Introduction
Human cytomegalovirus (HCMV; Human Herpesvirus 5), a member of the β-herpesvirus subfamily, is an extremely widespread human pathogen. The virus contains a linear dsDNA genome of approximately 250 kb that encodes over 200 and perhaps as many as 751 [1] protein products and fourteen micro RNAs (miRNA) [2]. The enveloped virion contains a 105nm icosahedral nucleocapsid surrounded by a proteinaceous tegument layer that assembles in the nucleus. The lipid bilayer contains nine virally encoded glycoproteins that are variably necessary for cellular entry. By all measures HCMV is an evolutionarily successful pathogen with levels of seroprevalence varying between 40% and 100% in a manner that closely relates with socioeconomic conditions [reviewed in [3]]. Like other herpesviruses [e.g. Varicella Zoster virus (VZV), Herpes Simplex viruses (HSV), Epstein-Barr virus (EBV)], HCMV infects hosts for life and cannot be cleared by immunological processes or antiviral drugs. Thus the virus is able to establish a latent infection involving limited viral gene expression during which no viral progeny are synthesized. However, spontaneous reactivation of productive replication can occur in response to immune abnormalities such as an immunocompromised state or pro-inflammatory signals. While latency hematopoietic myeloid tissues represent a key site of latency (and reactivation occurs from the derivative macrophages and dendritic cells), the virus is capable of replication in numerous and diverse cellular types including fibroblasts, endothelial cells, smooth muscle cells, neuronal cells, trophoblasts, epithelial cells, and hepatocytes. Importantly, cytomegaloviruses are highly species-specific and as a result no animal model supports HCMV replication.
Acute and chronic infection with HCMV is largely asymptomatic in healthy hosts. This effect derives from millions of years of co-evolutionary fine-tuning that have molded the virus into one that employs a low virulence life cycle and thus a protracted potential transmission period. However, when hosts are immunologically underdeveloped or otherwise impaired (especially during tissue transplantation and HIV-associated immunodeficiency), the virus can cause serious disease. Congenital HCMV infection affects up to 40,000 children per year in the U.S. and is the leading infectious cause of deafness and other neurodevelopmental disorders including microcephaly and cerebral palsy [see[4]]. In immunecompromised patients HCMV can cause severe opportunistic infections. In HIV-infected individuals, primary HCMV can lead to numerous acute conditions including hepatitis, esophagitis, gastritis, encephalitis, pneumonitis, enterocolitis [5], a pathological diversity related to the virus’ broad tissue tropism. Importantly, HCMV retinitis including retinal necrosis is especially common during HIV disease, accounting for ~85% of all such cases [6]. Immunosuppression associated with and donor/recipient serostatus during solid organ and hematopoietic stem cell transplantation is similarly linked to tissue invasive HCMV diseases. HCMV is significantly correlated with diminished graft survival during solid organ transplantation [7] as well as artery stenosis [8]. Allogeneic hematopoietic stem cell transplantation is associated with reactivating productive infections as well as late developing HCMV disease. Numerous studies tie HCMV to multiple chronic inflammatory disease states of healthy hosts including cardiovascular disease [9], cancer [10], cognitive decline [11], and functional impairment [12].
The immune response to HCMV is broad with respect to the immune cells and processes involved and continuously active for the duration of the infection. Antiviral host responses to HCMV begin quickly after virus-cell contact with pre-existing cellular factors such as nuclear domain 10-associated proteins (e.g. PML, Sp100, hDaxx, ATRX) acting to prevent initial viral gene expression [reviewed in [13]]. Virus-cell contact involves interaction with a growing list of pattern recognition receptors (PRRs; discussed below) that initiate intracellular signaling and consequent expression and secretion of numerous immunologically active cytokines and chemokines [14]. Humoral responses involving production of neutralizing antibodies (predominantly targeting virion envelope glycoprotein B) invariably occur during infection although evidence supporting their importance in control of cytomegalovirus is largely from non-HCMV animal models [15,16]. It is also widely accepted that T-cell-mediated immunity constitutes the primary mechanism by which HCMV infection is controlled [see [17]], a fact that is consistent with pathogenic manifestations described in an immunocompromised state. This control includes the activities of both CD8+ and CD4+ T-cells; remarkably, the proportion of these cells in peripheral blood directed toward HCMV-specific antigens can be up to 40% in aged hosts [18,19]. The impact of this multi-pronged attack on HCMV replication is the employment of diverse immune evasion phenotypes to counteract the antiviral effects thus ensuring persistent infection [see [20]]. The large genome size of the virus has enabled it to acquire, optimize, and implement a multi-faceted, often redundant array of immune evasion strategies that target virtually all examined functions of the antiviral immune response.
Interferon and the Antiviral Response
Interferons are cytokines that are crucial for limiting viral replication at the site of infection and for coordinating adaptive responses that lead to the development of specific, long lasting immunity. IFNs are comprised of three physiologically distinct types (I, II, III). Type I IFNs include IFNβ, thirteen IFNα subtypes, IFNτ, IFNκ and IFNω that are primarily responsible for generating tissue states refractory to virus replication via paracrine induction of antiviral effector genes (IFN-stimulated genes; ISGs). They also coordinate functions of cellular immune constituents such as natural killer (NK) cells, dendritic cells (DC), T cells, and B cells and promote neutrophil survival, NK-cell activation, DC maturation, T-cell proliferation, and B-cell differentiation [21,22,23,24,25]. Type I IFN also enhances the expression of molecules important for directing inflammatory and adaptive immunity such as MHC class I, CD38, interleukins (BLyS, IL-6, IL-10 and IL-15) and multiple chemokines [26,27,28,29,30]. As such, type I IFNs assume both direct and indirect roles in antiviral immunity.
IFNα/β activate downstream signaling pathways (discussed below) by binding to the IFNα receptor 1 and 2 complex (IFNAR1 and IFNAR2), which is ubiquitously expressed on all cell types [31]. IFNγ is the sole Type II IFN and while it is capable of upregulation of directly antiviral genes its broader function involves immune cell (NK cell, macrophage, B cell) activation. Type III IFNs, including IFN-λ1 (IL-29), IFN-λ2 (IL-28A) and IFN-λ3 (IL-28B), activate signaling pathways that are highly similar to type I IFNs but through a distinct receptor complex (IL-28R1 or IFN-λR1 and IL10R1) [32,33]. This review will focus largely on the directly antiviral effects of type I IFNs, their relevance for replication of HCMV, and the strategies employed by the virus to evade, withstand, or co-opt these responses.
The importance of IFNs combating cytomegalovirus and herpesviral infection is exemplified in reports of animals and humans carrying inactivating mutations in the IFN receptors or associated signaling molecules. Mice deficient in IFN responses exhibit lethal susceptibility to murine cytomegalovirus (MCMV) infection [34]. Moreover, homozygous mutations in the signaling molecule STAT1, which is an essential downstream factor in the IFNα/β- and IFNγ-signaling pathways (see below), is associated with lethal outcomes from infection with multiple herpesviruses including with HSV-1 [35], HCMV [36], and Epstein-Barr virus [37]. Importantly, recombinant IFNα has also been used therapeutically to successfully control HCMV-induced retinitis during AIDS [38] and to control viremia following congenital infections [39]. These observations demonstrate the importance of properly functioning type I IFN-dependent physiological effects for controlling cytomegalovirus infection.
In Vitro Antiviral Activity of Type I IFN
Type I IFN stimulates an antiviral state in tissues that has evolved to be inherently broad-spectrum with respect to the diversity of virus types affected. Although susceptibility may vary significantly between viral strains [40], it is interesting to note that HCMV is conspicuously insensitive to the effects of IFN exposure relative to other, even related virus types such as HSV [41]. The molecular bases for HCMV resistance necessitates more thorough investigation to be understood, however, the virus exhibits multiple, diverse phenotypes directed at inhibiting or co-opting the physiological effects of IFN exposure. In addition, IFN pre-treatment of cells prior to infection is necessary to observe any antiviral effects and IFN-mediated inhibition is decreased to near absence when added after infection has significantly progressed [42]. It is also interesting to note that HCMV replication is more severely blocked in the presence of both type I and type II IFN, a combination that confers an obvious synergistic inhibitory effect [43]. In addition, other cytokines such as interleukin 1β (IL-1β) may also contribute to IFN's effects via enhancement of expression of IFN itself [44] or other antiviral genes. Intriguingly, HCMV entry strongly triggers the IFN-independent (mostly IRF3-dependent) expression of numerous genes classified as ISGs [45], many of which have had antiviral phenotypes attributed to them [46,47,48,49,50]. Thus the virus does not require a host cell entry program that prevents ISG induction as seen for other herpesviruses including KSHV [51], RhCMV [52]; and HSV-1 [53].
IFN Signaling
Fundamentally, IFNα/β act by inducing the expression of immunological and antiviral effector genes (ISGs). This occurs following signal transduction initiated by binding of the proteins to their receptor complex, triggering dimerization of IFNAR1 and IFNAR2 and leading to activation of the constitutively associated Janus kinases Jak1 and tyrosine kinase Tyk2. Autophosphorylation of Janus kinases is followed by phosphorylation of tyrosine residues in the cytoplasmic tail of IFNAR1 and 2 creating a docking site for the transcription factors signal transducer and activator of transcription (STAT) 1 and 2. Subsequent phosphorylation of STAT tyrosine residues by Jak1 and Tyk2 drives the dimerization of STAT1 and STAT2 via Src-homology 2 (SH2) domains. STAT1/2 dimers form a stable complex with IFN regulatory factor 9 (IRF9). This complex (termed ISGF3) shuttles to the nucleus where it drives the expression of genes containing IFN-stimulated response elements (ISRE) in their promoter [reviewed in [54]]. STAT1, STAT2 and IRF9 themselves represent ISGs that are expressed upon JAK-STAT signaling, thereby establishing a positive feedback amplification loop [55,56]. The JAK-STAT pathway is negatively regulated by cellular proteins such as suppressor of cytokine signaling (SOCS), various members of the protein inhibitor of activated STAT (PIAS) family, and protein tyrosine phosphatases (PTPs) [54]. STAT1 is actually shared by the type I and type II IFN signaling pathways in that IFNγ stimulation leads to formation of STAT1 homodimers, which trigger expression of a distinct set of promoters containing a γ-activated sequence (GAS). Interestingly, unphosphorylated STAT1 and STAT2 have also been shown to be capable of nuclear localization and gene transcription [57,58] and even to confer antiviral effects [59]. In addition, phosphorylated STATs are implemented in negative regulation of ISGs [58], indicating that JAK-STAT pathway is more complex than initially reported. The relevance of this activity for viral clearance remains largely unknown, however.
IFNα/β Synthesis in Response to HCMV Infection
Expression and secretion of type I IFNs and pro-inflammatory cytokines and chemokines occur in response to cellular detection of virus binding and entry. IFNα/β is induced during infection of mice with murine CMV as early as 6h post-exposure [60] and includes a second wave at 36-48h [61]. In vitro induction of IFNα/β by HCMV infection has been repeatedly shown for multiple cell types including fibroblasts [62], neuronal cells [63], smooth muscle cells [64], dendritic cells [65,66], endothelial cells [67], lymphocytes [62], and pro-monocytic cells [68].
IFN secretion is preceded by synthesis of the corresponding mRNA and protein. This is especially well-characterized for pathogen-mediated expression of IFNβ [69,70], which requires activation of pivotal transcription factors such as IFN regulatory factors (IRF) and nuclear factor κB (NFκB). Indeed, IRF3 and NFκB are activated in response to HCMV infection in vitro [46,49,64,71,72,73,74,75]. Moreover, IRF3 has been demonstrated to be required for IFNβ synthesis during HCMV infection of fibroblasts [49]. While the essentiality of NFκB for IFN induction has not been firmly established it is likely that the protein acts as an enhancer of IFNβ mRNA expression [48,76,77].
IRF3 and NFκB activation result from signaling cascades initiated by the sensing of pathogen-associated molecular patterns (PAMPs) by germline-encoded pattern recognition receptors (PRRs). The list of IFN-terminal PRRs involved in detecting viral infection is long and expanding [see [78]]. This includes diverse groups of proteins that can share subcellular localization, amino acid similarity, or molecular agonist. Surprisingly, the number of PRRs that have been implicated in HCMV-mediated IFN induction is also substantial and growing. Initially, Toll-like receptor 2 (TLR2) was shown to be involved in the cellular innate response to both HCMV infection [79,80] and exposure to a soluble form of HCMV gB [80,81]. However, TLR2 engagement leads to NFκB but not IRF3 activation and thus the molecule is not required for IFNβ transcription [80]. Oddly, however, while soluble gB does induce patterns of IFN-associated gene expression that resemble those triggered by HCMV [47] the virus-triggered IFN response actually more closely resembles that induced by cytoplasmic dsDNA [82,83]. In fact, data are now accumulating that demonstrate a central role for innate cellular detection of HCMV-associated DNA in both the IFN [68,82] and inflammatory responses (our unpublished observations). In fibroblasts, HCMV-mediated IRF3 activation and IFN expression is eliminated following RNAi-mediated depletion of the cytoplasmic dsDNA sensor Z-DNA binding protein 1 (ZBP1; also known as DAI) [82]. HCMV infection of pro-monocytic THP-1 cells failed to trigger IFNβ transcription after depletion of the dsDNA receptor IFNγ-inducible protein 16 (IFI16) [68]. Other work has also identified essentiality of TLR3 in IFNβ transcription in these cells [84]. Interestingly, Kuenzel and colleagues uncovered a novel role for nucleotide-binding oligomerization domain-like receptor containing a CARD 5 (NLRC5) in HCMV-induced expression of IFNα [85]. The authors did not examine IRF status and it is possible that NLRC5 may be involved in activation of IRF7, a transcription factor more closely tied to amplification of IFNα subtypes [see [86]. The seemingly contradictory identification of so many essential HCMV-specific, IFN-terminal PRRs represents a puzzle that will begs for a more satisfying explanation. This may involve a more intricate understanding of the tissue and cellular distribution of PRRs and associated signaling molecules.
IFN Stimulated Genes
IFNα/β-induced JAK/STAT signaling is known to affect the transcription of a large assortment of cellular genes. Using oligonucleotide arrays Der and colleagues studied mRNA expression of 6800 human genes following stimulation with IFNα, IFNβ or IFNγ and found over 300 to be upregulated [56]. While functions (antiviral and otherwise) have been attributed to a growing number of ISGs [see [87,88]], the roles of the vast majority of these genes remain to be described. However, among the best characterized are genes that encode proteins that directly impair the intracellular molecular processes essential for virus replication. ISG proteins also act directly on basic cell processes by inducing apoptosis, inhibiting transcription and promoting signaling through other pathways. In addition, ISG proteins are involved in the recruitment of lymphocytes and stimulate the development of adaptive immune responses by enhancing antigen presentation [reviewed in [89]]. Mechanistic details on the molecular and antiviral functions for a key group if ISG proteins have been reported and a more thorough understanding of their role in viral (including CMV) clearance is now emerging.
Among the most studied antiviral ISG proteins are those whose activity is dependent on cytoplasmic double stranded RNA (dsRNA), a molecule indicative of virus (including HCMV) infection. The IFN-inducible serine-threonine protein kinase R (PKR) contains a dsRNA binding domain that triggers protein dimerization and autophosphorylation after which it phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α). This process leads to a block in host and viral protein translation resulting in inhibition of virus replication [90]. 2’5’-oligoadenylate synthetases (OAS) are also induced by type I interferons [56]. These proteins synthesize oligoadenylates upon interaction with dsRNA. 2’5’-oligoadenylates activate the latent endoribonuclease L (RNase L), which indiscriminately cleaves mRNA and ribosomal RNA, thereby halting viral replication [91]. The PRR retinoic acid inducible gene I (RIG-I; discussed below) also represents a dsRNA-sensing ISG [92]. Activation of RIG-I leads to a conformational change exposing the two C-terminal caspase activation and recruitment domains (CARD) that interact with the CARD of the mitochondrial anti-viral signaling protein IFN promoter stimulator 1 (IPS-1, also known as MAVS, VISA, Cardif) and enhances IFNβ synthesis through IRF3/NFκB activation [92,93,94]. Other IFN-inducible PRRs have been identified and the upregulation of these molecules presumably results from a necessity to increase cellular sensitivity to viral entry and enhance secretion of antiviral cytokines during active infection. These include receptors of dsDNA such as IFI16, ZBP1/DAI, and AIM2 that have been implicated in cytomegalovirus detection [68,82,95]. As discussed below, a number of ISGs are additionally induced in an IFN-independent, yet IRF3-dependent manner [45]. These include genes for which antiviral functions have been clearly described including viperin [96], ISG54/ISG56 [97], and ISG15 [98]. Ultimately, replication of HCMV in the presence of ISG proteins requires viral mechanisms to evade, deactivate, or co-opt the functions of these molecules.
HCMV Inhibits IFN-Dependent Signaling and ISG Expression
Despite the numerous and multi-faceted antiviral effects of type I IFNs, HCMV is able to invade, multiply, and establish persistent infection in healthy human hosts. This capacity is a direct result of the strategies employed by the virus to block IFN-dependent functions, to impair IFN induction, and to manipulate the physiological outcomes of ISG proteins. The large coding capacity of beta-herpesviruses allows for highly specified proteins that target multiple processes of the host immune response and many of these proteins remain to be characterized.
Several reports have described impairment of IFNα/β-mediated gene expression in cells infected with HCMV [46,99,100]. Initially, Miller and colleagues found that IFNα-induced phosphorylation of IFNAR1, Jak1, Tyk2, STAT1 and STAT2 was blocked in fibroblasts and epithelial cells infected with the HCMV strain Towne [101]. In addition, they observed reduced levels of Jak1 and IRF9 expression [101]. This result was subsequently confirmed by Le and colleagues who reported a reduction in JAK1 levels as early as 24 h.p.i. [40]. Moreover, Miller and colleagues published that fibroblasts infected with Towne do not express IFNγ-dependent genes (e.g. MHCII or IRF1) and that this results from an inhibition of IFNγ-induced phosphorylation of IFNGR1, Jak1, Jak2, and STAT1 late in infection [102]. HCMV Towne was also found to induce proteasome-dependent degradation of Jak1 [102]. Since Jak1 is involved in both type I and type II signaling pathways and its activation drives the phosphorylation of the IFN receptors, the kinase itself and the STAT proteins, both pathways are perhaps inhibited by the same viral protein(s). Both studies suggest this phenotype is mediated by an immediate early or early protein [101,102] that has not yet been identified. A report from Baron and Davignon further confirmed Towne's resistance to IFNγ-stimulation by showing that Jak1 and STAT1 phosphorylation was blocked early in infection, yet without reduction in Jak1 levels [103]. The cellular tyrosine phosphatase SHP2 was shown to be involved in the dephopshorylation of Jak1 and STAT1 [104] as well as in HCMV-mediated inhibition of STAT1 dephosphorylation [103]. Together these data imply that at earlier times post infection HCMV manipulates IFN-induced phosphorylation of Jak1 by activating SHP2 and possibly other tyrosine phosphatases. At later times Jak1 and IRF9 overall protein levels are reduced, thereby counteracting the IFN-mediated induction of the members of the Jak-STAT pathway. As mentioned above, type I and the type II IFNs synergistically inhibit HCMV replication [105]. As such, Jak1 inhibition may confer a proviral effect by inactivating at least two antiviral pathways.
Interestingly, proteasome-dependent reduction in STAT2 levels (seen at 48 h.p.i.) has been demonstrated in MRC-5 cells infected with the HCMV strains AD169, TB40/E, UL1271 and UL1702, but not with Towne [40]. When comparing viral growth of AD169 and Towne in the presence of IFNα or IFNβ the authors also observed only minor titer differences, indicating that, unexpectedly, the reduction in STAT2 contributes little to the virus resistance to IFN [40]. While the M27 open reading frame was identified as the essential gene for MCMV-induced STAT2 degradation [106], deletion of the homologous HCMV UL27 coding region did not restore STAT2 levels during HCMV infection [40]. As such, the HCMV factor involved in this phenotype remains to be identified. At 24 h.p.i. Jak1 levels were greatly reduced and phosphorylation of STAT1 and STAT2 was blocked. The inhibition of STAT2 phosphorylation preceded STAT2 degradation in AD169-infected cells, so the inhibition of STAT phosphorylation likely results from diminished Jak1 expression.
Work by Paulus and colleagues demonstrates that deletion of HCMV IE1 gene renders HCMV hypersensitive to type I IFN [100]. Furthermore, overexpression of IE1 blocked the IFN-induced interaction between the ISGF3 complex and target DNA [100]. IE1 appears to interact with STAT1 and STAT2 and thereby relocalizing the molecules to nuclear promyelocytic leukemia (PML) bodies [100,107]. PML bodies are involved in DNA replication and transcription and IE1 is known to disrupt these structures [108]. Huh and colleagues demonstrated that the carboxyl-terminal acidic region of IE1 (amino acids 421-475) was required for its interaction with STAT2 [109]. This domain was further narrowed down to amino acids 373 to 420 that constitute the core domain necessary for the interaction between IE1 and STAT2 and the adjacent ancillary site comprising amino acids 421-445,, which is also required for an efficient interaction between the two proteins [107]. Deletion of these regions resulted in severely attenuated viral growth in the presence of type I IFNs [107,109]. However, infection with HCMV IE1Δ421-475, an IE1 mutant that is unable to interact with STAT2, did not fully restore IFNβ-induced ISG expression [109]. Nonetheless, Paulus and colleagues observed a complete restoration of IFN-induced ISG54 and Mx1 expression in cells infected with an IE1 deficient virus [100]. Together these data suggest that HCMV inhibition of type I IFN-induced signaling is solely dependent on IE1, which apparently can still affect STAT2-mediated signaling when an interaction is no longer detectable. In addition, HCMV IE1Δ421-475 that is unable to interact with and relocalize STAT2 to chromatin is still able to disrupt PML bodies, indicating that these represent separate phenotypes of IE1 [107]. Interestingly, IE1 has also have been implicated in the induction of certain type II ISGs. The protein appeared to promote the expression of IFIT2, IRF1, and IDO1 independently of IFNγ-signaling, though IFNγ-treatment enhanced the expression of these ISGs. The induction requires phosphorylation of STAT1 and the authors suggest that IE1 either phosphorylates STAT1 or it traps naturally occurring phosphorylated STAT1 in the nucleus by preventing dephosphorylation of the protein [110].
Thus far only IE1 has been identified as a modulator of type I and type II IFN responses, sequestering STAT2 to block type I ISGs expression but stimulating expression of type II ISGs by altering the phosphorylation status of STAT1. The modulation of Jak1 phosphorylation and expression appears to be an effective way to broadly affect IFN responses and the former is most likely dependent on SHP-2 activation, but the responsible ORF(s) remain unknown. The ORF(s) responsible for IRF9 and STAT2 reduction are not yet identified either, however STAT2 reduction is not observed in Towne-infected cells indicating that this phenotype is encoded on a region unique for the other HCMV strains.
Impairment of IFN Induction
Avoidance of the antiviral effects of type I IFN is obviously accomplished by preventing the synthesis or secretion of the molecule, a strategy employed by myriad virus types [reviewed in [111]]. The first indication that HCMV was capable of inhibiting IFN synthesis was uncovered in work by Browne and colleagues in which cellular transcriptome analysis following exposure of human fibroblasts to HCMV was performed [46]. In this study cells were exposed to live HCMV, virus rendered transcriptionally inactive by UV irradiation, or live HCMV in the presence of the protein synthesis inhibitor cycloheximide. These experiments revealed that transcriptionally and translationally viable virus induced levels of IFNβ expression that were remarkably lower than when these processes were inactive. In addition, ISGs were found to exhibit a similar pattern and multiple later studies confirmed these results [48,76,112,113]. These observations strongly suggested that HCMV-encoded proteins were capable of actively inhibiting infection-induced expression of IFNβ although whether this inhibition is specifically responsible for the subsequent decrease in ISG transcription has yet to be rigorously demonstrated.
Investigation aimed at identifying the responsible HCMV protein originally focused on the abundant viral tegument protein pp65 encoded by the ORF UL83 [114]. This protein has been shown to modulate other immune responses including antigen presentation [115] and NK cell cytotoxicity [116]. While contrasting results initially appeared regarding the importance of pp65 for HCMV-mediated IFNβ induction (one group attributed this to impairment of NFκB but not IRF3 [76], another group revealing the opposite [48]) it was eventually shown that a pp65 deletion virus used in these studies was actually defective for expression of other, off-target proteins [113]. This discovery led to uncovering of an NFκB-inhibitory function for one of these proteins, UL123-encoded IE2 [77,112,113]. It was also shown that this function blocked transcription by NFκB through the prevention of binding of NFκB to the IFNβ promoter, thereby inhibiting IFNβ expression as well as other cytokines [77]. The fact that NFκB is not believed to be strictly essential for IFNβ induction during HCMV infection as is IRF3 [49] may explain why some IFNβ secretion is detectable during live virus infection [82]. It is interesting to note, however, that the highly evolutionarily similar cytomegalovirus of Rhesus macaques (RhCMV) totally blocks induction of IFNβ transcription through inhibition of IRF3 phosphorylation via an unknown mechanism [52]. Also noteworthy is an observation made for HCMV pp65 in which the protein was shown to directly interact with the PRR IFI16 (discussed below) [117]. A role for this interaction in viral gene expression was described, however, it is highly curious that IFI16 has also been implicated in HCMV-mediated IFNβ transcription [68]. Thus the exact basis for HCMV-associated inhibition of IFNβ expression is likely not fully elucidated.
Viral interlukin-10 (cmvIL-10)
Human IL-10 is a pleiotropic, anti-inflammatory cytokine that limits innate and adaptive immune responses and thus controls damage to the host from immunopathology and autoimmunity [118]. HCMV has evolved to take advantage of these traits by acquiring an IL-10 coding region from the host genome and subsequently adapting the protein (cmvIL-10) to function as a proviral factor. CmvIL-10 binds with high affinity to the human IL-10 receptor, thereby eliciting immune regulatory effects that are similar to, or even more potent than, the responses induced by IL-10 itself [119,120,121]. Stimulation of PBMCs with CpG normally triggers secretion of IFNα and this response is greatly diminished when the PBMCs are treated with media from HCMV-infected cells [19]. When a viral mutant lacking cmvIL-10 is used, however, the media's inhibitory effect is lost. Interestingly, purified cmvIL-10 is able to directly inhibit IFNα secretion by plasmacytoid dendritic cells when cellular isolates are exposed to CpG, and this inhibition is as effective as when human IL-10 is used [122]. While the mechanism of inhibition has not been elucidated, these results indicate that the virus employs an extracellular, paracrine method of IFN inhibition that likely takes place in uninfected cells. HCMV might have employed this mechanism to limit exposure of systemic tissues to new IFN prior to infection with progeny virus.
Inhibition of ISG Protein Function
Type I IFN triggers the expression of hundreds of cellular genes. Many of these encode proteins that behave as antiviral effectors, directly impairing biochemical and molecular processes required for virus replication. While not all ISG proteins confer direct antiviral effects and anti-HCMV impacts have not been attributed to every one so characterized, inhibitory phenotypes directed at specific ISG proteins have been described for HCMV. Moreover, given the observed replication of the virus in the presence of multiple antiviral host proteins, it is probable that additional ISG-inhibitory phenotypes are to be uncovered for HCMV.
Double-stranded RNA activated protein kinase (PKR)
As described above PKR is one of the most well-characterized and pleiotropic ISGs [reviewed in [123]]. Like all other examined viruses, HCMV produces PKR-activating dsRNA during infection and thus evolved to inhibit the PKR pathway [124]. HCMV-mediated inhibition of PKR activity was shown by Adam Geballe's group. They observed that the function of the PKR inhibitory protein E3L of Vaccinia virus is restored during infection with a deletion mutant (VVΔE3L) by wild type HCMV and that this is dependent on early HCMV gene expression [125]. Importantly, the rescue is accompanied by reestablishment of protein translation and eIF-2α dephosphorylation. Ultimately, an ORF library screen was used to determine that two HCMV coding regions, TRS1 and IRS1 are able to rescue the PKR inhibition phenotype of VVΔE3L [126]. The rescuing ability was likewise found to be effective for HSV-1 lacking the PKR-inhibiting protein γ134.5 [127]. Either protein was subsequently shown to be dispensable (redundant) for HCMV replication and a double knock out mutant to be replication deficient and no longer inhibit PKR-mediated translation shut off [128,129,130]. Studies examining mechanisms of action [131,132,133] have revealed that TRS1 possesses two functions necessary for inhibiting PKR activation that are separable within the protein [131,132,133]. The amino portion of the protein specifically binds dsRNA following direct exposure and during HCMV infection [131,133]. The carboxy terminal portion binds PKR and sequesters it in the nucleus in an inactivated (unphosphorylated) form [132], suggesting that TRS1 physically maintains the protein away from the site of substrate recognition and activation.
2’,5’-Oligodenylate Synthetase (OAS)
OAS is also an enzyme that is activated upon dsRNA binding and for which activity results in global inhibition of protein synthesis [134]. Activated OAS synthesizes ATP molecules into 2’,5’-oligodenylate products that activate the latent ribonuclease RNase-L, which degrades viral and cellular RNAs thereby preventing corresponding protein expression. Early studies show that HCMV infection of fibroblasts and endothelial cells inhibits the IFNα signaling including IFNα–induced OAS upregulation [101]. Using the HCMV and VVΔE3L co-infection system described above, Child and colleagues demonstrated that HCMV can inhibit RNase-L-induced ribosomal RNA degradation [125]. While TRS1 and IRS1 are able to inhibit the OAS/RNase-L pathway induced by VVΔE3L infection [126], this is not the case in the context of HCMV infection as the double knockout virus (ΔIRS1/ΔTRS1) maintains the inhibitory phenotype [124] indicating that another viral protein was responsible. Accordingly, a role of HCMV ORF94 in the modulation of OAS has been identified [135]. Ectopic expression of ORF94 in HeLa cells reduces OAS1 mRNA and protein levels, even during treatment with IFNα or IFNγ. ORF94 expression was also able to directly impair OAS function during in vitro assays. Moreover, infection of human fibroblasts with recombinant virus unable to express ORF94 shows enhanced OAS1 mRNA levels compared to wild type virus and, importantly, the mutant virus demonstrates heightened susceptibility to IFNγ, suggesting an important role of ORF94 in tolerance of the IFN response. The mechanism by which ORF94 inhibits OAS expression and function and whether this occurs during latency remains unknown.
Retinoic acid inducible gene I (RIG-I)
RIG-I is an IRF3- and IFN-inducible cytoplasmic dsRNA sensor that signals through the mitochondrial adaptor IPS1/MAVS to activate NF-κB and IRF-3 and thus synthesis of type I interferons and pro-inflammatory cytokines [92,94]. In this way the molecule can enable a positive feedback loop that augments pathogen detection and antiviral cytokine production. A role for IPS1/MAVS (and thus RIG-I) in HCMV-induced IFN synthesis is unlikely since virus-mediated IRF3 activation and IFNβ synthesis occur in the molecule's absence [82]. However, this observation is admittedly curious since HCMV infection involves synthesis of dsRNA and cytoplasm-exposed dsDNA (a template for dsRNA synthesis by RNA POL3) [68,82], both of which can trigger IPS1/MAVS responses [136]. It was reported that IPS1/MAVS is cleaved and inactivated by caspases during pro-apoptotic cellular states [137,138], a strategy used by some viruses to impair antiviral responses [138,139]. Paradoxically, the HCMV-encoded inhibitor vMIA [140,141] impairs caspase-dependent cleavage of IPS1/MAVS and RIG-I degradation during apoptosis, perhaps a strategy for manipulating viral release. However, during HCMV infection of human fibroblasts, no IPS1/MAVS cleavage occurs yet RIG-I is degraded at later times (48-72h) post-infection, independently of apoptosis [142]. While the role of RIG-I in HCMV infection is not clear, these results further demonstrate the virus’ targeting of dsRNA-associated PRRs. However, the importance of RIG-I and its degradation for HCMV replication remains to be clarified.
HCMV “Re-Purposing” of ISG Proteins
Numerous IFN-induced proteins exhibit directly inhibitory effects on virus replication [87,88]. Consequently, viruses of disparate origins have evolved mechanisms to tolerate these responses. These vary in theme from the prevention of IFN and ISG induction to targeted impairment of individual ISG proteins. While employment of such approaches is known for HCMV, the virus also implements other extraordinary and perhaps unique stratagems for avoiding the inhibitory effects of ISG proteins. In recent years observations have accumulated demonstrating utilization of ISG protein functions as facilitators of HCMV replication. HCMV appears to have evolved to both allow (perhaps even enhance) cellular expression of certain ISGs and subsequently take advantage of their physiological effects. In this way the virus not only negates the antiviral effects of ISG proteins but actually “re-purposes” the molecules such that their activities confer proviral outcomes. Below we discuss the known instances in which this has been demonstrated, however, given the number of ISGs induced during HCMV infection it is highly likely that additional situations will be identified.
Interferonγ-inducible protein 16 (IFI16)
IFI16 is a member of the PYHIN protein group that contains a homotypic protein-protein binding (PYD) and DNA binding domain (HIN 200) [143]. As such, IFI16 is able to detect dsDNA and initiate innate immune reactions including IRF3 activation [68,144]. Thus far, IFI16 has been shown to exhibit at least two contrasting roles during HCMV infection. Cristea and colleagues found that IFI16, through an interaction with the viral tegument protein pp65, binds to the major immediate-early promoter (MIEP) and stimulates expression of essential HCMV transcripts [117]. Knockdown of IFI16 was subsequently found to diminish viral gene expression at low multiplicity and delay viral growth in fibroblasts thus indicating a pro-viral role for the protein [117]. In contrast, Gariano and colleagues actually demonstrate increased virus replication in the absence of IFI16 [145]. IFI16 overexpression is shown to actually inhibit expression of early and late kinetic classes of viral genes, and to specifically diminish UL54 (DNA polymerase) promoter activation by sequestering the required cellular factor Sp1 [145]. The results of Cristea et al. suggest that the protein modulates viral gene expression in a manner dependent on the promoter context. Accordingly, it could be speculated that the protein behaves as pro-viral early in infection and anti-viral at later points post-infection. The fact that pp65 (a protein contained in the infecting virion) interacts with IFI16 and therein promotes immediate early viral gene expression suggests that the virus actively re-directs its function towards a pro-viral activity. Moreover, this interaction may impair the PRR's ability to induce an IFN response and, interestingly, HCMV lacking pp65 expression is more susceptible to interferon treatment than is wild type virus [117]. Given the diversity of observations an understanding of the exact role of IFI16 in HCMV replication will clearly require more investigation.
Bone marrow stromal cell antigen 2 (BST2)
BST2 (or tetherin) is a type II transmembrane protein with a C-terminal glycosylphosphatidylinisotol (GPI) anchor whose expression is induced by IFNα/β exposure and hematopoietic differentiation [146,147,148]. Its primary anti-viral function described thus far is the tethering of progeny enveloped viruses to host cell membranes [reviewed in [149]]. Multiple viruses have thus developed diverse ways to overcome BST2 function [150]. For example, KSHV induces BST2 degradation through the viral ubiquitin ligase K5/MIR2 [151]. Interestingly, Viswanathan and colleagues studied the role of BST2 during HCMV infection and made the striking observation that the protein actually enhances rather than restricts virus replication [152]. Overexpression of BST2 in human fibroblasts yielded significantly more infectious particles than in untreated cells. In addition, siRNA-mediated BST2 silencing decreased HCMV entry into fibroblasts, THP-1 cells, and primary human monocytes [152]. The latter is particularly relevant since these cells play an important role in HCMV latency and reactivation [153]. Intriguingly, naturally non-permissive HEK293 cells actually acquired permissivity for HCMV replication following ectopic expression of BST2 [152]. The authors hypothesize that the mechanism by which BST2 enhances viral entry is by its tethering function, facilitating virus capture to the plasma membrane. In fact, BST2 is actually found in the HCMV virion, which is thought to be a pre-requisite for BST2 tethering function [152].
Viperin
The IFN- and IRF3-inducible protein viperin (also known as cig5) was originally described as being transcribed in response to in vitro HCMV infection [50] and its overexpression was found to impair HCMV replication [96]. The protein has had multiple antiviral molecular mechanisms described for it [see [154,155]. Subsequent work by the Cresswell research group revealed that while pre-expression of viperin does indeed block HCMV replication, when the gene is knocked down in host cells the virus actually grows to significantly lower titers [154]. This is attributed to an interaction later in the infection process between viperin and the HCMV apoptosis inhibitory protein vMIA, which directs the protein to the mitochondria. There it enhances generation of ATP and thus disrupts the actin cytoskeleton, a process that facilitates transport of virion components and ultimately virus assembly and release. Originally this process was attributed to vMIA [156] yet it is now likely that this is an indirect effect whereby the virus has acquired a method to achieve proviral cytoskeletal changes via the activity of an HCMV-induced cellular (assumed antiviral) protein.
Cyclooxygenase 2 (Cox2)
Cyclooxygenase 2 (COX2; Prostaglandin-Endoperoxide Synthase 2) is an IFN- and cytokine-inducible protein that is also triggered by infection with multiple herpesviruses including HCMV [46,157]. COX2 is an enzyme that catalyzes conversion of arachidonic acid to prostaglandin H2 and is thus a component of the eicosanoid synthetic pathway [see [158]]. In addition to inducing IFN-independent expression of COX2, HCMV infection also leads to an increase in prostaglandin E2, the synthetic end product of COX2 activity [159]. Importantly, exposure of cells to synthetic COX2 inhibitors severely impairs HCMV replication and cell-to-cell spread, an effect that is reversed upon supplementation of prostaglandin E2 [159,160]. COX2 activity is linked with expression of critical HCMV genes and viral protein localization. While directly antiviral phenotypes have not been described for COX2, it is clear that cytomegaloviruses have co-opted the protein's function to facilitate replication. Intriguingly, the genome of Rhesus macaque CMV has actually acquired from the host a functional COX2 ortholog that plays a role in cell tropism [161].
Conclusions and Future Directions
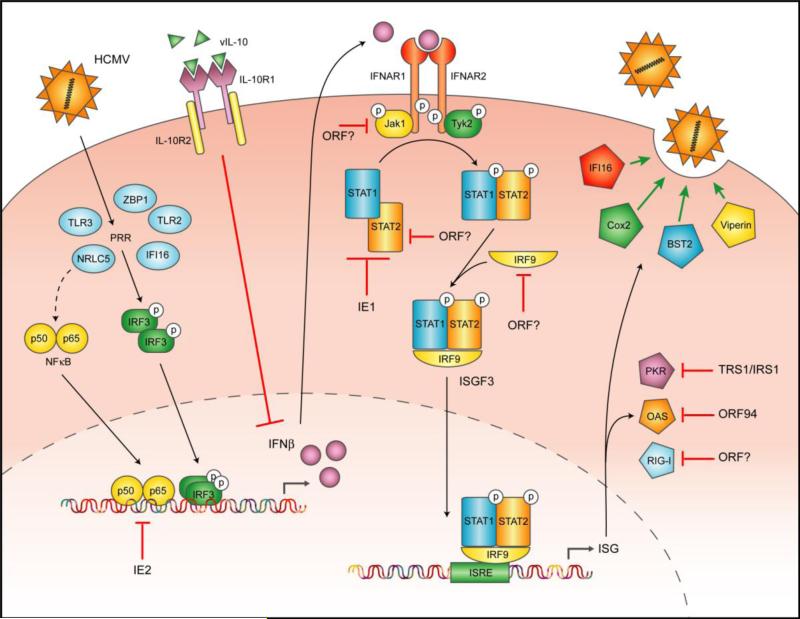
In line with the multi-phenotype, multi-target strategy employed to evade host T-cell-mediated responses [reviewed in [162]], HCMV likewise employs diverse mechanisms to withstand the type I IFN response. A summary of these is illustrated graphically in Figure 1. Unlike other virus types that are more susceptible to IFN (especially RNA-based viruses), HCMV possesses an extremely large genome with numerous protein and miRNA coding regions. This reality allows the virus to correspondingly utilize multiple and diverse methods to target all tiers of the IFN response including induction of the molecule, its intracellular signaling, and the functions of its induced factors. Given the number of predicted viral coding regions and the number of IFN-evasion phenotypes to which no gene(s) has been attributed, a great deal of work still remains to be undertaken. HCMV has also adopted an even more radical strategy by re-purposing the physiological functions of many ISG proteins to actually act as enablers of replication. Whether this is unique to this virus is currently unknown. In addition, given the number of explicitly antiviral genes induced during infection (e.g. ISG54, ISG56, ISG15, Zn-finger antiviral protein 1), it appears probable that HCMV has co-opted the functions of other such molecules. Finally, the importance of IFN/ISG induction and evasion for in vivo viral biology including latency and reactivation, pathogenesis, and inflammation is a crucial area of inquiry that will require the use of increasingly clever models and experimental approaches to address.
Figure 1.
Graphical summary of the IFN-associated targets and viral evasion molecules employed by HCMV.
Highlights.
Human cytomegalovirus is an extremely immunogenic pathogen that infects hosts for life.
Lifelong infection requires numerous sophisticated mechanisms of immune evasion.
The type I interferon system represents the first line of defense against a broad array of virus types including cytomegalovirus.
Human cytomegalovirus has evolved multiple phenotypes to counteract, withstand, or coopt physiological responses induced by type I interferons.
Acknowledgements
This work was supported by an American Heart Association Predoctoral Fellowship (L.A.) and NIH grants U54 AI081680-04 and R01 AI070890. We apologize to our colleagues whose work could not be cited due to length limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, et al. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20:202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 4.Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev. 2013;26:86–102. doi: 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steininger C, Puchhammer-Stockl E, Popow-Kraupp T. Cytomegalovirus disease in the era of highly active antiretroviral therapy (HAART). J Clin Virol. 2006;37:1–9. doi: 10.1016/j.jcv.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Yust I, Fox Z, Burke M, Johnson A, Turner D, et al. Retinal and extraocular cytomegalovirus end-organ disease in HIV-infected patients in Europe: a EuroSIDA study, 1994-2001. Eur J Clin Microbiol Infect Dis. 2004;23:550–559. doi: 10.1007/s10096-004-1160-2. [DOI] [PubMed] [Google Scholar]
- 7.Grattan MT, Moreno-Cabral CE, Starnes VA, Oyer PE, Stinson EB, et al. Cytomegalovirus infection is associated with cardiac allograft rejection and atherosclerosis. JAMA. 1989;261:3561–3566. [PubMed] [Google Scholar]
- 8.Pouria S, State OI, Wong W, Hendry BM. CMV infection is associated with transplant renal artery stenosis. QJM. 1998;91:185–189. doi: 10.1093/qjmed/91.3.185. [DOI] [PubMed] [Google Scholar]
- 9.Sorlie PD, Nieto FJ, Adam E, Folsom AR, Shahar E, et al. A prospective study of cytomegalovirus, herpes simplex virus 1, and coronary heart disease: the atherosclerosis risk in communities (ARIC) study. Arch Intern Med. 2000;160:2027–2032. doi: 10.1001/archinte.160.13.2027. [DOI] [PubMed] [Google Scholar]
- 10.Harkins L, Volk AL, Samanta M, Mikolaenko I, Britt WJ, et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–1563. doi: 10.1016/S0140-6736(02)11524-8. [DOI] [PubMed] [Google Scholar]
- 11.Aiello AE, Haan M, Blythe L, Moore K, Gonzalez JM, et al. The influence of latent viral infection on rate of cognitive decline over 4 years. J Am Geriatr Soc. 2006;54:1046–1054. doi: 10.1111/j.1532-5415.2006.00796.x. [DOI] [PubMed] [Google Scholar]
- 12.Schmaltz HN, Fried LP, Xue QL, Walston J, Leng SX, et al. Chronic cytomegalovirus infection and inflammation are associated with prevalent frailty in community-dwelling older women. J Am Geriatr Soc. 2005;53:747–754. doi: 10.1111/j.1532-5415.2005.53250.x. [DOI] [PubMed] [Google Scholar]
- 13.Tavalai N, Stamminger T. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta. 2008;1783:2207–2221. doi: 10.1016/j.bbamcr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Isaacson MK, Juckem LK, Compton T. Virus entry and innate immune activation. Curr Top Microbiol Immunol. 2008;325:85–100. doi: 10.1007/978-3-540-77349-8_5. [DOI] [PubMed] [Google Scholar]
- 15.Bratcher DF, Bourne N, Bravo FJ, Schleiss MR, Slaoui M, et al. Effect of passive antibody on congenital cytomegalovirus infection in guinea pigs. J Infect Dis. 1995;172:944–950. doi: 10.1093/infdis/172.4.944. [DOI] [PubMed] [Google Scholar]
- 16.Jonjic S, Pavic I, Polic B, Crnkovic I, Lucin P, et al. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J Exp Med. 1994;179:1713–1717. doi: 10.1084/jem.179.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev. 2009;22:76–98. doi: 10.1128/CMR.00034-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sester M, Sester U, Gartner B, Kubuschok B, Girndt M, et al. Sustained high frequencies of specific CD4 T cells restricted to a single persistent virus. J Virol. 2002;76:3748–3755. doi: 10.1128/JVI.76.8.3748-3755.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noriega V, Redmann V, Gardner T, Tortorella D. Diverse immune evasion strategies by human cytomegalovirus. Immunol Res. 2012;54:140–151. doi: 10.1007/s12026-012-8304-8. [DOI] [PubMed] [Google Scholar]
- 21.Banchereau J, Pascual V, Palucka AK. Autoimmunity through cytokine-induced dendritic cell activation. Immunity. 2004;20:539–550. doi: 10.1016/s1074-7613(04)00108-6. [DOI] [PubMed] [Google Scholar]
- 22.Mailliard RB, Son YI, Redlinger R, Coates PT, Giermasz A, et al. Dendritic cells mediate NK cell help for Th1 and CTL responses: two-signal requirement for the induction of NK cell helper function. J Immunol. 2003;171:2366–2373. doi: 10.4049/jimmunol.171.5.2366. [DOI] [PubMed] [Google Scholar]
- 23.Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14:432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 24.Seya T, Kasamatsu J, Azuma M, Shime H, Matsumoto M. Natural killer cell activation secondary to innate pattern sensing. J Innate Immun. 2011;3:264–273. doi: 10.1159/000326891. [DOI] [PubMed] [Google Scholar]
- 25.Tovey MG, Lallemand C, Thyphronitis G. Adjuvant activity of type I interferons. Biol Chem. 2008;389:541–545. doi: 10.1515/bc.2008.051. [DOI] [PubMed] [Google Scholar]
- 26.Biron CA, Brossay L. NK cells and NKT cells in innate defense against viral infections. Curr Opin Immunol. 2001;13:458–464. doi: 10.1016/s0952-7915(00)00241-7. [DOI] [PubMed] [Google Scholar]
- 27.Bussfeld D, Nain M, Hofmann P, Gemsa D, Sprenger H. Selective induction of the monocyte-attracting chemokines MCP-1 and IP-10 in vesicular stomatitis virus-infected human monocytes. J Interferon Cytokine Res. 2000;20:615–621. doi: 10.1089/107999000414781. [DOI] [PubMed] [Google Scholar]
- 28.Pogue SL, Preston BT, Stalder J, Bebbington CR, Cardarelli PM. The receptor for type I IFNs is highly expressed on peripheral blood B cells and monocytes and mediates a distinct profile of differentiation and activation of these cells. J Interferon Cytokine Res. 2004;24:131–139. doi: 10.1089/107999004322813372. [DOI] [PubMed] [Google Scholar]
- 29.Sakamoto E, Hato F, Kato T, Sakamoto C, Akahori M, et al. Type I and type II interferons delay human neutrophil apoptosis via activation of STAT3 and up-regulation of cellular inhibitor of apoptosis 2. J Leukoc Biol. 2005;78:301–309. doi: 10.1189/jlb.1104690. [DOI] [PubMed] [Google Scholar]
- 30.Salazar-Mather TP, Hokeness KL. Calling in the troops: regulation of inflammatory cell trafficking through innate cytokine/chemokine networks. Viral Immunol. 2003;16:291–306. doi: 10.1089/088282403322396109. [DOI] [PubMed] [Google Scholar]
- 31.Wang BX, Fish EN. The yin and yang of viruses and interferons. Trends Immunol. 2012;33:190–197. doi: 10.1016/j.it.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 33.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 34.Presti RM, Pollock JL, Dal Canto AJ, O'Guin AK, Virgin HWt. Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J Exp Med. 1998;188:577–588. doi: 10.1084/jem.188.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 36.Vairo D, Tassone L, Tabellini G, Tamassia N, Gasperini S, et al. Severe impairment of IFN-gamma and IFN-alpha responses in cells of a patient with a novel STAT1 splicing mutation. Blood. 2011;118:1806–1817. doi: 10.1182/blood-2011-01-330571. [DOI] [PubMed] [Google Scholar]
- 37.Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176:5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 38.Wen Y, Liu P. Interferon successfully inhibited refractory cytomegalovirus infection and resulted in CD4+ T-cells increase in a patient with AIDS. HIV Clin Trials. 2011;12:118–120. doi: 10.1310/hct1202-118. [DOI] [PubMed] [Google Scholar]
- 39.Emodi G, O'Reilly R, Muller A, Everson LK, Binswanger U, et al. Effect of human exogenous leukocyte interferon in cytomegalovirus infections. J Infect Dis. 1976;133(Suppl):A199–204. doi: 10.1093/infdis/133.supplement_2.a199. [DOI] [PubMed] [Google Scholar]
- 40.Le VT, Trilling M, Wilborn M, Hengel H, Zimmermann A. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J Gen Virol. 2008;89:2416–2426. doi: 10.1099/vir.0.2008/001669-0. [DOI] [PubMed] [Google Scholar]
- 41.Glasgow LA, Hanshaw JB, Merigan TC, Petralli JK. Interferon and cytomegalovirus in vivo and in vitro. Proc Soc Exp Biol Med. 1967;125:843–849. doi: 10.3181/00379727-125-32220. [DOI] [PubMed] [Google Scholar]
- 42.Holmes AR, Rasmussen L, Merigan TC. Factors affecting the interferon sensitivity of human cytomegalovirus. Intervirology. 1978;9:48–55. doi: 10.1159/000148920. [DOI] [PubMed] [Google Scholar]
- 43.Sainz B, Jr., LaMarca HL, Garry RF, Morris CA. Synergistic inhibition of human cytomegalovirus replication by interferon-alpha/beta and interferon-gamma. Virol J. 2005;2:14. doi: 10.1186/1743-422X-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwata M, Vieira J, Byrne M, Horton H, Torok-Storb B. Interleukin-1 (IL-1) inhibits growth of cytomegalovirus in human marrow stromal cells: inhibition is reversed upon removal of IL-1. Blood. 1999;94:572–578. [PubMed] [Google Scholar]
- 45.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmen KA, Singh J, Luukkonen BG, Lopper M, Bittner A, et al. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc Natl Acad Sci U S A. 2001;98:7140–7145. doi: 10.1073/pnas.121177598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abate D, Watanabe S, Mocarski E. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol. 2004;78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeFilippis VR, Robinson B, Keck TM, Hansen SG, Nelson JA, et al. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J Virol. 2006;80:1032–1037. doi: 10.1128/JVI.80.2.1032-1037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu H, Cong JP, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci U S A. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci U S A. 2002;99:5573–5578. doi: 10.1073/pnas.082420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeFilippis VR, Früh KJ. Rhesus cytomegalovirus particles prevent activation of interferon regulatory factor 3. J Virol. 2005;79:6419–6431. doi: 10.1128/JVI.79.10.6419-6431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol. 2004;78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y, Wang S, Gobl A, Oberg K. Interferon alpha induction of Stat1 and Stat2 and their prognostic significance in carcinoid tumors. Oncology. 2001;60:330–338. doi: 10.1159/000058529. [DOI] [PubMed] [Google Scholar]
- 56.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Testoni B, Vollenkle C, Guerrieri F, Gerbal-Chaloin S, Blandino G, et al. Chromatin dynamics of gene activation and repression in response to interferon alpha (IFN(alpha)) reveal new roles for phosphorylated and unphosphorylated forms of the transcription factor STAT2. J Biol Chem. 2011;286:20217–20227. doi: 10.1074/jbc.M111.231068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kraus TA, Lau JF, Parisien JP, Horvath CM. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J Biol Chem. 2003;278:13033–13038. doi: 10.1074/jbc.M212972200. [DOI] [PubMed] [Google Scholar]
- 60.Grundy JE, Trapman J, Allan JE, Shellam GR, Melief CJ. Evidence for a protective role of interferon in resistance to murine cytomegalovirus and its control by non-H-2-linked genes. Infect Immun. 1982;37:143–150. doi: 10.1128/iai.37.1.143-150.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider K, Loewendorf A, De Trez C, Fulton J, Rhode A, et al. Lymphotoxin-mediated crosstalk between B cells and splenic stroma promotes the initial type I interferon response to cytomegalovirus. Cell Host Microbe. 2008;3:67–76. doi: 10.1016/j.chom.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lun MT, Lorino G, Gaeta A, Dessy P, Cipriani P, et al. In vitro interferon production: induction in human leukocytes after exposure to cytomegalovirus antigens. Microbiologica. 1986;9:399–404. [PubMed] [Google Scholar]
- 63.van den Pol AN, Robek MD, Ghosh PK, Ozduman K, Bandi P, et al. Cytomegalovirus induces interferon-stimulated gene expression and is attenuated by interferon in the developing brain. J Virol. 2007;81:332–348. doi: 10.1128/JVI.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gravel SP, Servant MJ. Roles of an IkappaB kinase-related pathway in human cytomegalovirus-infected vascular smooth muscle cells: a molecular link in pathogen-induced proatherosclerotic conditions. J Biol Chem. 2005;280:7477–7486. doi: 10.1074/jbc.M410392200. [DOI] [PubMed] [Google Scholar]
- 65.Renneson J, Dutta B, Goriely S, Danis B, Lecomte S, et al. IL-12 and type I IFN response of neonatal myeloid DC to human CMV infection. Eur J Immunol. 2009;39:2789–2799. doi: 10.1002/eji.200939414. [DOI] [PubMed] [Google Scholar]
- 66.Kvale EO, Dalgaard J, Lund-Johansen F, Rollag H, Farkas L, et al. CD11c+ dendritic cells and plasmacytoid DCs are activated by human cytomegalovirus and retain efficient T cell-stimulatory capability upon infection. Blood. 2006;107:2022–2029. doi: 10.1182/blood-2005-05-2016. [DOI] [PubMed] [Google Scholar]
- 67.Sedmak DD, Chaiwiriyakul S, Knight DA, Waldmann WJ. The role of interferon beta in human cytomegalovirus-mediated inhibition of HLA DR induction on endothelial cells. Archives of Virology. 1995;140:111–126. doi: 10.1007/BF01309727. [DOI] [PubMed] [Google Scholar]
- 68.Horan KA, Hansen K, Jakobsen MR, Holm CK, Soby S, et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagarajan U. Induction and function of IFNbeta during viral and bacterial infection. Crit Rev Immunol. 2011;31:459–474. doi: 10.1615/critrevimmunol.v31.i6.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sin WX, Li P, Yeong JP, Chin KC. Activation and regulation of interferon-beta in immune responses. Immunol Res. 2012;53:25–40. doi: 10.1007/s12026-012-8293-7. [DOI] [PubMed] [Google Scholar]
- 71.Boehme KW, Singh J, Perry ST, Compton T. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J Virol. 2004;78:1202–1211. doi: 10.1128/JVI.78.3.1202-1211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yurochko AD, Kowalik TF, Huong SM, Huang ES. Human cytomegalovirus upregulates NF-kappa B activity by transactivating the NF-kappa B p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, et al. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon- stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Preston CM, Harman AN, Nicholl MJ. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J Virol. 2001;75:8909–8916. doi: 10.1128/JVI.75.19.8909-8916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yurochko AD, Hwang ES, Rasmussen L, Keay S, Pereira L, et al. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-kappaB during infection. J Virol. 1997;71:5051–5059. doi: 10.1128/jvi.71.7.5051-5059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Browne EP, Shenk T. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci U S A. 2003;100:11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor RT, Bresnahan WA. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression. J Virol. 2006;80:10763–10771. doi: 10.1128/JVI.01195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Compton T, Kurt-Jones EA, Boehme KW, Belko J, Latz E, et al. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J Virol. 2003;77:4588–4596. doi: 10.1128/JVI.77.8.4588-4596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juckem LK, Boehme KW, Feire AL, Compton T. Differential initiation of innate immune responses induced by human cytomegalovirus entry into fibroblast cells. J Immunol. 2008;180:4965–4977. doi: 10.4049/jimmunol.180.7.4965. [DOI] [PubMed] [Google Scholar]
- 81.Boehme KW, Guerrero M, Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. 2006;177:7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- 82.DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Fruh K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol. 2010;84:585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeFilippis VR, Sali T, Alvarado D, White L, Bresnahan W, et al. Activation of the interferon response by human cytomegalovirus occurs via cytoplasmic double-stranded DNA but not glycoprotein B. J Virol. 2010;84:8913–8925. doi: 10.1128/JVI.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yew KH, Carpenter C, Duncan RS, Harrison CJ. Human cytomegalovirus induces TLR4 signaling components in monocytes altering TIRAP, TRAM and downstream interferon-beta and TNF-alpha expression. PLoS One. 2012;7:e44500. doi: 10.1371/journal.pone.0044500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- 86.Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu SY, Sanchez DJ, Cheng G. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol. 2011;23:57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Veer MJ, Holko M, Frevel M, Walker E, Der S, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 90.Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, et al. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 91.Zhou A, Paranjape J, Brown TL, Nie H, Naik S, et al. Interferon action and apoptosis are defective in mice devoid of 2',5'-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 93.Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- 94.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-kappaB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 95.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao C, Collins MN, Hsiang TY, Krug RM. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol. 2013;21:181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miller DM, Zhang Y, Rahill BM, Waldman WJ, Sedmak DD. Human cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. J Immunol. 1999;162:6107–6113. [PubMed] [Google Scholar]
- 100.Paulus C, Krauss S, Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci U S A. 2006;103:3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miller DM, Zhang Y, Rahill BM, Waldman WJ, Sedmak DD. Human cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. Journal of Immunology. 1999;162:6107–6113. [PubMed] [Google Scholar]
- 102.Miller DM, Rahill BM, Boss JM, Lairmore MD, Durbin JE, et al. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baron M, Davignon JL. Inhibition of IFN-gamma-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. J Immunol. 2008;181:5530–5536. doi: 10.4049/jimmunol.181.8.5530. [DOI] [PubMed] [Google Scholar]
- 104.You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol Cell Biol. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sainz B, Jr., Lamarca HL, Garry RF, Morris CA. Synergistic inhibition of human cytomegalovirus replication by interferon-alpha/beta and interferon-gamma. Virol J. 2005;2:14. doi: 10.1186/1743-422X-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, et al. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-{gamma} signaling and antiviral responses. J Exp Med. 2005;201:1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krauss S, Kaps J, Czech N, Paulus C, Nevels M. Physical requirements and functional consequences of complex formation between the cytomegalovirus IE1 protein and human STAT2. J Virol. 2009;83:12854–12870. doi: 10.1128/JVI.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 109.Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, et al. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol. 2008;82:10444–10454. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Knoblach T, Grandel B, Seiler J, Nevels M, Paulus C. Human cytomegalovirus IE1 protein elicits a type II interferon-like host cell response that depends on activated STAT1 but not interferon-gamma. PLoS Pathog. 2011;7:e1002016. doi: 10.1371/journal.ppat.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Versteeg GA, Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Taylor RT, Bresnahan WA. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J Virol. 2005;79:3873–3877. doi: 10.1128/JVI.79.6.3873-3877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taylor RT, Bresnahan WA. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J Virol. 2006;80:920–928. doi: 10.1128/JVI.80.2.920-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Odeberg J, Plachter B, Branden L, Soderberg-Naucler C. Human cytomegalovirus protein pp65 mediates accumulation of HLA-DR in lysosomes and destruction of the HLA-DR alpha-chain. Blood. 2003;101:4870–4877. doi: 10.1182/blood-2002-05-1504. [DOI] [PubMed] [Google Scholar]
- 116.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 117.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, et al. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, et al. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 119.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci U S A. 2000;97:1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, et al. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci U S A. 2002;99:9404–9409. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chang WL, Barry PA. Attenuation of innate immunity by cytomegalovirus IL-10 establishes a long-term deficit of adaptive antiviral immunity. Proc Natl Acad Sci U S A. 2010;107:22647–22652. doi: 10.1073/pnas.1013794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chang WL, Barry PA, Szubin R, Wang D, Baumgarth N. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology. 2009;390:330–337. doi: 10.1016/j.virol.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Munir M, Berg M. The multiple faces of proteinkinase R in antiviral defense. Virulence. 2013;4:85–89. doi: 10.4161/viru.23134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marshall EE, Bierle CJ, Brune W, Geballe AP. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol. 2009;83:4112–4120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Child SJ, Jarrahian S, Harper VM, Geballe AP. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J Virol. 2002;76:4912–4918. doi: 10.1128/JVI.76.10.4912-4918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Child SJ, Hakki M, De Niro KL, Geballe AP. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J Virol. 2004;78:197–205. doi: 10.1128/JVI.78.1.197-205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cassady KA. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J Virol. 2005;79:8707–8715. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dunn W, Chou C, Li H, Hai R, Patterson D, et al. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A. 2003;100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Blankenship CA, Shenk T. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J Virol. 2002;76:12290–12299. doi: 10.1128/JVI.76.23.12290-12299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jones TR, Muzithras VP. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. Journal of Virology. 1992;66:2541–2546. doi: 10.1128/jvi.66.4.2541-2546.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hakki M, Geballe AP. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol. 2005;79:7311–7318. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hakki M, Marshall EE, De Niro KL, Geballe AP. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J Virol. 2006;80:11817–11826. doi: 10.1128/JVI.00957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bierle CJ, Semmens KM, Geballe AP. Double-stranded RNA binding by the human cytomegalovirus PKR antagonist TRS1. Virology. 2013;442:28–37. doi: 10.1016/j.virol.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peisley A, Hur S. Multi-level regulation of cellular recognition of viral dsRNA. Cell Mol Life Sci. 2013;70:1949–1963. doi: 10.1007/s00018-012-1149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tan JC, Avdic S, Cao JZ, Mocarski ES, White KL, et al. Inhibition of 2',5'-oligoadenylate synthetase expression and function by the human cytomegalovirus ORF94 gene product. J Virol. 2011;85:5696–5700. doi: 10.1128/JVI.02463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scott I, Norris KL. The mitochondrial antiviral signaling protein, MAVS, is cleaved during apoptosis. Biochem Biophys Res Commun. 2008;375:101–106. doi: 10.1016/j.bbrc.2008.07.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rebsamen M, Meylan E, Curran J, Tschopp J. The antiviral adaptor proteins Cardif and Trif are processed and inactivated by caspases. Cell Death Differ. 2008;15:1804–1811. doi: 10.1038/cdd.2008.119. [DOI] [PubMed] [Google Scholar]
- 139.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, et al. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A. 1999;96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, et al. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A. 2001;98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Scott I. Degradation of RIG-I following cytomegalovirus infection is independent of apoptosis. Microbes Infect. 2009;11:973–979. doi: 10.1016/j.micinf.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Veeranki S, Choubey D. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: Regulation of subcellular localization. Mol Immunol. 2012;49:567–571. doi: 10.1016/j.molimm.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012;8:e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cao W, Bover L, Cho M, Wen X, Hanabuchi S, et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med. 2009;206:1603–1614. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, et al. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 148.Goto T, Kennel SJ, Abe M, Takishita M, Kosaka M, et al. A novel membrane antigen selectively expressed on terminally differentiated human B cells. Blood. 1994;84:1922–1930. [PubMed] [Google Scholar]
- 149.Neil SJ. The antiviral activities of tetherin. Curr Top Microbiol Immunol. 2013;371:67–104. doi: 10.1007/978-3-642-37765-5_3. [DOI] [PubMed] [Google Scholar]
- 150.Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, et al. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 2010;6:e1000913. doi: 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, et al. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol. 2009;83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Viswanathan K, Smith MS, Malouli D, Mansouri M, Nelson JA, et al. BST2/Tetherin enhances entry of human cytomegalovirus. PLoS Pathog. 2011;7:e1002332. doi: 10.1371/journal.ppat.1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Reeves M, Sinclair J. Shenk T, Stinski MF, editors. Aspects of Human Cytomegalovirus Latency and Reactivation. Current Topics in Microbiology and Immunology. 2008. pp. 297–313. [DOI] [PubMed]
- 154.Seo JY, Yaneva R, Hinson ER, Cresswell P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science. 2011;332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- 155.Mattijssen S, Pruijn GJ. Viperin, a key player in the antiviral response. Microbes Infect. 2012;14:419–426. doi: 10.1016/j.micinf.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 156.Poncet D, Pauleau AL, Szabadkai G, Vozza A, Scholz SR, et al. Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J Cell Biol. 2006;174:985–996. doi: 10.1083/jcb.200604069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Reynolds AE, Enquist LW. Biological interactions between herpesviruses and cyclooxygenase enzymes. Rev Med Virol. 2006;16:393–403. doi: 10.1002/rmv.519. [DOI] [PubMed] [Google Scholar]
- 158.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 159.Zhu H, Cong JP, Yu D, Bresnahan WA, Shenk TE. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci U S A. 2002;99:3932–3937. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schroer J, Shenk T. Inhibition of cyclooxygenase activity blocks cell-to-cell spread of human cytomegalovirus. Proc Natl Acad Sci U S A. 2008;105:19468–19473. doi: 10.1073/pnas.0810740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rue CA, Jarvis MA, Knoche AJ, Meyers HL, DeFilippis VR, et al. A cyclooxygenase-2 homologue encoded by rhesus cytomegalovirus is a determinant for endothelial cell tropism. J Virol. 2004;78:12529–12536. doi: 10.1128/JVI.78.22.12529-12536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Basta S, Bennink JR. A survival game of hide and seek: cytomegaloviruses and MHC class I antigen presentation pathways. Viral Immunol. 2003;16:231–242. doi: 10.1089/088282403322396064. [DOI] [PubMed] [Google Scholar]