [DOI] [PubMed] [Google Scholar]
- Joyce IM, Pendola FL, O'Brien M, Eppig JJ. Regulation of prostaglandin-endoperoxide synthase 2 messenger ribonucleic acid expression in mouse granulosa cells during ovulan. Endocrinology. 2001;142:3187–3197. doi: 10.1210/endo.142.7.8268. [DOI] [PubMed] [Google Scholar]
- Kaivo-oja N, Jeffery LA, Ritvos O, Mottershead DG. Smad signalling in the ovary. Reprod. Biol. Endocrinol. 2006;4:21. doi: 10.1186/1477-7827-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. Runx2, a multifunctional transcription factor in skeletal development. J. Cell Biochem. 2002;87:1–8. doi: 10.1002/jcb.10276. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and maintenance by Runx2. Front Biosci. 2008;13:898–903. doi: 10.2741/2730. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010;339:189–195. doi: 10.1007/s00441-009-0832-8. [DOI] [PubMed] [Google Scholar]
- Lamour V, Detry C, Sanchez C, Henrotin Y, Castronovo V, Bellahcene A. Runx2- and histone deacetylase 3-mediated repression is relieved in differentiating human osteoblast cells to allow high bone sialoprotein expression. J. Biol. Chem. 2007;282:36240–36249. doi: 10.1074/jbc.M705833200. [DOI] [PubMed] [Google Scholar]
- Li F, Liu J, Jo M, Curry TE., Jr. A role for nuclear factor interleukin-3 (NFIL3), a critical transcriptional repressor, in down-regulation of periovulatory gene expression. Mol. Endocrinol. 2011;25:445–459. doi: 10.1210/me.2010-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Liu J, Park ES, Jo M, Curry TE., Jr. The B cell translocation gene (BTG) family in the rat ovary: hormonal induction, regulation, and impact on cell cycle kinetics. Endocrinology. 2009;150:3894–3902. doi: 10.1210/en.2008-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, Dey SK. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell. 1997;91:197–208. doi: 10.1016/s0092-8674(00)80402-x. [DOI] [PubMed] [Google Scholar]
- Liu J, Park ES, Curry TE, Jr., Jo M. Periovulatory expression of hyaluronan and proteoglycan link protein 1 (Hapln1) in the rat ovary: hormonal regulation and potential function. Mol. Endocrinol. 2010;24:1203–1217. doi: 10.1210/me.2009-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Park ES, Jo M. Runt-related transcription factor 1 regulates luteinized hormone-induced prostaglandin-endoperoxide synthase 2 expression in rat periovulatory granulosa cells. Endocrinology. 2009;150:3291–3300. doi: 10.1210/en.2008-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Hiebert SW. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene. 2000;245:223–235. doi: 10.1016/s0378-1119(00)00014-7. [DOI] [PubMed] [Google Scholar]
- Natraj U, Richards JS. Hormonal regulation, localization, and functional activity of the progesterone receptor in granulosa cells of rat preovulatory follicles. Endocrinology. 1993;133:761–769. doi: 10.1210/endo.133.2.8344215. [DOI] [PubMed] [Google Scholar]
- Ochsner SA, Day AJ, Rugg MS, Breyer RM, Gomer RH, Richards JS. Disrupted function of tumor necrosis factor-alpha-stimulated gene 6 blocks cumulus cell-oocyte complex expansion. Endocrinology. 2003;144:4376–4384. doi: 10.1210/en.2003-0487. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- Ongeri EM, Verderame MF, Hammond JM. Follicle-stimulating hormone induction of ovarian insulin-like growth factor-binding protein-3 transcription requires a TATA box-binding protein and the protein kinase A and phosphatidylinositol-3 kinase pathways. Mol. Endocrinol. 2005;19:1837–1848. doi: 10.1210/me.2004-0487. [DOI] [PubMed] [Google Scholar]
- Otto F, Lubbert M, Stock M. Upstream and downstream targets of RUNX proteins. J. Cell Biochem. 2003;89:9–18. doi: 10.1002/jcb.10491. [DOI] [PubMed] [Google Scholar]
- Park ES, Choi S, Muse KN, Curry TE, Jr., Jo M. Response gene to complement 32 expression is induced by the luteinizing hormone (LH) surge and regulated by LH-induced mediators in the rodent ovary. Endocrinology. 2008;149:3025–3036. doi: 10.1210/en.2007-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park ES, Lind AK, Dahm-Kahler P, Carletti MZ, Christenson LK, Curry TE, Jr., Jo M. RUNX2 transcription factor regulates gene expression in luteinizing granulosa cells of rat ovaries. Mol. Endocrinol. 2010;24:846–858. doi: 10.1210/me.2009-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25:589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Richards JS. Genetics of ovulation. Semin. Reprod. Med. 2007;25:235–242. doi: 10.1055/s-2007-980217. [DOI] [PubMed] [Google Scholar]
- Richards JS, Russell DL, Robker RL, Dajee M, Alliston TN. Molecular mechanisms of ovulation and luteinization. Mol. Cell Endocrinol. 1998;145:47–54. doi: 10.1016/s0303-7207(98)00168-3. [DOI] [PubMed] [Google Scholar]
- Rini D, Calabi F. Identification and comparative analysis of a second runx3 promoter. Gene. 2001;273:13–22. doi: 10.1016/s0378-1119(01)00579-0. [DOI] [PubMed] [Google Scholar]
- Robert C, Gagne D, Lussier JG, Bousquet D, Barnes FL, Sirard MA. Presence of LH receptor mRNA in granulosa cells as a potential marker of oocyte developmental competence and characterization of the bovine splicing isoforms. Reproduction. 2003;125:437–446. [PubMed] [Google Scholar]
- Robker RL, Russell DL, Espey LL, Lydon JP, O'Malley BW, Richards JS. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4689–4694. doi: 10.1073/pnas.080073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers RJ, Waterman MR, Simpson ER. Levels of messenger ribonucleic acid encoding cholesterol side-chain cleavage cytochrome P-450, 17 alpha-hydroxylase cytochrome P-450, adrenodoxin, and low density lipoprotein receptor in bovine follicles and corpora lutea throughout the ovarian cycle. Mol. Endocrinol. 1987;1:274–279. doi: 10.1210/mend-1-3-274. [DOI] [PubMed] [Google Scholar]
- Russell DL, Robker RL. Molecular mechanisms of ovulation: co-ordination through the cumulus complex. Hum. Reprod. Update. 2007;13:289–312. doi: 10.1093/humupd/dml062. [DOI] [PubMed] [Google Scholar]
- Schwall RH, Erickson GF. A new in vitro model system for the study of luteinizing hormone receptor down-regulation. J. Biol. Chem. 1983;258:3442–3445. [PubMed] [Google Scholar]
- Shimada M, Yanai Y, Okazaki T, Yamashita Y, Sriraman V, Wilson MC, Richards JS. Synaptosomal-associated protein 25 gene expression is hormonally regulated during ovulation and is involved in cytokine/chemokine exocytosis from granulosa cells. Mol. Endocrinol. 2007;21:2487–2502. doi: 10.1210/me.2007-0042. [DOI] [PubMed] [Google Scholar]
- Shimizu E, Selvamurugan N, Westendorf JJ, Olson EN, Partridge NC. HDAC4 represses matrix metalloproteinase-13 transcription in osteoblastic cells, and parathyroid hormone controls this repression. J. Biol. Chem. 2010;285:9616–9626. doi: 10.1074/jbc.M109.094862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois J, Richards JS. Transcriptional regulation of the rat prostaglandin endoperoxide synthase 2 gene in granulosa cells. Evidence for the role of a cis-acting C/EBP beta promoter element. J. Biol. Chem. 1993;268:21931–21938. [PubMed] [Google Scholar]
- Sirois J, Sayasith K, Brown KA, Stock AE, Bouchard N, Dore M. Cyclooxygenase-2 and its role in ovulation: a 2004 account. Hum. Reprod. Update. 2004;10:373–385. doi: 10.1093/humupd/dmh032. [DOI] [PubMed] [Google Scholar]
- Sirois J, Simmons DL, Richards JS. Hormonal regulation of messenger ribonucleic acid encoding a novel isoform of prostaglandin endoperoxide H synthase in rat preovulatory follicles. Induction in vivo and in vitro. J. Biol. Chem. 1992;267:11586–11592. [PubMed] [Google Scholar]
- Sirotkin AV. Transcription factors and ovarian functions. J. Cell Physiol. 2010;225:20–26. doi: 10.1002/jcp.22248. [DOI] [PubMed] [Google Scholar]
- Speck NA, Stacy T, Wang Q, North T, Gu TL, Miller J, Binder M, Marin-Padilla M. Core-binding factor: a central player in hematopoiesis and leukemia. Cancer Res. 1999;59:1789s–1793s. [PubMed] [Google Scholar]
- Spender LC, Whiteman HJ, Karstegl CE, Farrell PJ. Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene. 2005;24:1873–1881. doi: 10.1038/sj.onc.1208404. [DOI] [PubMed] [Google Scholar]
- Sriraman V, Sharma SC, Richards JS. Transactivation of the progesterone receptor gene in granulosa cells: evidence that Sp1/Sp3 binding sites in the proximal promoter play a key role in luteinizing hormone inducibility. Mol. Endocrinol. 2003;17:436–449. doi: 10.1210/me.2002-0252. [DOI] [PubMed] [Google Scholar]
- Sterneck E, Tessarollo L, Johnson PF. An essential role for C/EBPbeta in female reproduction. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer RL, Xu F, Duffy DM. Molecular control of ovulation and luteinization in the primate follicle. Front Biosci. 2007;12:297–307. doi: 10.2741/2065. [DOI] [PubMed] [Google Scholar]
- Sun X, Wei L, Chen Q, Terek RM. HDAC4 represses vascular endothelial growth factor expression in chondrosarcoma by modulating RUNX2 activity. J. Biol. Chem. 2009;284:21881–21890. doi: 10.1074/jbc.M109.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Vitolo MI, Anglin IE, Mahoney WM, Jr., Renoud KJ, Gartenhaus RB, Bachman KE, Passaniti A. The RUNX2 transcription factor cooperates with the YES-associated protein, YAP65, to promote cell transformation. Cancer Biol. Ther. 2007;6:856–863. doi: 10.4161/cbt.6.6.4241. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ. Transcriptional co-repressors of Runx2. J. Cell Biochem. 2006;98:54–64. doi: 10.1002/jcb.20805. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Hiebert SW. Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J. Cell Biochem. 1999;32–33(Suppl):51–58. doi: 10.1002/(sici)1097-4644(1999)75:32+<51::aid-jcb7>3.3.co;2-j. [DOI] [PubMed] [Google Scholar]
- Wong WF, Kurokawa M, Satake M, Kohu K. Down-regulation of Runx1 expression by TCR signal involves an autoregulatory mechanism and contributes to IL-2 production. J. Biol. Chem. 2011;286:11110–11118. doi: 10.1074/jbc.M110.166694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZS, Liu SG, Hinson TK, Quarles LD. Characterization of the upstream mouse Cbfa1/Runx2 promoter. J. Cell Biochem. 2001;82:647–659. doi: 10.1002/jcb.1192. [DOI] [PubMed] [Google Scholar]
- Yoshioka S, Ochsner S, Russell DL, Ujioka T, Fujii S, Richards JS, Espey LL. Expression of tumor necrosis factor-stimulated gene-6 in the rat ovary in response to an ovulatory dose of gonadotropin. Endocrinology. 2000;141:4114–4119. doi: 10.1210/endo.141.11.7784. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Jin L, Stilling GA, Ruebel KH, Coonse K, Tanizaki Y, Raz A, Lloyd RV. RUNX1 and RUNX2 upregulate Galectin-3 expression in human pituitary tumors. Endocrine. 2009;35:101–111. doi: 10.1007/s12020-008-9129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.