Abstract
Background and Aims
Few phylogeographic studies have been undertaken of species confined to narrow, linear coastal systems where past sea level and geomorphological changes may have had a profound effect on species population sizes and distributions. In this study, a phylogeographic analysis was conducted of Eucalyptus gomphocephala (tuart), a tree species restricted to a 400 × 10 km band of coastal sand-plain in south west Australia. Here, there is little known about the response of coastal vegetation to glacial/interglacial climate change, and a test was made as to whether this species was likely to have persisted widely through the Last Glacial Maximum (LGM), or conforms to a post-LGM dispersal model of recovery from few refugia.
Methods
The genetic structure over the entire range of tuart was assessed using seven nuclear (21 populations; n = 595) and four chloroplast (24 populations; n = 238) microsatellite markers designed for eucalypt species. Correlative palaeodistribution modelling was also conducted based on five climatic variables, within two LGM models.
Key Results
The chloroplast markers generated six haplotypes, which were strongly geographically structured (GST = 0·86 and RST = 0·75). Nuclear microsatellite diversity was high (overall mean HE 0·75) and uniformly distributed (FST = 0·05), with a strong pattern of isolation by distance (r2 = 0·362, P = 0·001). Distribution models of E. gomphocephala during the LGM showed a wide distribution that extended at least 30 km westward from the current distribution to the palaeo-coastline.
Conclusions
The chloroplast and nuclear data suggest wide persistence of E. gomphocephala during the LGM. Palaeodistribution modelling supports the conclusions drawn from genetic data and indicates a widespread westward shift of E. gomphocephala onto the exposed continental shelf during the LGM. This study highlights the importance of the inclusion of complementary, non-genetic data (information on geomorphology and palaeoclimate) to interpret phylogeographic patterns.
Keywords: Australian biogeography, climate change, coastal geomorphology, Eucalyptus gomphocephala, founder effects, Last Glacial Maximum, LGM, microsatellites, Myrtaceae, palaeodistribution modelling, phylogeography, southern hemisphere, south-western Australia, tuart
INTRODUCTION
For the arctic and temperate regions of the northern hemisphere, there is a consensus that post-Last Glacial Maximum (LGM) distributions of species were established via a complex mixture of migration over large distances from southern refugia, or short-range expansion from refugia whose current locations often conflict with present knowledge of tolerance range and models of past climatic conditions (Jackson and Overpeck, 2000; Hewitt, 2004; Beheregaray, 2008). In contrast, the impact of past climatic fluctuations in other regions and/or landscapes is less well understood (Beheregaray, 2008). For example, there are surprisingly few studies of the effect of historical climatic shifts and the subsequent changes in sea levels on the distribution of genealogical lineages within coastal landscapes (e.g. Kadreit et al., 2005; Arafeh and Kadreit, 2006; Molins et al., 2009). The world's coastlines are highly dynamic systems, where interactions between climate, substrate, water and wind have led to inundation, erosion, transportation and deposition of sediments, and subsequent geologically rapid changes in coastal geomorphology. These fluctuations have had significant impacts, particularly on currently vegetated, low-lying coastal dune systems. Critically, understanding the nature of these impacts can help inform on the capacity of species to respond to predicted rapid future climate change, and therefore their management and conservation (Oberle and Schaal, 2011).
The south-west of Western Australia (SWWA) is one of 34 internationally recognized biodiversity hotspots (Myers et al., 2000; Mittermeier et al., 2004), with a high level of diversity and endemism of animals and flowering plants (Hopper and Gioia, 2004). In contrast to central and northern Europe and much of North America, Pleistocene glaciation in Australia was restricted to small areas of the south-east, and absent from Western Australia (Hopper and Gioia, 2004). While currently experiencing a Mediterranean-type climate, average temperatures during the LGM were up to 6·5 °C cooler than at present (Hubbard, 1995). In contrast to south-eastern Australia, modelling studies have concluded that the precipitation regimes in SWWA did not collapse during the LGM (Wyroll et al., 2000) and that winter rainfall may have been higher than today (Rojas et al., 2009). However, there is limited evidence on the characteristics of glacial/interglacial climate change in SWWA and the response of vegetation, particularly in a coastal context. A palynological study by Pickett and Newsome (1997) suggests that coastal eucalypt woodland persisted across the LGM and may have migrated westward onto the exposed continental shelf, tracking a suitable maritime climate. Previous phylogeographic studies in the region have largely been focused on species from the semi-arid interior, and general conclusions are that the major biotic responses to climatic change involved persistence and resilience rather than large-scale migration (e.g. Byrne et al., 2002, 2003; Byrne and Hines, 2004). Therefore, it is largely unknown to what extent these findings apply to species from more mesic, coastal areas, which also sustained constant changes to local geomorphology.
This study is located on the Swan Coastal Plain (SCP), in SWWA. The SCP is a narrow, low relief coastal strip of alluvial and aeolian sands with a north–south extent of approx. 550 km, confined by the Indian Ocean to the west and the Darling Scarp to the east (Seddon, 2004). The geomorphology of the SCP was formed during successive changes in sea levels through the Pliocene and Quaternary, and its history is characterized by: (1) Quaternary dune building; (2) recent deposition of marine and riverine sediments; and (3) significant oceanic transgressions and regressions (Churchill, 1959; Playford et al., 1976; Semeniuk, 1997; Seddon, 2004). Large, mobile sand dunes built during a cooler, drier and probably windier climate would have had a marked impact on the SCP (Seddon, 2004). During the LGM, sea levels in the region were 120 m below current levels, and the SCP extended as far as 30 km westward onto the exposed continental shelf (Seddon, 2004). Sea level rises were most pronounced at the end of the Sangamon Interglacial [approx. 120 thousand years ago (kya), 4–6 m above current levels (Mercer, 1968)] and the middle of the Pliocene [approx. 3 million years ago (Mya), 25 m above current levels (Raymo et al., 2011)], where the Indian Ocean would have extended in places to the base of the Darling Scarp, approx. 20 km inland from its present position. As the SCP falls in elevation from north to south, sea level rises would have had a wider impact on low-lying areas such as the Yalgorup Plain, with multiple elevated features such as Reabold Hill (at 93 m, the highest point on the SCP) forming fragmented, offshore islands through the mid SCP during high sea levels.
Eucalyptus gomphocephala (tuart) is a model system to assess the influence of glacial/interglacial climate change, and changing sea levels and coastal geomorphology, on tree species confined to a linear, coastal system. Eucalyptus gomphocephala is endemic to calcareous soils on the SCP, on substrates of various ages. Most populations of E. gomphocephala occur on the Quindalup (nearest to the coast and the youngest formed <12 kya) or Spearwood (intermediate and formed approx. 40 kya) dune systems of the SCP, and rarely on the non-calcareous Bassendean (most easterly and oldest and formed approx. 800 000 kya) dune system or the Guildford formation of the Pinjarra Plain (Keighery et al., 2002). A priori it would be expected that Pleistocene climatic oscillations and changes in coastal geomorphology would have had a strong influence on the distribution of genetic diversity in the species.
We use chloroplast (Steane et al., 2005) and nuclear microsatellite markers (Brondani et al., 1998; Steane et al., 2001; Ottewell et al., 2005), combined with palaeodistribution modelling, to better understand historical distributions of the coastal tree species E. gomphocephala, and how the climatic and geomorphological changes of the Pleistocene may have shaped patterns of genetic differentiation and diversity. Specifically, this study tested whether E. gomphocephala persisted widely during the LGM or conforms to a post-LGM dispersal model of recovery from few refugia. We then interpreted these data in the context of historical responses to climate change, and how this informs conservation implications for future climate change.
MATERIALS AND METHODS
Study species
Eucalyptus gomphocephala is a medium to tall tree, endemic to the SCP, which is distributed along a narrow coastal strip (extending inland up to 10 km) (Fig. 1). Prior to European settlement in 1829, there were >111 600 ha (Hopkins et al., 1996) of E. gomphocephala woodlands, but now only approx. 30 000 ha remain (Tuart Response Group, 2003). Eucalyptus gomphocephala generally shows little morphological variation (Coates et al., 2002); however, it takes the form of a smaller tree or mallee in some northern and southern near-coastal populations (Ruthrof et al., 2002). A previous genetic study of E. gomphocephala examined allozyme variation for seven populations, and found generally low genetic differentiation (GST = 0·11) and similar levels of genetic diversity within each of the seven populations studied (Coates et al., 2002) .
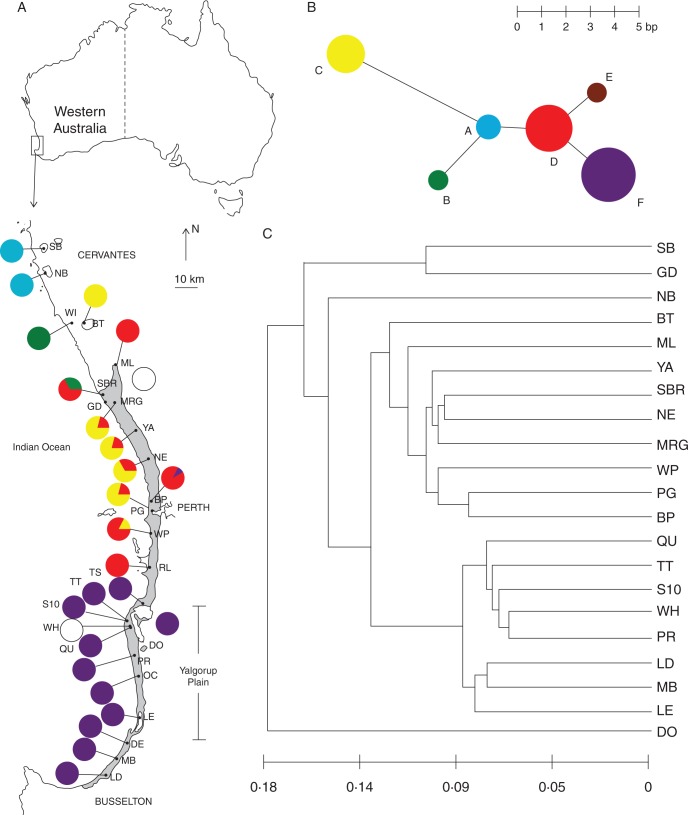
Fig. 1.
Study location and genetic analyses for Eucalyptus gomphocephala. (A) Geographic distribution of chloroplast microsatellite haplotypes. White circles indicate populations for which chloroplast data are not available. Section sizes are proportional to the number of individuals displaying that haplotype. Grey shading indicates the distribution of E. gomphocephala (map source Government of Western Australia, 2003). (B) Statistical parsimony tree of haplotypes based on four chloroplast microsatellites. Circle sizes indicate relative haplotype frequencies. Haplotypes present in ≤12 individuals are represented with a circle of the same size. Branch length indicates the number of nucleotides between haplotypes. (C) UPGMA phenograms of the relationship between populations based on nuclear SSR data. The scale bar represents Nei's (1972) genetic distance.
The mating system of E. gomphocephala is predominantly outcrossing, although the rate of outcrossing varies from complete selfing to complete outcrossing within and among populations and families, with an overall mean outcrossing rate of approx. 0·7 (Coates et al., 2002; Bradbury and Krauss, 2013). Pollination is primarily performed by non-specific insects; however, bird and mammal pollination are also expected (Ruthrof et al., 2002). While wind pollination is often discounted in eucalypts, airborne pollen of E. calophylla and E. marginata has been caught in traps 20 km off the coast of mainland Western Australia on Rottnest Island (Speck, 1953), where these species do not occur. Chloroplast DNA (cpDNA) is maternally inherited in most angiosperms, including eucalypts (Byrne et al., 1993; McKinnon et al., 2001), and thus is dispersed only by seed. Seed of eucalypts is dispersed primarily by gravity, which is largely within distances of two tree heights (Cremer, 1977), although secondary long-distance dispersal of seed-bearing fruit is possible by large birds such as Red-tailed Black Cockatoos (Calyptorhynchus banksii) and Carnaby's Black Cockatoo (C. latirostris).
Sampling
Leaf material was collected from 10–30 mature trees at each of 26 populations representing the entire geographic distribution of E. gomphocephala (Fig. 1A) (location co-ordinates Table 2). Samples from 21 of these populations were genotyped in the nuclear microsatellite study and from 24 in the chloroplast microsatellite study. The north–south extent of sampling was 350 km and included small, disjunct populations that are found in the northern part of the species range. The dune systems on which populations are located (Table 2) were identified by reference to geological maps at a scale of 1:50 000 compiled by the Geological Survey of Western Australia (GSWA available at http://geodocs.doir.wa.gov.au/document/document SearchCriteria.). Two populations were located on both Quindalup and Spearwood dunes (Table 2).
Table 2.
Genetic diversity parameters and chloroplast haplotypes for 26 populations of Eucalyptus gomphocephala included in this study
| Population | n | Latitude °S | Longtitude °E | AT | AR | S | HO | HE | F | Bottleneck | H |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SB (Q) | 28 | 30°25′50·0′′ | 115°7′17·0′′ | 59 | 7·63 | 1 | 0·67 | 0·76 | 0·12 | – | A(10) |
| NB (S) | 28 | 30°34′33·0′′ | 115°18′32·4′′ | 54 | 7·23 | 0 | 0·75 | 0·75 | 0·00 | – | A(9) |
| WI* (Q) | – | 30°52′19·8′′ | 115°18′32·4′′ | – | – | – | – | – | – | – | B(9) |
| BT (S) | 29 | 30°52′29·8′′ | 115°21 51·8′′ | 58 | 7·51 | 0 | 0·73 | 0·76 | 0·04 | – | C(10) |
| ML (S) | 30 | 31°07′22·1′′ | 115°33′13·3′′ | 57 | 7·49 | 0 | 0·78 | 0·76 | –0·04 | – | D(10) |
| SBR (Q) | 27 | 31°18′18·2′′ | 115°28′36·5′′ | 58 | 7·69 | 0 | 0·77 | 0·79 | 0·02 | – | B(3) D(7) |
| GD (Q) | 20 | 31°20′36·0′′ | 115°29′35·0′′′ | 56 | 7·85 | 0 | 0·72 | 0·76 | 0·04 | – | NUC |
| MRG (S) | 30 | 31°21′07·1′′ | 115°32′47·0′′ | 64 | 8·19 | 0 | 0·79 | 0·77 | –0·03 | – | C(8) D(2) |
| YA (S) | 30 | 31°30′58·0′′ | 115°40′11·2′′ | 62 | 8·05 | 0 | 0·75 | 0·78 | 0·04 | – | C(7) D(2) |
| NE (S) | 30 | 31°40′53·9′′ | 115°14′53·0′′ | 63 | 7·99 | 2 | 0·79 | 0·78 | –0·02 | – | D(3) C(7) |
| BP (Q and S) | 30 | 31°56′24·1′′ | 115°46′00·9′′ | 69 | 8·51 | 1 | 0·77 | 0·79 | 0·02 | – | F(1) D(10) |
| PG (S) | 27 | 31°59′51·2′′ | 115°46′16·9′′ | 59 | 7·54 | 0 | 0·67 | 0·76 | 0·12 | – | C(7) D(2) |
| WP (Q) | 24 | 32°07′44·7′′ | 115°45′48·3′′ | 55 | 7·37 | 0 | 0·74 | 0·75 | 0·01 | – | C(2) D(8) |
| RL (S) | 10 | 32°19′21·0′′ | 115°47′42·1′′ | – | – | – | – | – | – | – | D(10) |
| TS (S) | 10 | 32°31′18·7′′ | 115°44′43·6′′ | – | – | – | – | – | – | – | F(10) |
| TT (S) | 30 | 32°39′14·2′′ | 115°37′10·8′′ | 63 | 8·01 | 2 | 0·73 | 0·77 | 0·05 | – | F(10) |
| S10 (S) | 30 | 32°40′37·1′′ | 115°36′44·8′′ | 60 | 7·69 | 0 | 0·66 | 0·73 | 0·10 | – | F(10) |
| WH (S) | 30 | 32°41′22·0′′ | 115°37′09·0′′ | 60 | 7·45 | 0 | 0·67 | 0·72 | 0·06 | – | NUC |
| QU (S) | 30 | 32°41′49·6′′ | 115°38′32·6′′ | 56 | 7·29 | 0 | 0·66 | 0·70 | 0·04 | – | F(10) |
| DO (G) | 23 | 32°49′05·5′′ | 115°44′03·8′′ | 34 | 4·79 | 0 | 0·66 | 0·67 | 0·02 | P = 0·016 | F(10) |
| PR (S) | 30 | 32°51′02·5′′ | 115°39′53·9′′ | 65 | 8·29 | 4 | 0·72 | 0·75 | 0·01 | – | F(10) |
| OC* (S) | 10 | 32°57′49·0′′ | 115°43′34·3′′ | – | – | – | – | – | – | – | F(10) |
| LE (Q) | 29 | 33°13′58·7′′ | 115°41′53·8′′ | 54 | 7·01 | 0 | 0·68 | 0·69 | 0·01 | – | F(10) |
| DE* (Q) | 10 | 33°23′40·8′′ | 115°36′49·9′′ | – | – | – | – | – | – | – | F(10) |
| MB (S) | 30 | 33°28′34·3′′ | 115°33′31·4′′ | 61 | 7·87 | 0 | 0·72 | 0·73 | 0·00 | – | D(10) |
| LD (S) | 30 | 33°34′42·6′′ | 115°29′41·9′′ | 61 | 7·72 | 2 | 0·72 | 0·72 | –0·01 | – | E(10) |
| Grand mean | 59 | 7·64 | 0·57 | 0·72 | 0·75 | 0·03 |
n, number of individuals sampled per population; AT, total number of alleles observed across all loci; AR, allelic richness; S, total number of private alleles observed; HO, observed heterozygosity; HE, expected heterozygosity; F, fixation index; Bottleneck: ‘–’, no evidence of bottleneck under TPM, two-tailed P-values of significant excess of gene diversity under the TPM; H, the haplotypes found in a population; numbers in parentheses indicate the number of genotyped individuals in a population with a particular haplotype; NUC, nuclear data only.
*Population with chloroplast data only.
The dune systems on which populations are located are indicated by the following codes in parentheses: Q, Quindalup; S, Spearwood; and G, Guilderton formation.
Molecular analyses
DNA extraction
DNA was extracted at the Australian Genome Research Facility (AGRF Plant Genomics Centre, Adelaide, Australia) from freeze-dried leaf material using Nucleospin 96 Plant II with buffer set PL2/3 (Macherey-Nagel GmbH & Co., Düren, Germany), as per the manufacturer's instructions.
Chloroplast microsatellite genotyping
Primers designed for Eucalyptus species (Steane et al., 2005) were used to amplify chloroplast microsatellites and genotype 238 individuals from 24 populations. Initially, ten primer pairs (EMCRC59cp, EMCRC60cp, EMCRC62cp, EMCRC65cp, EMCRC67cp, EMCRC74cp, EMCRC84cp, EMCRC85cp, EMCRC86cp and EMCRC90cp) were screened against seven E. gomphocephala individuals, selected from across the species range, using the polymerase chain reaction (PCR) protocol described in Steane et al. (2005). From the initial ten primer pairs, all but two (EMCRC62cp and EMCRC86cp) produced strong and reproducible amplification profiles. Of the remaining eight primers, EMCRC74cp, EMCRC84cp and EMCRC90cp were monomorphic and, therefore, not included in this study.
Multiplex PCR was performed using EMCRC59cp, EMCRC60cp, EMCRC65cp, EMCRC67cp and EMCRC85cp primer pairs and the QIAGEN PCR Multiplex Kit (QIAGEN, Hilden, Germany) in 12·5 µL reactions as follows: 1× Multiplex PCR Master Mix, 2 µm each primer, dH2O and 20 ng of DNA. Forward primers were labelled with D2, D3, D4 WELLRED (Sigma Aldrich, Castle Hill, Australia) fluorescent dyes. The multiplex PCR followed the manufacturer's protocol and the following profile was used: an initial denaturation for 5 min at 95 °C; 28 cycles of 30 s at 95 °C, 1 min 30 s at an annealing temperature of 55 °C, and an extension of 30 s at 72 °C; with a final extension of 30 min at 60 °C. The PCR products were diluted 1:40 in deionized formamide and separated by electrophoresis using a CEQ8800 (Beckman-Coulter, Fullerton, CA, USA). Fragment analysis and sizing was performed using CEQ8800 Genetic Analysis System software version 9·0·25 (Beckman-Coulter). We genotyped 237 individuals from 24 populations (mean n = 10).
Chloroplast microsatellite allele sequencing
Each unique allele was sequenced to confirm that size variants occurred in the simple sequence repeat (SSR), and to rule out the presence of mutations in the flanking regions. Genomic DNA was amplified using the above protocols in a 50 µL total volume. The products were run on a 1 % agarose gel and visualized under UV light. The PCR products were purified prior to sequencing using a QIAquick PCR Purification Kit (QIAGEN), following the manufacturer's instructions. Sequencing reactions were performed with forward and reverse primers in 10 µL volumes, using the ABI Big-dye V3·1 Ready-Reaction Kit (Applied Biosystems, Foster City, CA, USA), following the manufacturer's instructions. Labelled sequence fragments were separated and detected using a 3730XL DNA Analyser (Applied Biosystems) at AGRF (Perth Node). Sequences were edited and aligned using Codon Code Aligner (CodonCode Corp., Centerville, MA, USA) and checked by eye for mutations in the microsatellite repeat and flanking regions.
Nuclear microsatellite genotyping
Nineteen nuclear microsatellite markers developed for eucalypt species by Steane et al (2001) (EMCRC 1a, EMCRC2, EMCRC3, EMCRC4, EMCRC5, EMCRC6), Brondani et al (1998) (EMBRA1, EMBRA2, EMBRA3, EMBRA4, EMBRA5, EMBRA6, EMBRA7, EMBRA9, EMBRA10 and EMBRA18) and Ottewell et al. (2005) (EL07, EL13 and EL16), were screened for product amplification and polymorphism. Seven loci (Table 1) produced reproducible, scorable and polymorphic products, and were used to obtain measures of nuclear genetic diversity and genetic differentiation in E. gomphocephala. Multiplex PCR was performed using the QIAGEN PCR Multiplex Kit (QIAGEN) in 12·5 µL reactions as follows: 1× Multiplex PCR Master Mix, 0·5× Q solution, 0·2 µm each primer, dH2O and 5–30 ng of DNA. Forward primers were labelled with D2, D3, D4 WellRED (Sigma Aldrich) fluorescent dyes. The multiplex PCR followed the manufacturer's protocol with some modifications, and the following profile was used: an initial denaturation for 15 min at 95 °C; 30 cycles of 30 s at 94 °C, 1 min 30 s at an annealing temperature of 55 °C, and an extension of 60 s at 72 °C; with a final extension of 10 min at 72 °C. The PCR products were diluted 1:40 in deionized formamide and separated by electrophoresis using a CEQ8800 (Beckman-Coulter). Fragment analysis and sizing was performed using CEQ8800 Genetic Analysis System software version 9·0·25 (Beckman-Coulter). We genotyped 595 individuals from 21 populations (mean n = 28).
Table 1.
Genetic parameters for each of the seven nuclear microsatellite loci used in this study of Eucalyptus gomphocephala
| Locus | Allele size range | AT | HO | HE | FIS | FST |
|---|---|---|---|---|---|---|
| EMBRA5 | 91–133 | 17 | 0·79 ± 0·02 | 0·80 ± 0·01 | 0·010 ± 0·019 | 0·051 |
| EL13 | 171–189 | 10 | 0·73 ± 0·02 | 0·73 ± 0·01 | 0·010 ± 0·021 | 0·068 |
| EMCRC2 | 158–198 | 15 | 0·69 ± 0·02 | 0·82 ± 0·01 | 0·159 ± 0·030 | 0·072 |
| EMBRA10 | 114–146 | 16 | 0·87 ± 0·01 | 0·83 ± 0·01 | –0·048 ± 0·018 | 0·060 |
| EMBRA6 | 125–187 | 27 | 0·76 ± 0·03 | 0·84 ± 0·02 | 0·095 ± 0·027 | 0·087 |
| EMCRC3 | 113–139 | 11 | 0·71 ± 0·03 | 0·70 ± 0·02 | –0·011 ± 0·031 | 0·080 |
| EMCRC6 | 157–181 | 8 | 0·52 ± 0·03 | 0·51 ± 0·03 | –0·016 ± 0·021 | 0·055 |
| Mean | 14·9 ± 2·4 | 0·72 ± 0·04 | 0·75 ± 0·04 | 0·028 ± 0·028 | 0·068 ± 0·01 |
AT, total number of observed alleles; HO, observed heterozygosity; HE, expected heterozygosity; FIS, inbreeding in individuals relative to their population; FST, inbreeding in populations relative to the total species.
Data analysis
Chloroplast microsatellites
A haplotype can be defined as a distinct combination of alleles for a set of chloroplast loci. The haplotype composition and number of haplotypes were calculated for each population. Alleles at microsatellite loci can be analysed as unordered where comparisons do not account for variation in allele size, and as ordered where the assumed number of mutational steps between alleles provides additional information. Mean within-population genetic diversity, species total genetic diversity and population differentiation were calculated, treating alleles as unordered (hS, hT and GST) and ordered (vS, vT and RST) following the method of Pons and Petit (1996) using PERMUT. The differentiation statistics GST and RST were compared: RST > GST means that more closely related haplotypes occur in the same population, indicating phylogeographic structure (Pons and Petit, 1996). The geographic distributions of chloroplast haplotypes were mapped and a haplotype tree showing the number of base pair differences between haplotypes was calculated in Network 4·6·0·0 (available at http://www.fluxus-engineering.com/sharepub.htm#a10) using the median-joining network algorithm (Bandelt et al., 1999). The relative influences of seed- and pollen-mediated gene flow can be estimated using inbreeding statistics calculated from chloroplast and nuclear markers in the following equation (eqn 5a of Ennos, 1994):
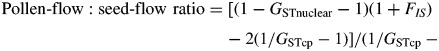 |
Nuclear microsatellites
Nuclear genetic diversity
The genetic diversity of 595 individuals of E. gomphocephala was analysed at seven microsatellite loci using GenAlEx version 6·41 (Peakall and Smouse, 2006) (available at: http://biology.anu.edu.au/GenAlEx/Welcome.html). The total number of alleles (A), number of private alleles (S), and observed and expected heterozygosity (HO and HE) were calculated and averaged for each locus and population. Alleles were deemed private if they occurred in one population and did not occur in any other population (Slatkin, 1985). Wright's fixation index (F) (Wright, 1931) was also calculated for each population. Deviation from Hardy–Weinberg equilibrium and linkage disequilibrium between loci in each pair of populations (with Bonferroni correction) was tested using GENEPOP 4·1 (Raymond and Rousset, 1995) (available at http://kimura.univ-montp2.fr/~rousset/Genepop.htm). Allelic richness (AR) was calculated for each population using FSTAT 2·9·3·2 (Goudet, 2002) (available at http://www2.unil.ch/popgen/softwares/fstat.htm). Estimates were adjusted for variation in sample size using the rarefaction method of El Mousadik and Petit (1996).
BOTTLENECK version 1·2·02 (Cornuet and Luikart, 1997) (available at http://www.ensam.inra.fr/URLB/bottleneck/bottleneck.html) was used to test for evidence of a genetic bottleneck (recent reduction in effective population size) in each population. Of the three available tests, the Wilcoxon sign-rank test was applied, because (1) the sign test has low statistical power; and (2) the standardized differences test requires data from ≥20 loci (Cornuet and Luikart, 1997). We used the two-phase mutation model (TPM), which is intermediate between the stepwise mutation model (SMM) and the infinite allele model (IAM), because few microsatellite loci follow the strict (one-step) SMM (Di Rienzo et al., 1994). We ran the TPM simulation as 90 % one-step mutations and 10 % multistep changes.
Geographic structuring of nuclear genetic variation
The significance of the relationship between Nei's (1972) genetic distance and geographical distance between populations was tested using the Mantel procedure (Mantel, 1967) with 1000 permutations of the correlation coefficient r in GenAlEx 6·41 (Peakall and Smouse, 2006). Analysis of molecular variance (AMOVA) was performed to partition total genetic variation into within- and among-population components using GenAlEx 6·41 (Peakall and Smouse, 2006). The significance of the AMOVA was assessed with 1000 permutations of the data. Significant differentiation between all populations was determined by obtaining population pairwise FST values using FSTAT version 2·9·3·2 (Goudet, 2002) with P-values determined by 4200 (20× number of comparisons) random permutations. Within FSTAT, the significance of each P-value was assessed using the log-likelihood statistic G, and Bonferroni correction was applied for multiple testing. Relationships between populations were visualized with an unweighted pair group method with arithmetic mean (UPGMA) dendrogram, based on Nei's (1972) genetic distance using GDA version 1·1 (Lewis and Zaykin, 2001).
Spatial genetic structure was further examined using the Bayesian clustering approach STRUCTURE 2·0 (Pritchard et al., 2000; Falush et al., 2003) (available at: http://pritch.bsd.uchicago.edu/software). STRUCTURE assigns individuals probabilistically to user-defined K populations so as to achieve Hardy–Weinberg and linkage disequilibrium within populations. STRUCTURE was run using the LOCPRIOR model as a sub-set of the admixture model, and assuming correlated allele frequencies. The LOCPRIOR model has the ability to use the sampling location information to assist clustering, and is recommended when the signal of structuring in data sets is too weak to be detected by the standard STRUCTURE models (as suggested by preliminary runs that showed poor assignment proportions in some populations) (Hubisz et al., 2009). STRUCTURE was run with 100 000 Markov chain Monte Carlo iterations after a burn-in period of 100 000 iterations and modelled with K = 1–21, with ten iterations of each K (Gilbert et al., 2012). Structure Harvester (Earl and vonHoldt, 2012) (available at http://users.soe.ucsc.edu/~dearl/software/struct_harvest/) was used to infer an optimal K based on the method of Evanno et al. (2005) and observing the slope of the plot of lnP(K). We also ran STRUCTURE excluding the DO (Doman Road) population using the same parameters, as the evidence for a genetic bottleneck in this population (see the Results) may bias the result.
Dune genetic diversity and structure
We examined whether nuclear genetic diversity and structure were significantly different in populations growing on younger Holocene Quindalup dune substrates compared with populations growing on older Pleistocene Spearwood dune substrates. This scenario is expected if the populations on Quindalup dunes have experienced founder effects due to recent colonization. We excluded the populations DO and BP, because they occurred on neither dune system, or occurred at a complex junction of both dune systems. Populations were allocated to one of two ‘dune system’ groups – Quindalup (Q) or Spearwood (S) (Table 2). First, mean AR (using a rarefaction procedure to account for differing sample sizes), HO, HE, fixation index (FIS) and FST were calculated for each group. The significance of the difference between groups was determined by 1000 random permutations (randomly allocating each population to each group) and P-values were taken as the proportion of randomized data sets where the difference between the groups was greater than the observed data set. Group analyses were performed using FSTAT version 2·9·3·2 (Goudet, 2002). We also assessed whether chloroplast diversity and structure differ among the two dune systems by calculating mean within-population genetic diversity, total genetic diversity and population differentiation treating alleles as unordered (hS, hT and GST) and ordered (vS, vT and RST) (Pons and Petit, 1996). Finally, we also compared chloroplast haplotype richness (HR) with ADZE, version 1·0 (Szpiech et al., 2008) using a rarefaction procedure to account for differing sample sizes. Differences in chloroplast diversity and structure values were tested using t-tests.
Palaeodistribution modelling
Modelling present species distributions
Presence data for E. gomphocephala were sourced from the Atlas of Living Australia (http://www.ala.org.au). Exclusion of dubious species records resulted in 251 viable records for E. gomphocephala. Modelling of the present range of the species was undertaken using the maximum entropy algorithm implemented in Maxent version 3·3·3a (Phillips et al., 2006). The default parameter settings were used (maximum number of background points 10 000; regularization multiplier 1; auto features; maximum iterations 500; convergence threshold 0·00001; and duplicate records deleted) as suggested by Phillips and Dudík (2008). Logistic output was chosen, ranging from zero at the lowest likelihood of presence to one, the strongest prediction for presence (Phillips, 2008). Modern climate variables were obtained from the Worldclim database (Hijmans et al., 2005) at the 30 arc-s resolution. In preliminary investigations, models using all 19 bioclimatic variables produced the most accurate predictions of the modern range, but suffered from overfitting (Beaumont et al., 2007) when projecting onto LGM climates. To reduce the likelihood of overfitting, the data were restricted based upon variables known to be important for the distribution of temperate plants in general (Katz, 2002; Svenning et al., 2011) and based upon their representative importance in the preliminary model. The constrained model consisted of two temperature variables (temperature seasonality and annual temperature range), and three precipitation variables (precipitation seasonality, precipitation of the warmest quarter and precipitation of the coldest quarter).
Palaeodistribution modelling
To model the palaeodistribution of E. gomphocephala, we projected models of climatic niche based on species’ current ranges onto two different global circulation models (GCMs) for the LGM: CCSM3·0 (Community Climate System Model 3·0; Collins et al., 2006) and MIROC 240 3·2 (Model for Interdisciplinary Research on Climate 3·2; Hasumi and Emori, 2004), both available from the Worldclim database in 2·5 arc-min resolution. The two GCMs varied considerably in the predicted temperature and rainfall depression for SWWA during the LGM.
Choosing thresholds
Although there is no consensus in the literature on the optimal approach, it is generally accepted that thresholds are required to convert the logistic values output by SDMs into presence and absence data. We applied the tenth percentile training presence (10 %), which underpredicts the known range so that the 10 % most extreme presence observations are predicted as being absent. Predictions based on this threshold can be considered to represent the ‘core of the species present range’, which excludes ephemeral populations, or populations in unusual microclimatic conditions within a cell (Morueta-Holme et al., 2010).
RESULTS
Chloroplast microsatellites
Chloroplast microsatellite diversity
In four of the five loci, sequencing revealed that fragment length differences were the result of variation in microsatellite regions. Primer EMCRC85 contained a multistate indel and variation at multiple, distinct, microsatellite regions, and was thus excluded from analyses. The remaining four primers, EMCRC59cp, EMCRC60cp, EMCRC65cp and EMCRC67cp, produced three, two, three and three alleles, respectively, giving a total of 11 different alleles. Several of the alleles differed by 1 bp; however, with the inclusion of size standards, there was generally no difficulty in differentiating between these alleles. Wherever allele scores were ambiguous, samples were re-amplified (between 1 and 5 % of PCRs). If allele scoring remained ambiguous after re-amplification (<1 % of reactions), the samples were excluded from the study. The 11 different alleles produced a total of six haplotypes (Supplementary Data Table S1). Haplotype frequencies varied considerably, with the most common haplotype, F, being present in 39 % of individuals, while haplotype E accounted for only 4·2 % of individuals.
Haplotype relationships
The statistical parsimony tree showed that most haplotypes were connected to only one other haplotype, with the exception of haplotypes D and A which were internal to the network and each connected to three other haplotypes (Fig. 1B). In general, there was only a 1 or 2 bp difference between haplotypes; however, haplotype C was separated by 5 bp from the next closest haplotype.
Geographic distribution of haplotypes
The six haplotypes found in E. gomphocephala showed geographic structure and generally occupied specific regions throughout the species range (Fig. 1A). Overall, one private haplotype was found, in the most southerly population of Ludlow (LD, haplotype E). Haplotype A was unique to the two disjunct, most northerly populations (SB and NB), and fixed in all sampled individuals. Haplotype B was found in the northern populations WI and SBR. In populations at the centre and mid north of the species range, two haplotypes were identified in each population (combinations of haplotypes B, C, D and F), whereas all populations located on and immediately south, north and east of the Yalgorup Plain (populations TS, TT, S10, QU, PR, OC, LE, DO and DE; n = 100) were fixed for haplotype F. Haplotype D was the most geographically widespread of the haplotypes (Fig. 1A). The most divergent haplotype (haplotype C) was found in several populations at the centre and mid north of the species range (populations BT, MRG, YA, NE, BP and WP), and commonly in association with haplotype D (Fig. 1A). Overall, there was low within-population diversity (hS = 0·11, vS = 0·20), high total diversity (hT = 0·77, vT = 0·77) and strong differentiation among populations (GST = 0·86, RST = 0·75). A test for phylogeographic structure showed no significant difference between GST and RST (P = 0·05). This indicates that, generally, across the range of E. gomphocephala, ‘related’ (via stepwise mutation) haplotypes are no more likely than ‘unrelated’ haplotypes to be found together at a locality. Using the GST and FIS values derived from the chloroplast and nuclear microsatellite data, we calculated that the ratio of pollen-mediated to seed-mediated gene flow was 200:1. This suggests that gene flow in E. gomphocephala is affected predominantly by pollen.
Nuclear microsatellites
Nuclear microsatellite diversity within populations
All seven loci were highly variable, with the number of alleles detected per locus ranging from eight to 27 (mean 15; Table 1). Gene diversities were high (HE ranging from 0·51 to 0·84; overall mean HE 0·75; see Table 1). FST values were positive for all loci (0·05–0·09) and there was strong variation in FIS for all loci (Table 1). After applying the Bonferroni correction, tests of Hardy–Weinberg deviation and linkage disequilibrium were not significant. However, EMBRA5 showed significant heterozygote deficit in SB, and EMCRC2 showed significant heterozygote deficit in GD, PG, TT, S10, PR, LE and LD.
With the exception of the eastern outlier DO population, within-population genetic diversity was very similar across all populations assessed (Table 2). The total number of alleles (mean AT 59), AR (mean AR 7·58) and HE (mean HE 0·75) per population were lowest at DO (Table 2). In nearly all populations, HO was lower than HE (Table 2). Hence, small positive F-values (Wrights fixation index) were found in most populations. Private alleles were found at five out of 21 populations (SB, NE, BP, TT and PR), and there was no apparent geographic pattern in their distribution. The DO population was the only population to show significant evidence of genetic bottlenecks.
Genetic structure
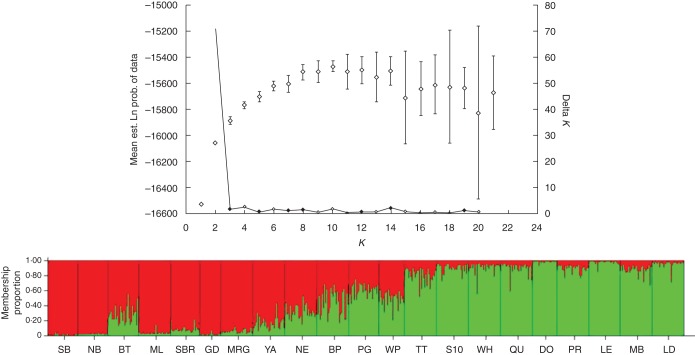
Pairwise FST values varied from 0·006 (between PG and BP) to 0·142 (between DO and SB), and most pairwise comparisons were significantly greater than 0 (Supplementary Data Table S2). An AMOVA of hierarchical genetic diversity revealed that of the total genetic variation, a significant (P < 0·01) amount (5 %) was partitioned among populations (Supplementary Data Table S3). Overall, there was significant isolation by distance, with a positive relationship found between genetic differentiation and geographical distance (r2 = 0·362, P = 0·001). The pattern of branching on the UPGMA very closely reflected the geographic location of populations (with the exception of the positions of DO and GD; Fig. 1C). The STRUCTURE analysis identified K = 2 as the optimal number of groups (Fig. 2). The same result was achieved whether the DO population was included or excluded from analyses. Individuals from the north (SB to MRG, with the exception of BT) and south (TT to LD) of the species range were strongly allocated to their respective genetic groups, while substantial admixture was found for geographically intermediate populations (YA to WP).
Fig. 2.
The estimated membership proportions of sampled Eucalyptus gomphocephala individuals at each site in the two genetic clusters inferred with STRUCTURE (Pritchard et al., 2000) based on nuclear SSR data. Top: Structure Harvester – Delta K peaks at K = 2. Bottom: bar chart of relative proportional membership of each individual (single bars) within each sampling location/population (solid lines) to one of two genetic clusters (red, green).
Dune diversity and structure
Nuclear genetic diversity and differentiation parameters were not significantly different among the two dune systems (Table 3). There were also no significant differences in chloroplast genetic diversity and structure between the two dune systems (Table 3).
Table 3.
Comparison of Quindalup and Spearwood dune systems of Eucalyptus gomphocephala for the mean values of nuclear and chloroplast genetic diversity and differentiation parameters
| Quindalup | Spearwood | P-value | |
|---|---|---|---|
| AR | 7·51 | 7·74 | 0·58(NS) |
| HO | 0·72 | 0·73 | 0·70(NS) |
| HE | 0·77 | 0·76 | 0·87(NS) |
| FIS | 0·06 | 0·05 | 0·48(NS) |
| FST | 0·07 | 0·05 | 0·15(NS) |
| hS/vS | 0·21/0·05 | 0·12/0·03 | 0·49(NS)/0·51(NS) |
| hT/vT | 0·90/0·95 | 0·80/0·81 | 0·22(NS)/0·66(NS) |
| GST/RST | 0·77/0·95 | 0·84/0·96 | 0·65(NS)/0·77(NS) |
| HR | 5 | 5 | – |
Nuclear parameters: AR, allelic richness; HO, observed heterozygosity; HE, expected heterozygosity; FIS, fixation index. Chloroplast parameters: ordered and unordered alleles, respectively: hS, vS, within-population genetic diversity; hT, vT, total genetic diversity; population differentiation, GST, RST.
HR, haplotype richness.
–, P-value not calculable.
Palaeodistribution modelling
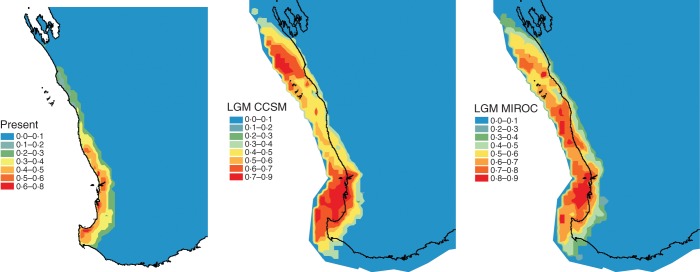
The modelled range of E. gomphocephala for present climates encompassed the known extent of occurrence of E. gomphocephala, but overpredicted 172 % of the estimated extent of E. gomphocephala. The 10 % threshold made predictions outside the known distribution in some areas, extending the potential distribution north along the coast to the southern reaches of Shark Bay, and also expanding the distribution throughout the lower south-west. Although the predicted LGM distributions differed slightly between the GCM models, both models suggest that E. gomphocephala had a widespread distribution during the LGM (Fig. 3). Both model scenarios strongly predicted that E. gomphocephala extended west of the present coastline, and as far north as present Shark Bay. However, CCSM predicted poor suitability from PG to north of the current distribution of E. gomphocephala.
Fig. 3.
Maximum entropy correlative distribution models of Eucalyptus gomphocephala under present climate conditions (present), and two models of the last glacial maximum (LGM CCSM and LGM MIROC). Values represent the propotional likelihood of occurrence (0·0–1·0). The present model represents a 172 % overestimation of known distributions.
DISCUSSION
Molecular evidence for a widespread geographic distribution during the LGM
Coastal ecosystems will be strongly impacted by anthropogenic-induced climate change, with sea level rises likely to cause the salinization of wetlands, inundation of low-lying areas and fragmentation of landscapes. Nevertheless, there are surprisingly few examples of phylogeographic studies of coastal plant species. In our study, the general nature and extent of the geographic distribution of microsatellite variation for chloroplast and nuclear DNA suggest that the SCP endemic tree E. gomphocephala has persisted at multiple locations across a widespread distribution during the climatic extremes of the LGM. The significant genetic structure and equivalent levels of within-population genetic variation for nuclear microsatellites, and the high total diversity and generally high level of differentiation among populations at these chloroplast markers also do not support a scenario of range-wide expansion from one or a few refugia after the LGM. Palaeodistribution modelling corroborates the genetic evidence for a widespread geographic distribution of the species during the LGM, with suitable climate predicted hundreds of kilometres north and south of its current distribution. The palaeodistribution of tuart would have been ultimately limited within this climatic envelope by substrate (sandy coastal plain) and sea level influenced by topography.
The slow mutation rate of chloroplast microsatellite regions (between 3·2 × 10−5 and 7·9 × 10−5) (Provan et al., 1999, 2001) suggests that differences observed between haplotypes are unlikely to have arisen since the last glacial period. However, shallow divergences between haplotypes (generally 1–4 bp) suggest that past sea transgressions did not produce deep vicariance signatures. Nuclear and chloroplast patterns generally coincided, although there is a strong disparity between levels of chloroplast (GST = 0·86) and nuclear differentiation (FST = 0·05) (ratio of pollen to seed gene flow approx. 200). This is consistent with studies of other eucalypt species (e.g. Wheeler and Byrne, 2006) and can be explained by the different dispersal distances of pollen and seeds, and modes of inheritance and mutation rates of the two genomes.
Our data are consistent with general conclusions drawn from studies of chloroplast and/or nuclear variation in eucalypt species in south-western (Byrne and Macdonald, 2000; Byrne and Hines, 2004; Wheeler and Byrne, 2006; Byrne and Hopper, 2008) and south-eastern Australia (Steane et al., 1998; Jackson et al., 1999; Freeman et al., 2001; Nevill et al., 2010; Bloomfield et al., 2011), which demonstrate that geographic stasis, and/or complex patterns of short-range expansion and contraction were typical responses of taxa to climatic conditions during the LGM in Australia. This is in strong contrast to the general response (but not always) of arctic and temperate northern hemisphere tree species impacted by glaciations, where relatively simple, large-scale, northwards range expansion movements following restriction to one or a few southern refugia during the LGM are apparent (Hewitt, 2000, 2004; Beheregaray, 2008).
While persistence over a broad geographic range is suggested by the data, persistence does not appear to have been uniform or continuous, and some areas may have been more impacted by past climate or geomorphologic changes than others. For example, we found the fixation of a single common haplotype (F) in all sampled individuals over a 100 km range from the Yalgorup region (Fig. 1C). This is in contrast to the patchy distribution of distinct haplotypes found across most of the species range. Chloroplast haplotype fixation over a number of geographically proximate populations is indicative of either a genetic bottleneck arising from relatively recent (re-)colonization of an area, or in situ persistence and a severe reduction in population size (Hewitt, 1996). The stratigraphical sequence suggests relatively dramatic coastal change on the Yalgorup Plain associated with rising and falling Holocene sea levels (Semeniuk, 1995) and where many of the current sites occupied by E. gomphocephala are at <10 m above current sea levels. Similar cpDNA patterns and inferred bottleneck events have been identified in other eucalypt studies (e.g. Byrne and Macdonald, 2000; Freeman et al., 2001; Nevill et al., 2010), and in many northern hemisphere species No evidence of genetic bottlenecks at nuclear markers was found in the Yalgorup Plain populations (Table 2); however, empirical tests of bottleneck detection at nuclear markers have been equivocal (Williamson-Natesan, 2005; Marshall et al., 2009; Hundertmark and Van Daele, 2010). Also, a nuclear DNA bottleneck is not necessarily expected, not only for technical reasons and because it is dependent on many demographic factors, but cpDNA diversity could also be more limited because of recurrent patterns of contraction throughout the Quaternary, rather than a single LGM event.
The genetic data suggest that populations located at the centre of the species range (from WP in the south to YA in the north) are composed of admixed genotypes from distinct gene pools. In these populations, haplotype C (only found in the north and centre of the species range) is commonly associated with haplotype D (predominantly found in the south and centre of the species range). The haplotype network indicates that these haplotypes were not directly derived from each other, or closely related. The co-occurrence of genetically distinct haplotypes is good evidence for convergence of two independently evolved lineages at a suture zone (Remington, 1968; Hewitt, 1996). Alternatively, haplotype C may have been acquired from another species through hybridization and introgression. Interspecific hybridization is common in eucalypts, with numerous studies suggesting that reticulate evolution is likely to have played an important role in the evolution of the genus (Steane et al., 1998; Jackson et al., 1999; McKinnon et al., 1999, 2001, 2004; Pollock et al., 2013). Admixture in this region is further supported by the nuclear data, where the STRUCTURE analysis shows distinct northern and southern groups, with a cline of relative membership at the centre of the species range. Palaeodistribution modelling supports (but not conclusively) that some of the area where admixed genotypes occur was less suitable for E. gomphocephala during the LGM, requiring (re-)colonization since the LGM (Fig. 3). While recognizing that other explanations are possible, the palaeodistribution modelling does provide independent support for the hypothesis of a historical separation between northern and southern distributions.
Comparison of genetic patterns on Quindalup and Spearwood dune systems
We found similar levels of genetic diversity and differentiation at both chloroplast and nuclear microsatellites for populations on the Holocene formed Quindalup and older Quaternary Spearwood dune systems (Table 3). Population genetic diversity at colonized locations relative to sites where there has been longer persistence depends on a number of factors (reviewed in Pannell and Dorken, 2006) including: (1) the number of founders that arrive at a site prior to the original colonizers reaching reproductive maturity and fully occupying the habitat (Austerlitz et al., 2000); (2) colonization of an area from multiple, genetically distinct refugia (Zaneto and Kremer, 1995; Hewitt, 1996); and (3) hybridization with co-colonizing species. First, E. gomphocephala does not produce appreciable seed crops until approx. 10 years of age (Ruthrof et al., 2002), and there may have been multiple cycles of regeneration, accompanied by invasion, by increasing numbers of founders before sites were fully occupied by the species. Secondly, colonization from multiple, genetically distinct refugia appears unlikely given that most populations located on Quindalup dunes contained the same haplotypes as populations located on adjacent Spearwood dunes. Thirdly, across its range, E. gomphocephala is sympatric with nine other eucalypt species, and hybridization has been recorded between E. gomphocephala and three of these (E. decipiens, E. cornuta and E. rudis) (Coates et al., 2002). Therefore, the opportunity for hybridization with co-colonizing species is likely to have been present. A final explanation is extensive pollen-mediated gene flow both during and after colonization. Pollen-mediated gene flow in eucalypts is facilitated by insects, birds and small mammals, and, although its distribution is typically leptokurtic, extensive long-distance pollen flow events over many kilometres have been recorded in related species (Skabo et al., 1998; Mimura et al., 2009). These long-distance dispersal events would lead to the homogenization of the nuclear gene pool, which is reflected in the low overall and pairwise FST results.
Factors that have contributed to the persistence of E. gomphocephala
A maritime climate and/or short-range habitat tracking are likely to have been important factors in the wide persistence of tuart during the LGM. The Leeuwin Current along the western Australian coast is a major influence on the climate of coastal regions, and evidence of its continued presence, even during glacial periods (Spooner et al., 2010), supports conclusions of a relatively subdued local climate impact during the LGM (e.g. Wyroll et al., 2000; Rojas et al., 2009). Similarly benign palaeo-environmental inferences are drawn for other Mediterranean-type climatic regions, where species diversity is also high. For example, palynological and phylogeographic studies from the south-west Cape of southern Africa show surprisingly little change in vegetation communities during the LGM, despite climatic and environmental dynamism elsewhere (Meadows and Sugden, 1993: Linder et al., 2003). Secondly, the lowering of sea levels during the LGM led to an expansion of the SCP 30 km onto the exposed continental shelf and may have provided the opportunity, at least, for E. gomphocephala to track a suitable maritime climate. Picket and Newsome (1997) found strong evidence that vegetation zones on the SCP shifted westwards during the LGM, and suggested that a combination of soil type, depth to groundwater, topography, reduced continentality and the buffering effect of the sea on precipitation and temperature may have provided a suitable habitat for eucalypt woodland species on the expanding coastal plain. This is supported by our palaeodistribution modelling, with both scenarios showing strong evidence for occurrence of E. gomphocephala westward of the current coastline during the LGM.
Conclusions
This study provides a surprisingly rare example of the response of a coastal species to historical climate change and highlights the importance of other data (in this case geomorphology and palaeodistribution modelling) in the interpretation of phylogeographic patterns. As this is the first phylogeographic study of an endemic SCP plant species, studies of chloroplast variation in co-distributed species are required in order to identify if congruent patterns of the geographical distribution of genetic variation exist. A common response to Pleistocene climate fluctuations ultimately informs on the resilience of these diverse plant communities to future climate change. Biodiversity conservation has been traditionally based on maps of species distributions (Margules and Pressey, 2000), but there is increasing understanding of the dynamic nature of biodiversity, and the need to preserve ecological and evolutionary processes (Klein et al., 2009). Short-range habitat tracking onto the exposed continental shelf during the LGM demonstrates a capacity in E. gomphocephala to respond to, and persist through, changing climates. However, habitat fragmentation associated with urbanization may prove to be the key factor in increasing extinction risk by reducing the capacity to disperse. Ultimately, an understanding of the capacity for resilience that is harboured within population genetic variation may be critical for conservation in landscapes within which the potential for migration is negatively impacted.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Margaret Byrne, Kingsley Dixon and Vic Seminuik for their advice and discussions. Samples were collected under licence # SW013399 from the WA Department of Environment and Conservation. This research was supported in part by an Australian Research Council Linkage grant (LP0669757) to S.K.
LITERATURE CITED
- Arafeh R, Kadreit JW. Long-distance seed dispersal, clone longevity and lack of phylogeographical structure in the European distributional range of the coastal Calystegia soldanella (L.) R. Br (Convolvulaceae) Journal of Biogeography. 2006;33:1461–1469. [Google Scholar]
- Austerlitz F, Mariette S, Machon N, Gouyon PH, Godelle B. Effects of colonization processes on genetic diversity: differences between annual plants and tree species. Genetics. 2000;154:1309–1321. doi: 10.1093/genetics/154.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Beaumont LJ, Pitman A, Poulsen M, Hughes L. Where will species go? Incorporating new advances in climate modelling into projections of species distributions. Global Change Biology. 2007;13:1368–1385. [Google Scholar]
- Beheregaray LB. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Molecular Ecology. 2008;17:3754–3774. doi: 10.1111/j.1365-294X.2008.03857.x. [DOI] [PubMed] [Google Scholar]
- Bloomfield JA, Nevill P, Potts BM, Vaillancourt RE, Steane DA. Molecular genetic variation in a widespread forest tree species Eucalyptus obliqua (Myrtaceae) on the island of Tasmania. Australian Journal of Botany. 2011;59:226–237. [Google Scholar]
- Bradbury D, Krauss SL. Limited impact of fragmentation and disturbance on the mating system of tuart (Eucalyptus gomphocephala, Myrtaceae): implications for seed-source quality in ecological restoration. Australian Journal of Botany. 2013;61:148–160. [Google Scholar]
- Brondani R, Brondani C, Tarchini R, Grattapaglia D. Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla. Theoretical and Applied Genetics. 1998;97:816–827. [Google Scholar]
- Byrne M, Hines B. Phylogeographical analysis of cpDNA variation in Eucalyptus loxophleba (Myrtaceae) Australian Journal of Botany. 2004;52:459–470. [Google Scholar]
- Byrne M, Hopper S. Granite outcrops as ancient islands in old landscapes: evidence from the phylogeography and population genetics of Eucalyptus caesia (Myrtaceae) in Western Australia. Biological Journal of the Linnean Society. 2008;93:177–188. [Google Scholar]
- Byrne M, Macdonald B. Phylogeography and conservation of three oil mallee taxa, Eucalyptus kochii ssp kochii, ssp plenissima and E. horistes. Australian Journal of Botany. 2000;48:305–312. [Google Scholar]
- Byrne M, Moran G, Tibbits W. Restriction map and maternal inheritance of chloroplast DNA in Eucalyptus nitens. Journal of Heredity. 1993;84:218–220. [Google Scholar]
- Byrne M, MacDonald B, Coates D. Phylogeographical patterns in chloroplast DNA variation within the Acacia acuminata (Leguminosae: Mimosoideae) complex in Western Australia. Journal of Evolutionary Biology. 2002;15:576–587. [Google Scholar]
- Byrne M, Macdonald B, Brand J. Phylogeography and divergence in the chloroplast genome of Western Australian Sandalwood (Santalum spicatum) Heredity. 2003;91:389–395. doi: 10.1038/sj.hdy.6800346. [DOI] [PubMed] [Google Scholar]
- Byrne M, Elliot CP, Yates CJ, Coates DJ. Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia. Conservation Genetics. 2008;9:97–105. [Google Scholar]
- Churchill DM. Late Quaternary eustatic changes in the Swan River district. Journal of the Royal Society of Western Australia. 1959;42:53–55. [Google Scholar]
- Coates D, Keighery G, Broadhurst L. Genetic and morphological variation, and the mating system in Tuart. In: Keighery B, Longman V, editors. Tuart (Eucalyptus gomphocephala) and Tuart communities. Nedlands, Western Australia: Perth Branch Wildlife Society of Western Australia (Inc.); 2002. pp. 89–107. [Google Scholar]
- Collins WD, Bitz CM, Blackmon ML, et al. The Community Climate System Model version 3 (CCSM3) Journal of Climate. 2006;19:2122–2143. [Google Scholar]
- Cornuet J, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1997;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer KW. Distance of seed dispersal in eucalypts estimated from seed weights. Australian Forest Research. 1977;7:225–228. [Google Scholar]
- Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M. Mutational processes of simple sequence repeat loci in human populations. Proceedings of the National Academy of Sciences, USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetic Resources. 2012;4:359–361. [Google Scholar]
- El Mousadik A, Petit R. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa L. Skeels) endemic to Morocco. Theoretical and Applied Genetics. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Ennos RA. Estimating the relative rates of pollen and seed migration among plant populations. Heredity. 1994;72:250–259. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard K. Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JS, Jackson HD, Steane DA, et al. Chloroplast DNA phylogeography of Eucalyptus globulus. Australian Journal of Botany. 2001;49:585–596. [Google Scholar]
- Gilbert KJ, Andrew RL, Bock DG, et al. Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program STRUCTURE. Molecular Ecology. 2012;21:4925–4930. doi: 10.1111/j.1365-294X.2012.05754.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT version 2·9·3·2. Lausanne, Switzerland: Lausanne University; 2002. Department of Ecology and Evolution. [Google Scholar]
- Hasumi H, Emori S. K-1 coupled GCM (MIROC) description. 2004 Center for Climate System Research, University of Tokyo. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hopkins AJM, Coker J, Beeston GR, Bowan P, Harvey JM. Conservation status of vegetation types throughout Western Australia: Final Report. 1996 Australian Nature Conservation Agency National Reserves System Co-operative Research Program. [Google Scholar]
- Hopper SD, Gioia P. The Southwest Australian Floristic Region: evolution and conservation of a global hotspot of biodiversity. Annual Review of Ecology and Systematics. 2004;35:623–650. [Google Scholar]
- Hubbard NN. In search of regional palaeoclimates – Australia, 18,000 Yr BP. Palaeogeography, Palaeoclimatology, Palaeoecology. 1995;116:167–188. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark KJ, Van Daele LJ. Founder effect and bottleneck signatures in an introduced, insular population of elk. Conservation Genetics. 2010;11:139–147. [Google Scholar]
- Jackson HD, Steane DA, Potts BM, Vaillancourt RE. Chloroplast DNA evidence for reticulate evolution in Eucalyptus (Myrtaceae) Molecular Ecology. 1999;8:739–751. [Google Scholar]
- Jackson S, Overpeck J. Responses of plant populations and communities to environmental changes of the late Quaternary. Palaeobiology. 2000;26:194–220. [Google Scholar]
- Kadreit JW, Arafeh R, Somogyi G, Westbridge E. Terrestrial growth and marine dispersal? Comparative phylogeography of five coastal plant species at a European scale. Taxon. 2005;54:861–876. [Google Scholar]
- Katz RW. Techniques for estimating uncertainty in climate change scenarios and impact studies. Climate Research. 2002;20:167–185. [Google Scholar]
- Keighery B, Keighery G, Shepherd D. The distribution and conservation of Tuart and the community with which it lives. In: Keighery B, Longman V, editors. Tuart (Eucalyptus gomphocephala) and tuart communities. Nedland, Western Australia: Perth Branch Wildflower Society of Western Australia (Inc.); 2002. pp. 6–86. [Google Scholar]
- Klein C, Wilson K, Watts M, et al. Incorporating ecological and evolutionary processes into continental-scale conservation planning. Evolutionary Applications. 2009;19:206–217. doi: 10.1890/07-1684.1. [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D. Genetic Data Analysis Software, version 1·1. 2001 [Google Scholar]
- Linder HP, Eldenas P, Briggs BG. Contrasting patterns of radiation in African and Australian Restionaceae. Evolution. 2003;57:2688–2702. doi: 10.1111/j.0014-3820.2003.tb01513.x. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405:243–253. doi: 10.1038/35012251. [DOI] [PubMed] [Google Scholar]
- Marshall JC, Kingsbury BA, Minchella DJ. Microsatellite variation, population structure, and bottlenecks in the threatened copperbelly water snake. Conservation Genetics. 2009;10:465–476. [Google Scholar]
- McKinnon GE, Steane DA, Potts BM, Vaillancourt RE. Incongruence between chloroplast and species phylogenies in Eucalyptus subgenus Monocalyptus (Myrtaceae) American Journal of Botany. 1999;86:1038–1046. [PubMed] [Google Scholar]
- McKinnon GE, Vaillancourt RE, Jackson HD, Potts BM. Chloroplast sharing in the Tasmanian eucalypts. Evolution. 2001;55:703–711. doi: 10.1554/0014-3820(2001)055[0703:csitte]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- McKinnon GE, Jordan GJ, Vaillancourt RE, Steane DA, Potts BM. Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359:275–284. doi: 10.1098/rstb.2003.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows ME, Sugden JM. The late quaternary palaeoecology of a floristic kingdom: the southwestern Cape South Africa. Palaeogeography, Palaeoclimatology, Palaeoecology. 1993;101:271–281. [Google Scholar]
- Mercer JH. Antarctic ice and Sangamon sea level. International Association of Scientific Hydrology. 1968;79:217–225. [Google Scholar]
- Mimura M, Barbour RC, Potts BM, Vaillancourt RE, Watanabe KN. Comparison of contemporary mating patterns in continuous and fragmented Eucalyptus globulus native forests. Molecular Ecology. 2009;18:4180–4192. doi: 10.1111/j.1365-294X.2009.04350.x. [DOI] [PubMed] [Google Scholar]
- Mittermeier RA, Robles Gil P, Hoffman M, et al. Hotspots revisited. CEMEX: Garza Garcia N.L. Mexico; 2004. [Google Scholar]
- Molins A, Mayol M, Rosselló JA. Phylogeographical structure in the coastal species Senecio rodriguezii (Asteraceae), a narrowly distributed endemic Mediterranean plant. Journal of Biogeography. 2009;36:1372–1383. [Google Scholar]
- Morueta-Holme N, Fløjgaard C, Svenning JC. Climate change risks and conservation implications for a threatened small-range mammal species. PloS One. 2010;5 doi: 10.1371/journal.pone.0010360. pe10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. American Naturalist. 1972;106:283–292. [Google Scholar]
- Nevill P, Bossinger G, Ades P. Phylogeography of the world's tallest angiosperm Eucalyptus regnans: evidence for multiple isolated quaternary refugia. Journal of Biogeography. 2010;37:179–192. [Google Scholar]
- Oberle B, Schaal BA. Responses to historical climate change identify contemporary threats to diversity in Dodecatheon. Proceedings of the National Academy of Sciences, USA. 2011;108:5655–5660. doi: 10.1073/pnas.1012302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottewell KM, Donnellan SC, Moran GF, Paton DC. Multiplexed microsatellite markers for the genetic analysis of Eucalyptus leucoxylon (Myrtaceae) and their utility for ecological and breeding studies in other Eucalyptus species. Journal of Heredity. 2005;96:445–451. doi: 10.1093/jhered/esi057. [DOI] [PubMed] [Google Scholar]
- Pannell JR, Dorken ME. Colonisations as a common demoninator in plant metapopulations and range expansions: effects on genetic diversity and sexual systems. Landscape Ecology. 2006;21:837–848. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modelling of species geographic distributions. Ecological Modelling. 2006;190:231–259. [Google Scholar]
- Phillips SJ., Dudík M. Modelling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. [Google Scholar]
- Phillips SJ. Transferability, sample selection bias and background data in presence-only modelling: a response to Peterson et al. (2007) Ecography. 2008;31:272–278. [Google Scholar]
- Pickett EJ, Newsome JC. Eucalyptus (Myrtaceae) pollen and its potential role in investigations of Holocene environments in southwestern Australia. Review of Palaeobotany and Palynology. 1997;98:187–205. [Google Scholar]
- Playford PE, Cockbain AE, Low GH. Geology of the Perth Basin Western Australia. Geological Society of Western Australia, Bulletin 124. 1976 [Google Scholar]
- Pollock LJ, Bayly MJ, Nevill PG, Vesk PA. Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia. Journal of Biogeography. 2013;40:155–167. [Google Scholar]
- Pons O, Petit R. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Soranzo N, Wilson NJ, Goldstein DB, Powell W. A low mutation rate for chloroplast microsatellites. Genetics. 1999;153:943–947. doi: 10.1093/genetics/153.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Powell W, Hollingsworth PM. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends in Ecology and Evolution. 2001;16:142–147. doi: 10.1016/s0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Raymo ME, Mitrovica JX, O'Leary MJ, DeConto RM, Hearty PJ. Departures from eustasy in Pliocene sea-level records. Nature Geoscience. 2011;4:328–332. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1·2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Remington CL. Suture-zones of hybrid interaction between recently joined biotas. In: Dobzhansky T, Hecht MK, Steere WC, editors. Evolutionary biology. New York: Plenum; 1968. pp. 321–428. [Google Scholar]
- Rojas M, Moreno P, Kageyama M, et al. The Southern Westerlies during the Last Glacial Maximum in PMIP2 simulations. Climate Dynamics. 2009;32 p525e548. [Google Scholar]
- Ruthrof KX, Yates CJ, Loneragan WA. The biology of Tuart. In: Keighery BJ, Longman VM, editors. Tuart (Eucalyptus gomphocephala) and Tuart communities. Nedlands, Western Australia: Perth Branch Wildflower Society of Western Australia (Inc.); 2002. pp. 108–121. [Google Scholar]
- Seddon G. Sense of place: a response to an environment. The Swan coastal plain, Western Australia. Bloomings Books: Melbourne; 2004. [Google Scholar]
- Semeniuk V. New Pleistocene and Holocene stratigraphic units in the Yalgorup Plain area, southern Swan Coastal Plain. Royal Society of Western Australia. 1995;78:67–79. [Google Scholar]
- Semeniuk V. Pleistocene coastal palaeogeography in Southwestern Australia – carbonate and quartz sand sedimentation in cuspate forelands, barriers and ribbon shoreline deposits. Journal of Coastal Research. 1997;13:468–489. [Google Scholar]
- Skabo S, Vaillancourt R, Potts B. Fine-scale genetic structure of Eucalyptus globulus ssp. globulus forest revealed by RAPDS. Australian Journal of Botany. 1998;46:583–594. [Google Scholar]
- Slatkin M. Rare alleles as indicators of gene flow. Evolution. 1985;39:53–65. doi: 10.1111/j.1558-5646.1985.tb04079.x. [DOI] [PubMed] [Google Scholar]
- Speck NH. Atmospheric pollen in the city of Perth and environs. Journal of the Royal Society of Western Australia. 1953;37:119–125. [Google Scholar]
- Spooner MI, De Deckker P, Barrows TT, Fifield LK. The behaviour of the Leeuwin Current offshore NW Australia during the last five glacial–interglacial cycles. Global and Planetary Change. 2010;75:119–132. [Google Scholar]
- Steane DA, Byrne M, Vaillancourt RE, Potts BM. Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae) Australian Systematic Botany. 1998;11:25–40. [Google Scholar]
- Steane D, Vaillancourt R, Russell J, Powell W, Marshall D, Potts B. Development and characterisation of microsatellite loci in Eucalyptus globulus (Myrtaceae) Silvae Genetica. 2001;50:89–91. [Google Scholar]
- Steane DA, Jones RC, Vaillancourt RE. A set of chloroplast microsatellite primers for Eucalyptus (Myrtaceae) Molecular Ecology Notes. 2005;5:538–541. [Google Scholar]
- Svenning JC., Fløjgaard C, Marske KA, Nógues-Bravo D, Normand S. Applications of species distribution modelling to palaeobiology. Quaternary Science Reviews. 2011;30:2930–2947. [Google Scholar]
- Szpiech ZA, Jakobsson M, Rossenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24:2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuart Response Group. An atlas of Tuart woodlands on the Swan Coastal Plain in Western Australia. Perth, Australia: Government of Western Australia; 2003. [Google Scholar]
- Wheeler MA, Byrne M. Congruence between phylogeographic patterns in cpDNA variation in Eucalyptus marginata (Myrtaceae) and geomorphology of the Darling Plateau, south-west of Western Australia. Australian Journal of Botany. 2006;54:17–26. [Google Scholar]
- Williamson-Natesan EG. Comparison of methods for detecting bottlenecks from microsatellite loci. Conservation Genetics. 2005;6:551–562. [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrwoll KH, Dong B, Valdes P. On the position of the southern hemisphere westerlies at the Last Glacial Maximum: an outline of AGCM simulation results and evaluation of their implication. Quaternary Science Reviews. 2000;19:881–898. [Google Scholar]
- Zaneto A, Kremer A. Geographical structure of gene diversity in Quercus petraea (Matt) Liebl. 1. Monolocus patterns of variation. Heredity. 1995;75:506–517. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.