Abstract
Background and Aims
Zingiberales comprise a clade of eight tropical monocot families including approx. 2500 species and are hypothesized to have undergone an ancient, rapid radiation during the Cretaceous. Zingiberales display substantial variation in floral morphology, and several members are ecologically and economically important. Deep phylogenetic relationships among primary lineages of Zingiberales have proved difficult to resolve in previous studies, representing a key region of uncertainty in the monocot tree of life.
Methods
Next-generation sequencing was used to construct complete plastid gene sets for nine taxa of Zingiberales, which were added to five previously sequenced sets in an attempt to resolve deep relationships among families in the order. Variation in taxon sampling, process partition inclusion and partition model parameters were examined to assess their effects on topology and support.
Key Results
Codon-based likelihood analysis identified a strongly supported clade of ((Cannaceae, Marantaceae), (Costaceae, Zingiberaceae)), sister to (Musaceae, (Lowiaceae, Strelitziaceae)), collectively sister to Heliconiaceae. However, the deepest divergences in this phylogenetic analysis comprised short branches with weak support. Additionally, manipulation of matrices resulted in differing deep topologies in an unpredictable fashion. Alternative topology testing allowed statistical rejection of some of the topologies. Saturation fails to explain observed topological uncertainty and low support at the base of Zingiberales. Evidence for conflict among the plastid data was based on a support metric that accounts for conflicting resampled topologies.
Conclusions
Many relationships were resolved with robust support, but the paucity of character information supporting the deepest nodes and the existence of conflict suggest that plastid coding regions are insufficient to resolve and support the earliest divergences among families of Zingiberales. Whole plastomes will continue to be highly useful in plant phylogenetics, but the current study adds to a growing body of literature suggesting that they may not provide enough character information for resolving ancient, rapid radiations.
Keywords: Tropical gingers, Zingiberales, plastome, next-generation sequencing, Illumina, phylogeny, evolution, monocots, support, phylogenomics, ancient radiation, plastid gene set
INTRODUCTION
Phylogenetic patterns generated by ancient, rapid radiations are difficult to resolve (e.g. Soltis and Soltis, 2004; Baurain et al., 2007; Whitfield and Lockhart, 2007; Dunn et al., 2008; Philippe et al., 2011). Recently developed next-generation sequencing (NGS) technologies provide access to unprecedented amounts of genomic character data to aid in the resolution of deep phylogenetic relationships. For example, in monocotyledonous angiosperms, application of large plastid gene data sets generated by NGS technologies has been highly effective at resolving both deep and shallow nodes (e.g. Givnish et al., 2010; Steele et al., 2012; Barrett et al., 2013). The development of multiplex barcodes allows several accessions to be sequenced simultaneously and for the data corresponding to each accession to be sorted out informatically. Genome-scale alignments of orthologous genes across multiple taxa can now readily be assembled by pooling several barcoded samples (e.g. Cronn et al., 2008) in a few lanes of an Illumina HiSeq flow cell (Illumina Inc., San Diego, CA, USA).
Plastid genes have long been exploited for phylogenetic inference in angiosperm systematics, but until fairly recently most studies were restricted to a few genes, constrained by the technological and cost limitations of PCRs and Sanger sequencing reactions (e.g. Soltis et al., 2000; Davis et al., 2004; Chase et al., 2006; but see Soltis et al., 2011). Through advances in NGS technologies, the acquisition of complete plastid genomes or a large suite of plastid genes for any taxon is a realistic goal in modern plant systematics (e.g. Stull et al., 2013). Thus, limitations in data acquisition are becoming more exclusively centred around the ability to acquire taxa and build taxon-rich matrices than to generate data for the individual taxa sampled [e.g. compare the data matrices (Asparagales) of Seberg et al., 2012 and Steele et al., 2012]. Given current budget limitations and the cost of generating NGS data, researchers must strategically sample taxa to span as much phylogenetic diversity as possible within the target clade or taxon. One approach is to sample taxa such that the earliest divergence event within each representative sub-group (e.g. including at least one representative of each of three monophyletic subfamilies within a family of interest), while focusing more intensively on particular groups of interest (e.g. Poales in Givnish et al., 2010; Asparagales and Poales in Steele et al., 2012; commelinid monocots in Barrett et al., 2013).
In studies with orders-of-magnitude greater numbers of informative characters than numbers of taxa, researchers must cautiously evaluate factors that may influence topology and branch support. For example, these factors might include the intensity of taxon sampling (e.g. Zwickl and Hillis, 2002; Hillis et al., 2003; Heath et al., 2008; Philippe et al., 2011), choice of outgroups, inclusion/exclusion of other taxa outside the group of interest and data partitioning effects (e.g. Graham et al., 2006) and how all of these factors influence various phylogenetic reconstruction methods (distance, parsimony, likelihood or Bayesian). Due to the large numbers of characters typically generated in phylogenomic studies, spurious or misleading relationships may be assigned high branch support (discussed in Philippe et al., 2011). In data sets with strong phylogenetic signal and low conflict favouring a single topology, the influence of these factors is hypothesized to be weak; however, phylogenomic data sets constructed to address ‘deep’ relationships often do not conform to this scenario, and certain areas of the phylogenetic trees remain unresolved or with weak branch support (Jansen et al., 2007; Moore et al., 2007; Givnish et al., 2010; Barrett et al., 2013; Davis et al., 2013). The stability of a phylogenetic hypothesis can thus be assessed by varying the aforementioned matrix characteristics.
The monocotyledonous angiosperm order Zingiberales comprises a significant element of tropical forest ecosystems around the world and displays significant diversity in floral morphology (Tomlinson, 1969; Dahlgren et al., 1985; Kress, 1986, 1990; Kirchoff, 1993; Smith, 1993; Zomlefer, 1994; Rudall and Bateman, 2004; Kirchoff et al., 2009; Bartlett and Specht, 2010, 2011; Specht et al., 2012). Zingiberales are a clade of typically large herbs in eight monophyletic families: Musaceae, Strelitziaceae, Lowiaceae, Heliconiaceae, Cannaceae, Costaceae, Marantaceae and Zingiberaceae. These families together are pantropical and encompass approx. 95 genera and 2500 species (Kress et al., 2001, 2002). Members of Zingiberales are economically important as crop species (banana, plantain and ginger), ornamentals (heliconias, prayer plants, cannas and birds-of-paradise) and spices (cardamom and galangal). Zingiberales share a strongly supported sister relationship with Commelinales; collectively, these orders are sister to an expanded Poales (containing grasses, sedges, bromeliads and related families), sensu Angiosperm Phylogeny Group III (APG III, 2009). Along with Arecales (palms) and the unplaced Dasypogonaceae, Zingiberales, Commelinales and Poales comprise a well-supported commelinid clade, although relationships among the major clades of commelinids are less strongly supported (e.g. Davis et al., 2004; Chase et al., 2006; Givnish et al., 2010; Barrett et al., 2013).
While a well-supported phylogenetic hypothesis for Zingiberales remains elusive (e.g. Kress, 1986, 1990; Smith et al., 1993; Kress et al., 2001; 2002), a resolved phylogenetic tree for Zingiberales is critical for the interpretation of morphological character data, floral developmental evolution, pollinator and herbivore coevolution, biogeographic patterns, fossil and stratigraphic data, and divergence time estimates. Phylogenetic uncertainty is most pronounced at the base of the order, as evidenced by low branch support recovered in studies aiming to resolve deep relationships among families based on morphology and one or a few genes (Kress et al., 2001; Kress and Specht, 2006). The sequence of early branching events in Zingiberales represents one of the most recalcitrant areas yet to be resolved in the monocot tree of life.
Critical to resolving relationships at the base of Zingiberales is the placement of Heliconiaceae and Musaceae. Heliconia was initially included in Musaceae based on vegetative similarities (e.g. Petersen, 1889; Winkler, 1930). Morphological studies and recent phylogenetic analyses have confirmed the monophyly of Heliconiaceae as a separate lineage in Zingiberales (Nakai, 1941; Tomlinson, 1962; Dahlgren et al., 1985; Kress, 1986; Kress et al., 2001), but the placement of Musaceae and Heliconiaceae in the order is uncertain. One of three most parsimonious trees recovered from a combined analysis of morphology, plastid DNA (rbcL and atpB) and nuclear DNA (18S) placed Heliconiaceae as sister to the ginger clade (Kress et al., 2001), a lineage composed of ((Cannaceae, Marantaceae), (Costaceae, Zingiberaceae)). Musaceae were resolved as the earliest diverging lineage in the order, with (Lowiaceae, Strelitziaceae) sister to the clade of Heliconiaceae plus the ginger families (Kress et al., 2001). However, analysis of DNA sequences alone (in the same study) placed Heliconia as sister to (Strelitziaceae, Lowiaceae), also with weak support.
Fossil zingiberalean taxa date from the Cretaceous (Rodriguez-de la Rosa and Cevallos-Ferriz, 1994) on the basis of fruits, seeds, leaves, rhizomes and phytoliths. Zingiberales probably diverged from sister Commelinales approx. 120 million years ago (MYA) (Kress and Specht, 2006). The initial radiation of Zingiberales is hypothesized to have occurred rapidly, with recent fossil-calibrated molecular estimates ranging from 110 to 80 MYA (Late Cretaceous) for the diversification of primary lineages (Kress and Specht, 2006; see also Janssen and Bremer, 2004; Magallon and Castillo, 2009). This is illustrated in molecular phylogenetic analysis by short internal branches, which are often problematic for phylogenetic inference and have been observed in other monocot groups hypothesized to have undergone relatively ancient, rapid diversifications, such as arecoid palms (e.g. Baker et al., 2009) and epidendroid orchids (e.g. Freudenstein et al., 2004; van den Berg et al., 2005; Gorniak et al., 2010).
In the current study, nine whole-plastid gene sets for representative families of Zingiberales were generated by NGS and added to a matrix of coding regions of the monocots to address the following questions. (1) Do whole-plastid gene sets resolve and provide support for deep relationships among families of Zingiberales, and, if so, are these consistent with relationships recovered in previous studies? (2) What effect, if any, does variable sampling of monocot taxa outside Zingiberales have on relationships and support within the order? (3) What is the effect of using or excluding various data partitions (protein-coding + plastid rDNA, protein-coding only, first + second codon positions, or third codon positions)? (4) In a likelihood framework, does model partitioning influence relationships and/or support? (5) Is there evidence of substitution saturation in the protein-coding and non-protein-coding plastid gene data?
MATERIALS AND METHODS
Plant material, DNA isolation and sequencing
Plant material was collected for nine taxa of Zingiberales to complete a data set with at least one representative from each of the eight families, in addition to five previously sequenced plastomes: Musa acuminata, Heliconia collinsiana, Alpinia zerumbet, Renealmia alpinia and Zingiber spectabile (see Supplementary Data Accession Numbers). Fresh leaf material was dried in silica gel and stored at –20 °C for subsequent DNA extraction. Voucher specimens are listed in Table 1.
Table 1.
Voucher information, GenBank accession numbers and Illumina sequence statistics for newly sequenced plastomes of Zingiberales
| Family: Species; GenBank accession number; [Voucher information] | #reads | #mapped | %ptDNA | x-cov |
|---|---|---|---|---|
| Cannaceae: Canna indica L.; KF601570; [JLM 2013-001, Leebens-Mack et al., 2013-001 (GA)] | 4 378 843 | 977 898 | 22·33 | 579·83 |
| Costaceae: Costus pulerulentus Standl. & L.O.Williams; KF601573; [FTBG 2004-0330, Zomlefer et al. 2294 (FTG, NY)] | 3 519 114 | 47 657 | 1·35 | 28·26 |
| Costaceae: Monocostus uniflorus (Poepp. ex Petersen) Maas; KF601572; [FTBG 2000-894H, Zomlefer et al. 2337 (FTG, GA, NY)] | 2 394 189 | 32 705 | 1·37 | 19·39 |
| Lowiaceae: Orchidantha fimbriata Holttum; KF601569; [FTBG 2003-1178A, Zomlefer et al. 2296 (FTG, NY)] | 10 569 061 | 38 514 | 0·36 | 22·84 |
| Marantaceae: Maranta leuconeura E.Morren; KF601571; [NYBG 53/67, Stevenson et al., 53/67 (NY)] | 7 500 600 | 264 364 | 3·52 | 156·75 |
| Marantaceae: Thaumatococcus daniellii (Benn.) Benth. & Hook.f.; KF601575; [FTBG 83374A, Zomlefer et al. 2324 (FTG, NY)] | 1 341 401 | 42 371 | 3·16 | 25·12 |
| Musaceae: Musa textilis Née; KF601567; [FTG 2007-0825A, Zomlefer et al. 2349 (FTG, NY)] | 3 987 133 | 270 982 | 6·8 | 160·67 |
| Strelitziaceae: Ravenala madagascariensis Sonn.; KF601568; [P.1395G, Zomlefer et al. 2350 (FTG, NY)] | 3 386 460 | 56 706 | 1·67 | 33·62 |
| Zingiberaceae: Curcuma roscoeana Wall.; KF601574; [FTG 96-1594A, Zomlefer et al. 2299 (FTG, NY)] | 4 871 653 | 89 310 | 1·83 | 52·95 |
Vouchers correspond respectively to: [living collection numbers; herbarium voucher numbers; (herbaria where deposited)]. FTG, Fairchild Tropical Botanic Garden Herbarium; GA, University of Georgia Herbarium; NY, New York Botanical Garden Herbarium; FTBG, Fairchild Tropical Botanic Garden live plant accession; NYBG, New York Botanical Garden live plant accession.
#mapped, the number of raw reads mapped to the plastome of Heliconia collinsiana (GenBank accession number JX088660; Barrett et al., 2013) in the program BWA; %ptDNA, the percentage of raw reads mapped to the Heliconia reference in BWA/the total number of raw reads; x-cov, [the total number of reads mapped × read length]/161 907 bp of the Heliconia reference plastome.
Using the conservative default parameters in BWA (maximum number of differences allowed = 3) across divergent plastomes probably represents an underestimate of plastome coverage.
Genomic DNA was extracted as described in Barrett and Davis (2012) and Barrett et al. (2013), using either a modified cetyltrimethyl ammonium bromide (CTAB) protocol (Doyle and Doyle, 1987) or the Qiagen DNeasy Plant Mini Kit protocol (Qiagen, Inc., Valencia, CA, USA) to prepare DNA for Illumina library construction (Illumina, Inc., San Diego, CA, USA). DNA concentrations were quantified using a NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, USA), and DNA quality was assessed via electrophoresis on a 2 % agarose gel, in which isolations were verified for high molecular weight, non-degraded DNA. Isolations yielding >15 ng μL−1 of high-quality genomic DNA were then tested for relative plastid DNA concentration via quantitative PCR, by amplifying a 110 bp portion of the rbcL gene using SYBR® GreenER™ qPCR SuperMix (Invitrogen, Carlsbad, CA, USA) following the protocol of Barrett et al. (2013). Library preparation, multiplex barcoding and single-end sequencing on an Illumina GAIIx were completed at Cold Spring Harbor Laboratory (Woodbury, NY, USA), yielding a minimum of approx. 20× mean plastome coverage for each taxon.
Short-read quality trimming, plastome assembly and annotation
Read quality for 71 or 96 bp single-end reads were assessed using FastQC (S. Andrews, http://www.bioinformatics.babraham.ac.uk/projects/fastqc), and low-quality 3' ends were trimmed using the java script Trimmomatic 0·17 (A. Bolger and M. Giorgi, http://www.usadellab.org/cms/index.php?page=trimmomatic), specifically trimming ends with PHRED scores <26 (i.e. positions with greater than approx. 1/400 probability of containing an error since PHRED scores are based on a log10 scale), discarding reads with average PHRED scores <26 and those >25 bp long. Reads were then assembled de novo using Velvet 2·3 (Zerbino and Birney, 2008), employing a variety of hash lengths and coverage cut-off values. After preliminary testing of these parameters, a hash length of 51 and coverage cut-off of 10× (in ‘k-mer’ coverage, see Zerbino and Birney, 2008) were chosen as optimal based on maximum contig length, maximum coverage and N50 of contigs (data not shown). Minimum contig length was set to 300 bp (see Barrett and Davis, 2012; Barrett et al., 2013).
Reference-guided assembly was completed using the YASRA 2·32 assembly pipeline (Ratan, 2009), which aligns reads to a reference sequence with LASTZ (Harris, 2007) and then attempts to bridge gaps between contigs in an iterative process until no further improvements are possible. Contig read pileups for de novo and reference-guided assemblies were visually inspected in Tablet v1·11·05·03 (Milne et al., 2010) for putative rearrangements, assembly errors, and low coverage regions. Heliconia collinsiana (Heliconiaceae, GenBank accession no. JX088660) and Typha latifolia [Poales sensu APG III (2009), Typhaceae, NC_013823] were used as references for assembly. The rapid aligner BWA (Burroughs–Wheeler aligner, Li and Durbin, 2009) was used to align reads to Heliconia to obtain initial estimates of average plastome coverage. De novo and reference-guided contigs were joined into larger contigs in Sequencher (Gene Codes, Inc., Ann Arbor, MI, USA). Contig gaps were crossed and any differences between de novo and reference-guided contigs were corrected by searching the original read pool using the UNIX ‘grep’ function, and a final plastome sequence was completed following Barrett et al. (2013; see also Givnish et al., 2010). Plastomes were annotated in DOGMA (Wyman et al., 2004) and submitted to GenBank following Barrett and Davis (2012) and Barrett et al. (2013).
Plastid gene sequence alignment
Coding gene sets plus rDNA genes were deposited on an alignment server hosted by the University of Georgia, and initial multiple sequence alignments for each gene were accomplished in MUSCLE (Edgar, 2004) under default values, and mediated by custom PERL scripts (J. Kerry, University of Georgia-Athens) that automate the construction of a concatenated, partitioned NEXUS file. Because codon-based alignments were desired, coding loci were stripped of alignment gaps inferred by MUSCLE. Codon-based alignments were generated using the program MACSE, which allows preservation of reading frames in alignments that include incomplete sequences or errors from previous studies that cause apparent reading frame shifts (Ranwez et al., 2011). Alignments were generated individually for each of 83 protein-coding genes, manually adjusted and reconstituted into a single NEXUS file using SequenceMatrix (Vaidya et al., 2010). The file was then converted to FASTA in Mesquite (Maddison and Maddison, 2007), partitioned by first + second and third codon positions in MEGA5 (Tamura et al., 2011), and exported as two separate, partitioned matrices for downstream phylogenetic analyses (see below).
Taxon sets
To assess the influence of taxon sampling outside Zingiberales on the ingroup topology, four taxon sets were constructed: (1) a full monocot (FM) set of 56 taxa, including all representative orders of monocots (sensu APG III, 2009) with sampling focused on the commelinid clade; (2) a 45-taxon no-Poales monocot (NPM) set, identical to the FM matrix but excluding all representatives of Poales, which have extremely long branches and demonstrate rapid, heterogeneous evolutionary rates based on recent phylogenetic analyses of plastid-coding regions (e.g. Givnish et al., 2010; Barrett et al., 2013); (3) a no-Poales commelinid (NPC) sample including representatives of Zingiberales, Commelinales, Arecales and unplaced Dasypogonaceae; and (4) a 17-taxon Zingiberales–Commelinales (ZC) sample composed only of Zingiberales (14 taxa, eight families) + representative Commelinales [three families, consistently resolved as sister to Zingiberales (Davis et al., 2004; Chase et al., 2006; Graham et al., 2006; Givnish et al, 2010; Barrett et al., 2013; Davis et al., 2013)]. Acorus calamus was the outgroup in the FM and NPM sets, and Calamus (Arecales) and Xiphidium (Commelinales) were outgroup taxa in the NPC and ZC sets.
Data partitions
To assess the influence of different partitioning schemes among the plastome data, four data partitions were constructed: (1) an unpartitioned coding gene + rDNA (CR) matrix, including 83 protein-coding genes plus four rRNA genes; (2) an unpartitioned coding-only (CO) matrix, consisting of 83 protein-coding genes; (3) a matrix consisting of first and second codon positions (P1 + 2); and (4) a matrix consisting of codon position three (P3).
Phylogenetic analyses
Maximum parsimony (MP) and maximum likelihood (ML) analyses were conducted for each taxon set × data partition configuration. Parsimony analyses were run in TNT (Goloboff et al., 2008) using ten random addition sequences (RAS) + tree bisection–reconnection (TBR), saving 100 000 trees. This strategy was sufficient for rapidly finding the shortest tree, so more sophisticated heuristic searches were not necessary. Two thousand jackknife (Farris et al., 1996) pseudoreplicates were run for all parsimony analyses, under identical heuristic search parameters, with 37 % character removal probability (Farris et al., 1996). TNT utilizes a more conservative ‘strict consensus’ approach to resampling as opposed to a ‘frequency within replicates’ approach to summarize the results of each pseudoreplicate (see Davis et al., 2004; Freudenstein and Davis, 2010; Simmons and Freudenstein, 2011; Barrett et al., 2013). Both ‘absolute frequency’ and ‘GP/C frequency differences’ (Group Present/Contradicted; Goloboff et al., 2003) were used to summarize parsimony jackknife support. The latter metric measures the difference between the frequency with which a given group is recovered among the jackknife pseudoreplicate pool and the most frequent contradictory group. Support values calculated under GP/C are expected to be lower than absolute support values at nodes for which there is substantial conflict.
Likelihood analyses were implemented using RAxML (Stamatakis, 2006a) on the CIPRES 3·1 web server (Miller et al., 2010). Best-fit models were determined in MEGA5; GTR-Γ was the optimal model for first + second, third and all (combined) codon positions. The best-scoring ML tree was calculated under the GTR-Γ model, with the default number of rate categories (C = 25), initiating multiple searches from different starting seeds to check for any major stochastic discrepancies between runs. Branch support was assessed with 2000 non-parametric bootstrap pseudoreplicates, using the rapid bootstrap approach implemented in RAxML (Stamatakis et al., 2008); rapid bootstrapping uses the GTR-CAT approximation for resampling (Stamatakis, 2006b), with the final ML tree optimization under GTR-Γ. This approach was shown to yield support values that were consistent with the more computationally intensive standard RAxML bootstrap in a previous phylogenetic analysis with monocot plastid genomes (Barrett et al., 2013).
In addition to using an unpartitioned model, the CO matrix was analysed under both gene- and codon-partitioned models separately in RAxML, to assess whether model partitioning might affect topology and/or support. This allows base frequencies, the α-shape parameter and GTR substitution rates to be estimated separately for each partition. A gene × codon model analysis was not attempted, due to the risk of overparameterization (here, 2 codon partitions × 83 gene partitions = 166 site parameters) and associated parameter identifiability issues (Rannala, 2002; Lemmon and Moriarty, 2004; Sullivan, 2005).
Alternative topologies were evaluated with Shimodaira–Hasegawa (SH) and Approximately Unbiased (AU) tests (Shimodaira and Hasegawa, 1999) in the program CONSEL (Shimodaira and Hasegawa, 2001), to assess whether the data given each alternative tree had a significantly lower likelihood than the the data given the best-scoring tree. All tree topologies recovered from various analyses in the current study plus the topology from the total evidence analysis of Kress et al. (2001) were used as alternatives. A phi-test for recombination among sites within Zingiberales was conducted in SplitsTree v.4 (Huson and Bryant, 2006) for the ZC-CO matrix, with representative Commelinales removed.
To assess potential substitution saturation, pairwise GTR-corrected distances were plotted against rates of transitions and transversions for codon positions one and two, position three and all positions combined. These comparisons were made for the FM and ZC CO sets, to assess the presence of saturation among all sampled monocot plastid genes and among those sampled from representative Zingiberales. In addition to assessing saturation qualitatively, the statistical test of Xia et al. (2003) was used to compare the mean saturation among sites with that expected under an entropy model of full saturation, using an asymmetric tree model. All saturation analyses were conducted in DAMBE (Xia and Xie, 2001).
RESULTS
Matrix information
The full monocot matrix, including both coding loci and rDNA (FM-CR), has an aligned length of 81 409 nucleotides with 21 293 parsimony informative characters (PICs; Table 2, Supplementary Data Fig. S1). Removing rDNA (301 PICs) reduces this number to 76 722 total aligned nucleotides for protein-coding genes (25 574 aligned codons), yielding 20 992 PICs in the FM–CO matrix (Supplementary Data Fig. S2). Third positions account for approx. 54·3, 55·6, 55·5 and 57 % of all informative characters for the FM, NPM, NPC and ZC taxon sets, respectively; more than the number for positions 1 and 2 combined in all cases (Table 2). A total of 5786 characters are again informative in Zingerberales alone (ZC-CO matrix). A phi-test indicated no significant recombination signal (P = 1·0).
Table 2.
Details of the various taxon sample × data partition configurations analysed in this study
| Analysis | (Taxa data) |
# taxa | # PIC | MP length | # trees | CI | RI | Mean support (MP) |
ML (–lnL: GTR-G) |
Gene | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Code | Matrix | JK-abs | JK-GP/C | Unpart | Codon | ||||||||
| Full monocot | |||||||||||||
| 1 | FM-CR | Coding plus rDNA | 56 | 21 293 | 84 977 | 1 | 0·526 | 0·701 | 96·8 | 94·6 | –560 952 | ||
| 2 | FM-CO | Coding only | 56 | 20 992 | 83 950 | 1 | 0·524 | 0·700 | 96·8 | 94·8 | –545 677 | –539 379 | –539 112 |
| 3 | FM-P1 + 2 | Positions 1 and 2 | 56 | 9536 | 35 026 | 1 | 0·594 | 0·730 | 92·7 | 90·7 | –270 882 | ||
| 4 | FM-P3 | Position 3 | 56 | 11 456 | 48 858 | 1 | 0·475 | 0·681 | 94·1 | 92·5 | –267 553 | ||
| Non-Poales monocot | |||||||||||||
| 5 | NPM-CR | Coding plus rDNA | 45 | 18 036 | 67 429 | 1 | 0·573 | 0·693 | 97·6 | 96·2 | –471 970 | ||
| 6 | NPM-CO | Coding only | 45 | 17 843 | 66 745 | 1 | 0·571 | 0·692 | 97·4 | 95·7 | –459 020 | –453 556 | –452 988 |
| 7 | NPM-P1 + 2 | Positions 1 and 2 | 45 | 7912 | 28 251 | 2 | 0·633 | 0·718 | 96·2 | 95·5 | –233 252 | ||
| 8 | NPM-P3 | Position 3 | 45 | 9931 | 38 430 | 1 | 0·526 | 0·676 | 99·2 | 99·0 | –219 547 | ||
| Non-Poales commelinid | |||||||||||||
| 9 | NPC-CR | Coding plus rDNA | 26 | 11 099 | 32 570 | 1 | 0·719 | 0·768 | 96·2 | 93·8 | –292 830 | ||
| 10 | NPC-CO | Coding only | 26 | 10 972 | 32 161 | 1 | 0·716 | 0·767 | 96·0 | 93·6 | –282 498 | –278 978 | –277 717 |
| 11 | NPC-P1 + 2 | Positions 1 and 2 | 26 | 4887 | 14 584 | 1 | 0·756 | 0·782 | 92·6 | 91·7 | –156 108 | ||
| 12 | NPC-P3 | Position 3 | 26 | 6085 | 17 532 | 1 | 0·685 | 0·759 | 95·9 | 93·5 | –122 664 | ||
| Zing. + Comm. | |||||||||||||
| 13 | ZC-CR | Coding plus rDNA | 17 | 8440 | 23 970 | 1 | 0·787 | 0·731 | 95·1 | 94·2 | –239 458 | ||
| 14 | ZC-CO | Coding only | 17 | 8343 | 23 613 | 1 | 0·785 | 0·73 | 94·1 | 93·2 | –229 809 | –226 004 | –225 753 |
| 15 | ZC-P1 + 2 | Positions 1 and 2 | 17 | 3583 | 10 722 | 1 | 0·82 | 0·748 | 85·2 | 84·7 | –130 551 | ||
| 16 | ZC-P3 | Position 3 | 17 | 4760 | 12 854 | 1 | 0·758 | 0·721 | 93·5 | 88·1 | –112 327 | ||
# PIC, the number of potentially parsimony informative characters; MP L, parsimony tree length; # trees, the number of parsimonious trees recovered; CI, consistency index; RI, retention index; JK-abs, absolute (i.e. standard) parsimony jackknife support; JK-GP/C, group present/contradicted parsimony jackknife support; ‘Unpart’, ‘Codon’ and ‘Gene’, the likelihoods under unpartitioned, codon-partitioned and gene-partitioned models, respectively.
Phylogenetic analyses of the full monocot, coding-only matrix (FM-CO)
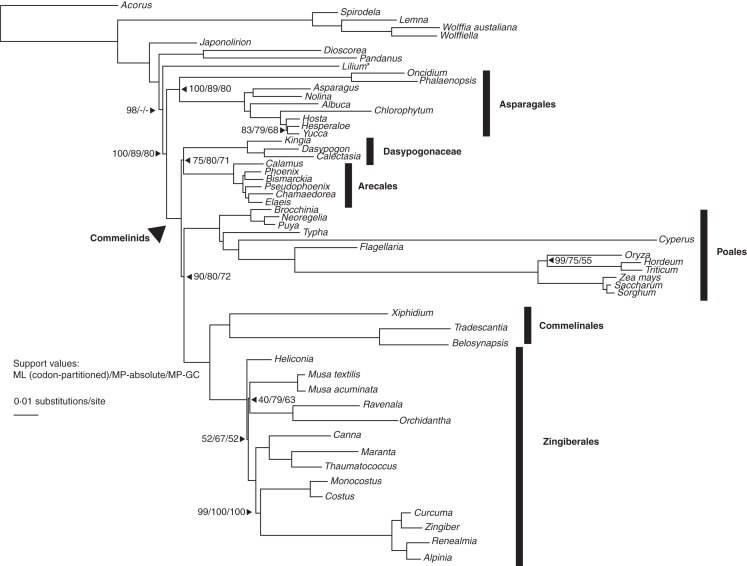
A single best-scoring ML topology was obtained with the codon-partitioned FM-CO matrix in all five randomly initiated searches (Fig. 1). Successively moving from the root of the tree, Acorus (outgroup) is sister to (Alismatales (Petrosaviales (Dioscoreales, Pandanales (Liliales (Asparagales, commelinid clade))))); deep monocot relationships generally received robust support. The only major difference between the MP and ML analyses is the placement of Lilium as sister to (Dioscoreales, Pandanales) in the former, with low to moderate support. In the commelinid clade, the unplaced Dasypogonaceae are sister to Arecales, collectively sister to (Poales (Zingiberales, Commelinales)). Commelinids are robustly supported as a clade, with moderate to strong support for relationships among the commelinid orders and Dasypogonaceae.
Fig. 1.
Best-scoring maximum-likelihood tree based on 83 protein-coding genes and the ‘full monocot’ taxon set, with the GTR-Γ model partitioned by codon position [FM-CO (codon); –lnL = –539 378·7814; see Table 2). The Zingiberales topology above corresponds to pattern 1 (see Fig. 3). Numbers adjacent to branches indicate support values (ML bootstrap/MP absolute jackknife/MP ‘group present–contradicted’ (i.e. GP/C) jackknife); branches with no values indicate 100 % support; the scale bar indicates a branch length of 0·01 substitutions per site. *Alternative placement of Lilium in the MP tree (Lilium (Pandanus, Dioscorea)).
In Zingiberales, the topology obtained corresponds to Pattern 1 (Figs 2 and 3; blue), the most commonly recovered of the ‘Heliconia-sister’ topologies. Heliconia is sister to the remaining Zingiberales, and Musa is sister to (Strelitziaceae, Lowiaceae), forming a clade that is collectively sister to the ‘ginger’ families ((Cannaceae, Marantaceae), (Costaceae, Zingiberaceae)). The deep relationships among the primary clades of Zingiberales, however, received only low support in the codon-partitioned ML boostrap analysis and low to moderate support for MP jackknife (Fig. 1). Moreover, GP/C jackknife values are lower by 15 % than absolute jackknife values at two of these critical nodes, the first uniting Musaceae, Strelitziaceae and Lowiaceae, and the second uniting the aforementioned clade with the clade of ginger families. The four ginger families form a well-supported clade, within which the four ginger families (Zingiberaceae, Costaceae) are sister to (Marantaceae, Cannaceae), all with robust support in both MP and ML analyses, including intrafamilial relationships. The monophyly of the ginger families and the robustness of the relationships in the ginger clade are consistent with previous phylogenetic results despite variability in taxon and character sampling.
Fig. 2.
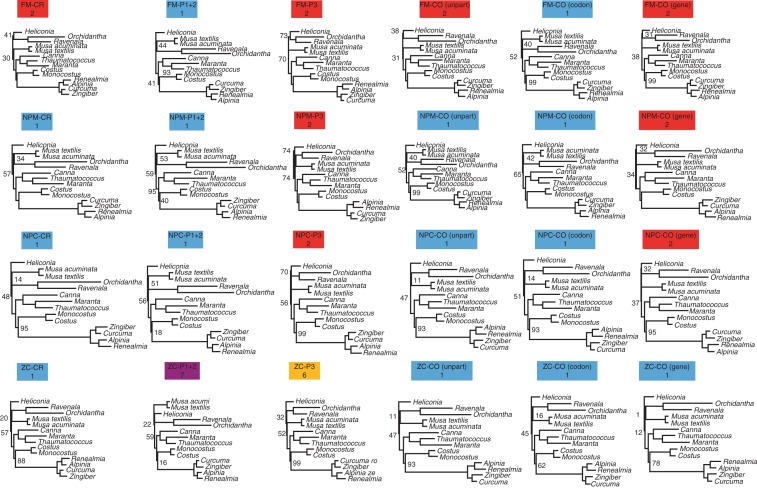
Maximum-likelihood phylogenetic trees for all taxon set × data partition configurations. Numbers adjacent to branches indicate bootstrap support values based on 2000 pseudoreplicates using the rapid bootstrap algorithm in RAxML. Taxon sample abbreviations: FM, full monocot; NPM, non-Poales monocot; NPC, non-Poales commelinid; ZC, Zingiberales–Commelinales. Data partition abbreviations: CR, coding + rDNA; CO, coding only; P1 + 2, codon positions 1 and 2; P3, codon position 3; unpart, codon and gene, unpartitioned, codon-partitioned and gene-partitioned ML models, respectively. Branch lengths are not to scale between analyses. Colours refer to the patterns observed (see Fig. 3).
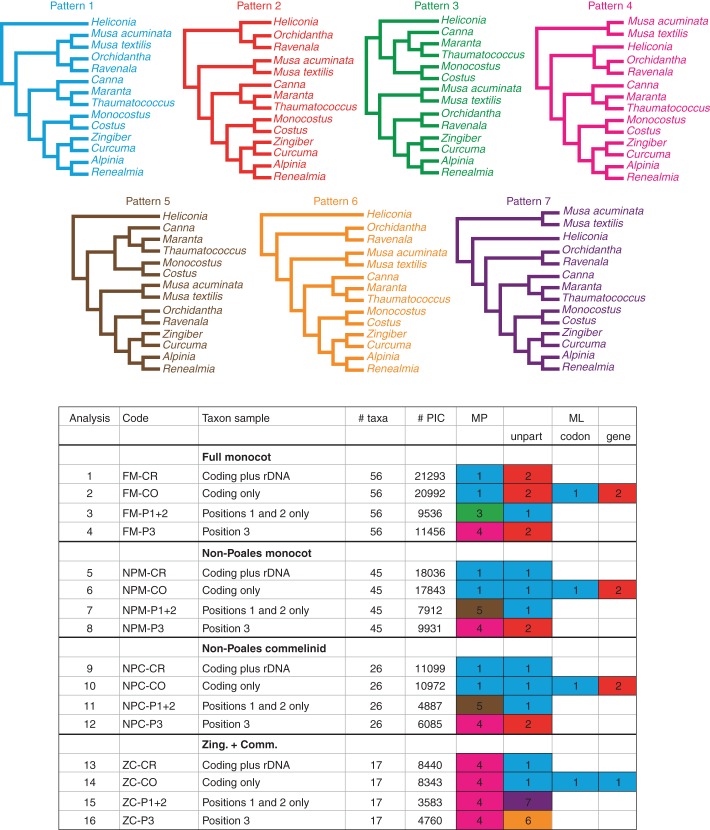
Fig. 3.
Top: cladograms recovered from various taxon set × data partition configurations. Taxon set abbreviations: FM, full monocot; NPM, non-Poales monocot; NPC, non-Poales commelinid; ZC, Zingiberales–Commelinales. Data partition abbreviations: CR. coding + rDNA; CO, coding only; P1 + 2, codon positions 1 and 2; P3, codon position 3; unpart, codon and gene, unpartitioned, codon-partitioned and gene-partitioned ML models, respectively. Colours of the observed patterns are replicated in boxes; e.g. blue = Pattern 1).
Effect of taxon sampling and data inclusion/exclusion
Changes in outgroup taxon sampling, inclusion/exclusion of process data partitions (i.e. codon positions) and ML model partitioning schemes all appeared to exert at least some influence on the internal topology of Zingiberales, with seven different topologies observed across all analyses (Table 3; Fig. 2; Fig. 3, Patterns 1–7). None of these topologies received robust, ‘deep’ support, regardless of the reconstruction method (Fig. 2). Internal support values outside the ginger families did not exceed 59 for ML bootstrap or 82 for parsimony ‘absolute’ jackknife values, considering the CR, CO and P1 + 2 matrices for all taxon samples. Deep support values were generally higher for topologies based on P3 matrices relative to those of positions one and two (Fig. 2).
Table 3.
Shimodaira–Hasegawa (SH) tests and Approximately Unbiased (AU) tests of the seven alternative topologies recovered from various analyses in this study relative to the best-scoring ML topology and using the ZC–CO (Zingiberales + Commelinales, coding-only) matrix
| Pattern | Topology | Likelihood | p(SH test) | p(AU test) | ΔlnL |
|---|---|---|---|---|---|
| 1* | (Hel((Mus(Str,Low)), Gingers)) | –229 809·0193 | NA | NA | NA |
| 2 | ((Hel(Low,Rav)), (Mus,Gingers)) | –229 852·1668 | 0·13 | 0·003 | 43·147517 |
| 3 | (Hel(Cos(Can,Mar)), (Mus(Zin(Str,Low)))) | –230 007·9099 | < 0·001 | < 0·001 | 198·890598 |
| 4 | (Mus(((Hel(Low,Str)), Gingers)) | –229 813·2730 | 0·847 | 0·522 | 4·253738 |
| 5 | (Hel((Cos(Can,Mar)), (Mus((Low,Str),Zin))) | –229 986·9456 | < 0·001 | < 0·001 | 177·926282 |
| 6 | (Hel((Low,Str)(Mus,Gingers))) | –229 811·2755 | 0·889 | 0·611 | 2·256203 |
| 7 | (Mus(Hel((Low,Str),Gingers))) | –229 820·8087 | 0·593 | 0·089 | 11·789418 |
| 8† | (Mus((Low,Str)(Hel,Gingers))) | –229 822·7744 | 0·534 | 0·035 | 13·755132 |
‘Pattern’ refers to the topology shown in Figs 2 and 3; ‘p(SH test)’ and p(AU test)' indicate P-values for each corresponding test (i.e. whether the alternative topology is significantly worse than the best-scoring ML tree – here pattern 1, in bold – and can thus be rejected).
Can, Cannaceae; Cos, Costaceae; Hel, Heliconiaceae; Low, Lowiaceae; Mar, Marantaceae; Mus, Musaceae; Str, Strelitziaceae; Zin, Zingiberaceae; ‘Gingers’, ((Can,Mar), (Cos,Zin)); NA, not assessed.
*The best-scoring ML topology.
†The same topology recovered in Kress et al. (2001; based on DNA + morphology) but only including taxa sampled in the present study.
Pattern 1 was the most frequently recovered topology (Fig. 3); for MP analyses, Pattern 4 was recovered slightly more frequently (Fig. 3). Pattern 2 only appeared among ML analyses, and Pattern 4 only appeared among MP analyses. These both represent a major proportion of recovered topologies within their respective reconstruction method. Considering only the unpartitioned ML analyses (MP topologies are also listed in Fig. 3), all matrix partitions yielded Pattern 2 for the FM taxon sample, except for P1 + 2 which yielded Pattern 1. Both of the ‘non-Poales’ matrices (NPM and NPC) gave identical ML topologies, in which the P3 matrix resulted in a Pattern 2 topology whereas all others resulted in Pattern 1. The ZC taxon sample resulted in a Pattern 1 topology for the CR and CO matrices, whereas the P1 + 2 matrix resulted in Pattern 7 and the P3 matrix resulted in Pattern 6; the latter two topologies were not observed in any other analyses (Figs 2 and 3). Partitioning the CO data (i.e. in ML partitioned model analyses) by gene or codon for the FM taxon sample resulted in a Pattern 1 topology, whereas the unpartitioned data yielded a Pattern 2 topology. For the NPM and NPC samples, both unpartitioned and codon-partitioned models gave Pattern 1, whereas gene partitioning gave Pattern 2; all partitioning schemes yielded Pattern 1 for the ZC samples. The MP analyses yielded Pattern 1 for CO and CR matrices in the FM, NPM and NPC taxon samples, and Pattern 4 for the ZC sample; a Pattern 4 topology was recovered for the P3 matrix in all taxon samples; and Pattern 3, 5, 5 and 4 topologies were recovered for the P1 + 2 matrix in the FM, NPM, NPC and ZC samples, respectively (Figs 2 and 3).
Tests of alternative topologies
Likelihood-based SH tests indicate that based only on two of the recovered topologies, Patterns 3 and 5, did the data yield significantly lower likelihood scores compared with that of the best-scoring tree; the others could not be rejected (Table 3). The common theme among the two rejected topologies in the SH tests is the non-monophyly of the clade of ginger families. Similarly, AU tests rejected topology Patterns 3 and 5, but in addition rejected Patterns 2 and 8. Notably, none of the rejected topologies under the AU test was observed for any of the ZC matrices, under either ML or MP (Table 3; Fig. 3).
Saturation analyses
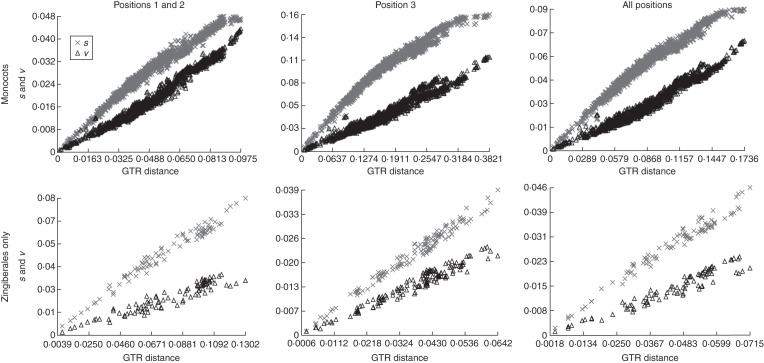
Plotting rates of transitions (s) and transversions (v) against GTR-corrected pairwise distances revealed, qualitatively, a weak pattern of saturation for transitions at first + second and third positions (and overall) in the FM taxon set, but no significant saturation was detected by the statistical test of Xia et al. (2003; Fig. 4; Table 4). This procedure compares a test statistic of saturation with a critical value at which noise begins to mislead phylogenetic inference; if the test statistic (ISS) is significantly less than the critical value (ISS.C), the interpretation is that there is no significant saturation. In fact, there were no significant instances of saturation among sequences in either the FM or ZC taxon sets which represent the two extremes of taxon sampling in this study.
Fig. 4.
Comparison of pairwise, uncorrected transition (s) and transversion (v) distances (y-axis) with GTR-corrected, pairwise distances (x-axis) for the full monocot (FM) and Zingiberales–Commelinales (ZC) taxon samples.
Table 4.
Tests for significant saturation of the coding-only (CO), codon positions 1 and 2 (P1 + 2) and codon position 3 (P3) matrices for the full-monocot (FM) and Zingiberales–Commelinales (ZC) taxon samples
| Taxon sample matrix | ISS | ISS.C | P (ISS < ISS.C) | Saturated? |
|---|---|---|---|---|
| FM-CO | 0·091 | 0·848 | <0·0001 | No |
| FM-P1 + 2 | 0·046 | 0·847 | <0·0001 | No |
| FM-P3 | 0·198 | 0·844 | <0·0001 | No |
| ZC-CO | 0·049 | 0·786 | <0·0001 | No |
| ZC-P1 + 2 | 0·036 | 0·675 | <0·0001 | No |
| ZC-P3 | 0·093 | 0·682 | <0·0001 | No |
ISS, test statistic measuring the average saturation in the matrix; ISS.C, entropy-based critical value of the data under complete saturation; P (ISS < ISS.C), significance of the saturation test of Xia et al. (2003); Saturated?, evidence for significant saturation.
DISCUSSION
Phylogenetic analysis of the CO matrix
Phylogenetic analysis of 83 coding genes of the plastid genome reveals robust support for the monophyly of: (1) (Zingiberales, Commelinales); (2) Zingiberales; (3) the four ‘ginger’ families (Cannaceae, Costaceae, Marantaceae and Zingiberaceae); (4) (Cannaceae, Marantaceae), and (Costaceae, Zingiberaceae); and (5) (Lowiaceae, Strelitziaceae) (Fig. 1). However, only weak support is recovered for deep relationships among families of Zingiberales (Fig. 1), despite using the most data-rich matrix to date for Zingiberales in terms of the number of phylogenetically informative characters. Familial relationships in this clade have historically been a phylogenetic challenge (Kress, 1990; Kress et al., 2001), exemplifying the pattern left behind by a rapid radiation (Kress and Specht, 2006). Specifically, the placement of Heliconiaceae as sister to all remaining families of Zingiberales in the present study contrasts with the provisionally accepted phylogenetic hypothesis of (Musaceae ((Lowiaceae, Strelitziaceae), (Heliconiaceae, ginger families))), but support for this relationship (Fig. 1) is weak. An AU test suggests that the data significantly reject the latter, provisionally accepted hypothesis (Table 3; Kress et al., 2001; APG III, 2009).
Implications for floral character evolution in Zingiberales (FM-CO matrix)
Zingiberales are a clade with much variation in floral morphology (Kirchoff, 1991; Kirchoff et al., 2009; Bartlett and Specht, 2010, 2011). If the result represented in Fig. 1 is a plausible hypothesis of relationships among major clades, then there are implications for the interpretations of floral evolution and diversification across the order. Based on the topology (Fig. 1; i.e. Pattern 1, Fig. 3), the ancestral zingiberalean flower would probably have had a relatively undifferentiated perianth, similar to prior reconstructions (Bartlett and Specht, 2010). Heliconia flowers, much like the flowers of Musaceae, have sepals and petals that differ only minimally in size, colour, shape and texture; the sepals and petals in both Heliconiaceae and Musaceae partly fuse to form a floral tube. In Heliconiaceae, adnation of sepals and petals is post-genital, indicating an independent derivation after its divergence from the remaining families. Fusion of the perianth in Musaceae is congenital and could have evolved independently in the Musaceae lineage, as this feature is not shared with other families in the order. The monomorphic perianth of Musaceae would also be interpreted as a derived feature in this phylogenetic context (Figs 1 and 3) and as an apomorphy for Musaceae.
Given a Heliconia-sister topology, however, the evolution of petaloidy in the stamen whorl would need to be reinterpreted. Heliconia flowers have a single small, petaloid staminode that is interpreted evolutionarily to be the first occurrence of petaloidy in the stamen whorl in Zingiberales, providing a synapomorphy of stamen whorl petaloidy uniting Heliconiaceae with the ginger clade (Kirchoff et al., 2009; Almeida et al., 2013; Specht et al., 2012). Given the sister position of Heliconiaceae to the remaining Zingiberales, staminode petaloidy either occurred early in the evolution of the order, with a subsequent loss in the ancestor of the remaining Zingiberales and regain in the ginger clade, or the petaloid staminode of Heliconiaceae evolved independently of the petaloid staminodes of the ginger lineage. The second hypothesis is more likely given the differences in size and morphology and the orientation (adaxial vs. abaxial) of the Heliconia staminode compared with staminodes in the ginger clade.
Another developmental trend across Zingiberales is the reduction in the number of fertile stamens from five or six in the early diverging lineages to one or one half in the ginger clade. Heliconia has five fertile stamens like most of the members of the early diverging lineages (Musaceae, Lowiaceae and Strelitziaceae); thus, the position of Heliconiaceae as sister to the remaining Zingiberales does not alter interpretations of the overall trends in fertile stamen reduction. Flowers of Musaceae, as in Heliconia, can develop a single staminode rather than displaying complete abortion of the infertile stamen whorl primordium; however, only in Heliconia does the infertile stamen become petaloid. Thus, based on the Heliconia-sister topology in Fig. 1, the reduction to a single fertile stamen is a synapomorphy for the ginger clade and the petaloidy of the single staminode in Heliconia evolved independently of the prominent petaloidy in the stamen whorls of the ginger clade.
The origin of symmetry is an important characteristic in the evolution of the zingiberalean flower (Kirchoff, 1988; Rudall and Bateman, 2004; Bartlett and Specht, 2011). Interpretations given a Heliconia-sister topology (Fig. 1; FM-CO matrix) would agree with previous hypotheses with corolla and stamen whorl zygomorphy as ancestral in Zingiberales (Bartlett and Specht, 2010), although the developmental processes leading to the bilateral symmetry may be interpreted differently. The Heliconia flower has a zygomorphic calyx at maturity due to the free adaxial (posterior) sepal, and the androecium is zygomorphic due to the suppression of fertility and laminar expansion of the posterior (oblique adaxial) member of the outer whorl (Kirchoff et al., 2009; Bartlett and Specht, 2011). Heliconia flowers are the only flowers of the order with oblique zygomorphy (Kirchoff et al., 2009; Bartlett and Specht, 2011). Thus, based on the Heliconia-sister topology, oblique zygomorphy would have been derived independently in Heliconiaceae after it diverged from the remaining Zingiberales. Zygomorphy of the androecium due to suppression of an adaxial stamen of the inner whorl would be a synapomorphy of the Musaceae/Lowiaceae/Strelitziaceae clade recovered in this analysis.
Character information and support for deep relationships in Zingiberales
Weak support for deep relationships among the major clades of Zingiberales is associated with short internal branch lengths for the deepest divergences among lineages (relative to those in the rest of the phylogenetic trees; Fig. 1). More than a quarter of the 20 992 PICs (5786, or 27·6 %) in the FM-CO matrix (i.e. among all monocots for protein-coding loci) are within Zingiberales (Table 2). A visual assessment of branch lengths in Zingiberales (Fig. 1) shows that only a small proportion of all potentially informative characters comprise the deepest internal branches of the tree, whereas branches leading to representatives of each major clade/family (e.g. Costaceae, Musaceae and Zingiberaceae) account for the majority. Thus, the entire coding portion of the plastome provides little phylogenetic information for the deep structure of Zingiberales.
Conflict among the plastome data
There is some evidence for conflict among the relatively few characters supporting relationships deep in the phylogenetic tree for Zingiberales. For example, a measure of resampling support that specifically accounts for conflict by incorporating information about the next most frequent contradictory group among replicates (GP/C jackknife support; Goloboff et al., 2003) reveals lower support values than standard, or ‘absolute’ jackknife values in some areas of the tree (Fig 1). Specifically, jackknife percentage differences between these two metrics (absolute vs. GP/C support) are 15 and 16 % for (Heliconiaceae, remaining Zingiberales) and (Musaceae (Lowiaceae, Strelitziaceae)), respectively. There is further evidence of conflict, in that the data could not reject most of the phylogenetic hypotheses from various analyses in this study (Table 3).
The combination of low information content of the coding portion of the plastome for deep branches of the phylogenetic tree for Zingiberales and conflict among the data prevents the recovery of a robustly supported plastid phylogenetic hypothesis. These findings illustrate the challenges associated with ancient (probably Cretaceous; Kress and Specht, 2006; Magallon and Castillo, 2009), rapid radiations that characterize many groups of monocot angiosperms and other highly diverse clades (Freudenstein et al., 2004; Baker et al., 2009; Steele et al., 2012; Barrett et al., 2013). These findings exemplify the limitations of protein-coding genes of the plastome to provide sufficient homoplasy-free data to resolve completely deep relationships of the plant tree of life. This counters the notion that sequencing massive amounts of data for a limited number of extant taxa will allow ‘true signal’ to swamp out conflict in difficult areas of the tree of life. For resolving short internal branches inherent to an ancient rapid radiation, evidence from the fossil record may be required to tease apart the order of divergence events.
Effects of taxon sampling and partitioning
Inclusion/exclusion of taxa outside Zingiberales, inclusion/exclusion of process data partitions (including different ML model partitioning schemes) and choice of reconstruction method all affected topology; moreover, they did so in a generally unpredictable manner (Figs 2 and 3). When all other variables were kept equal, manipulation of extra-zingiberalean taxon sampling and data partition inclusion/exclusion both resulted in differing topologies (i.e. within each ‘category’ there were two different topologies observed); analysing the data under MP vs. ML and under different ML partitioning schemes caused differences in topology in only some instances (Figs 2 and 3). In no instances was there robustly supported conflict among various data configurations, which would have been illustrated by alternative topologies having high bootstrap or jackknife values. No one form of manipulation (inclusion or exclusion of representative Poales, codon positions 1 and 2 vs. position 3, MP vs. ML, etc.) seemed to influence topology most heavily. The only clearly consistent pattern was for the codon-partitioned ML analysis of all four CO matrices that yielded topology Pattern 1 for all taxon sets (Figs 2 and 3).
Signal saturation among the data is not sufficient to explain differing topologies and lack of robust support for the deepest branches in Zingiberales (Table 4; Fig. 4). A pattern of saturation is expected to appear as a prominent ‘plateau’ when plotting model-corrected distances vs. uncorrected distances, especially in comparisons of the most divergent taxa, representing the point at which phylogenetic signal is overwhelmed by noise. Neither the FM-CO nor ZC-CO matrices strongly conformed to this expectation, and no significant patterns of saturation were detected.
These findings suggest complex interactions among taxon samples, data partitions (and partition models) and reconstruction methods. If support for deep relationships were strong, one would expect to see resilience in the face of these various data manipulations, but, instead, they have a definite, albeit difficult to quantify, effect on topology. One might expect this to be the case for areas of a tree with low support and short internal branches. These findings make a strong case for the need to investigate thoroughly the influence of various data configurations on topology and support, and the need for sampling additional representatives of each family. Inclusion of additional members of Heliconiaceae (i.e. other Heliconia spp.), Musaceae (Ensete, additional Musa, Musella), Strelitziaceae (Strelitzia, Phenakospermum) and Lowiaceae (additional Orchidantha) may improve resolution and support for the deepest nodes of Zingiberales. It is of interest to compare relatively complete and reduced taxon sets to assess the effects of limited taxon sampling on branch support for short internal internodes. Increased taxon sampling is also desirable for members of Commelinales, the closest set of outgroup taxa, which in Fig. 1 occupy relatively long terminal branches.
In addition to the primary goal of sampling plastomes for additional taxa in each family, future analyses in Zingiberales should include sequences from spacer and intron regions, insertion/deletion characters and whole-plastome characters (e.g. gene order or inversions). All of these could provide additional character information to bolster support for deep nodes. Recently developed analytical approaches, e.g. estimating the optimal number of evolutionary process partitions from the data and models incorporating heterotachy [Pagel and Meade, 2004; e.g. as implemented recently in Malpighiales (Xi et al., 2012)], may hold promise for some traditionally difficult nodes. Other approaches, such as coalescent-based analyses of multiple, unlinked nuclear loci (including the plastome as a single, linked molecule), may also be beneficial, if incomplete sorting of ancestral polymorphisms during the early radiation of the principal lineages of Zingiberales is a primary cause of conflict. Regardless, it is apparent that with increased taxon sampling, genome-scale nuclear data (transcriptomes, exomes, etc.) in such groups as Zingiberales should be used in combination with complete plastomes, mitochondrial genomes, morphology and data from the rich fossil record to gain a more holistic understanding of phylogeny and evolution of floral morphology in this group.
Conclusions
Plastid DNA data from 83 coding loci provided resolution and robust support for many clades within Zingiberales, but failed to provide support for the deepest nodes. Manipulation of taxon sets, data partitions and model configurations had substantial and unpredictable consequences for topology and support of deep nodes. The most likely explanation is a lack of character information for these nodes among the coding regions of the plastome. Plastid DNA has been and will continue to be extremely useful in plant phylogenetics (e.g. Soltis et al., 2000; Chase et al., 2006; Moore et al., 2010; Givnish et al., 2010; Parks et al., 2009, 2012; Steele et al., 2012; Xi et al., 2012; Barrett et al., 2013; Stull et al., 2013), and there will probably be a time when all extant (described) plant species will have at least one individual with a completely sequenced plastome. However, along with greatly advancing our understanding of plant phylogenetic relationships with robust support, a growing number of studies (e.g. Givnish et al., 2010; Moore et al., 2010; Xi et al., 2012; Barrett et al., 2013; this study) are revealing limitations of the aligned coding regions of the plastome to resolve completely and provide support for recalcitrant areas of the plant tree of life associated with short branches and conflict among data.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Eric Antonieu, Raj Ayyampalayam, Patrick Edger, Amanda Fisher, Elena Ghiban, Mary Guisinger, John Kerry, Dustin Mayfield, Michael McKain, J. Chris Pires and the staff at Cold Spring Harbor Laboratory for analytical assistance and advice. We also thank the staff at Fairchild Tropical Botanical Garden/Montgomery Botanical Center (Florida, USA) and the New York Botanical Garden for permission to collect specimens. Research was funded by the National Science Foundation (USA) [awards DEB 0830020 to J.D., DEB 0830009 to J.L.M. and W.Z., and DEB 0829762 to D.S.].
LITERATURE CITED
- APG III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Almeida AMR, Brown A, Specht CD. Tracking the development of the petaloid fertile stamen in Canna indica: insights into the origin of androecial petaloidy in the Zingiberales. AoB PLANTS. 2013;5:plt009. doi: 10.1093/aobpla/plt009. doi:10.1093/aobpla/plt009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker WJ, Savolainen V, Asmussen-Lange CB, et al. Complete generic-level phylogenetic analyses of palms (Arecaceae) with comparisons of supertree and supermatrix approaches. Systematic Biology. 2009;58:240–256. doi: 10.1093/sysbio/syp021. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. American Journal of Botany. 2012;99:1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI, Leebens-Mack J, Conran JG, Stevenson DW. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics. 2013;29:65–87. doi: 10.1111/j.1096-0031.2012.00418.x. [DOI] [PubMed] [Google Scholar]
- Bartlett ME, Specht CD. Evidence for the involvement of GLOBOSA-like gene duplications and expression divergence in the evolution of floral morphology in the Zingiberales. New Phytologist. 2010;187:521–541. doi: 10.1111/j.1469-8137.2010.03279.x. [DOI] [PubMed] [Google Scholar]
- Bartlett ME, Specht CD. Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. American Journal of Botany. 2011;98:227–243. doi: 10.3732/ajb.1000246. [DOI] [PubMed] [Google Scholar]
- Baurain D, Brinkmann H, Philippe H. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors? Molecular Biology and Evolution. 2007;24:6–9. doi: 10.1093/molbev/msl137. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Goldman DH, Freudenstein JV, Pridgeon AM, Cameron KM, Chase MW. An overview of the phylogenetic relationships within Epidendroideae inferred from multiple DNA regions and recircumscription of Epidendreae and Arethuseae (Orchidaceae) American Journal of Botany. 2005;92:613–624. doi: 10.3732/ajb.92.4.613. [DOI] [PubMed] [Google Scholar]
- Chase MW, Fay MF, Devey DS, et al. Multigene analyses of monocot relationships: a summary. Aliso. 2006;22:63–76. [Google Scholar]
- Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Research. 2008;36 doi: 10.1093/nar/gkn502. e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren RMT, Clifford HT, Yeo PF. The families of the monocotyledons. Berlin: Springer-Verlag; 1985. [Google Scholar]
- Davis JI, Stevenson DW, Petersen G, et al. A phylogeny of the monocots, as inferred from rbcL and atpA sequence variation, and a comparison of methods for calculating jackknife and bootstrap values. Systematic Botany. 2004;29:467–510. [Google Scholar]
- Davis JI, McNeal JR, Barrett CF, et al. Contrasting patterns of support among plastid genes and genomes for major clades of the monocotyledons. In: Wilkin P, Mayo SJ, editors. Early events in monocot evolution. Systematics Association Special Volume Series. Cambridge: Cambridge University Press; 2013. pp. 315–349. [Google Scholar]
- Doyle J, Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS, Albert VA, Kallersjo M, Lipscomb D, Kluge AG. Parsimony jackknifing outperforms neighbor-joining. Cladistics. 1996;12:99–124. doi: 10.1111/j.1096-0031.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- Freudenstein JV, Davis JI. Branch support via resampling: an empirical study. Cladistics. 2010;26:643–656. doi: 10.1111/j.1096-0031.2010.00304.x. [DOI] [PubMed] [Google Scholar]
- Freudenstein JV, van den Berg C, Goldman DH, Kores PJ, Molvray M, Chase MW. An expanded plastid DNA phylogeny of Orchidaceae and analysis of jackknife branch support strategy. American Journal of Botany. 2004;91:149–157. doi: 10.3732/ajb.91.1.149. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Ames M, McNeal JR, et al. Assembling the tree of the monocotyledons: plastome sequence phylogeny and evolution of Poales. Annals of the Missouri Botanical Garden. 2010;97:584–616. [Google Scholar]
- Goloboff PA, Farris JS, Källersjö M, Oxelman B, Ramírez MJ, Szumik CA. Improvements to resampling measures of group support. Cladistics. 2003;19:324–332. [Google Scholar]
- Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- Gorniak M, Paun O, Chase MW. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Molecular Phylogenetics and Evolution. 2010;56:784–795. doi: 10.1016/j.ympev.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Graham SW, Zgurski JM, McPherson MA, et al. Robust inference of monocot deep phylogeny using an expanded multigene plastid data set. Aliso. 2006;21:3–20. [Google Scholar]
- Harris RS. Improved pairwise alignment of genomic DNA. USA: Pennsylvania State University; 2007. PhD Thesis. [Google Scholar]
- Heath TA, Zwickl DJ, Kim J, Hillis DM. Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Systematic Biology. 2008;57:160–166. doi: 10.1080/10635150701884640. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Pollock DD, McGuire JA, Zwickl DJ. Is sparse taxon sampling a problem for phylogenetic inference? Systematic Biology. 2003;52:124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences, USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen T, Bremer K. The age of major monocot groups inferred from 800+ rbcL sequences. Botanical Journal of the Linnean Society. 2004;146:385–398. [Google Scholar]
- Kirchoff BK. Floral ontogeny and evolution in the ginger group of the Zingiberales. In: Leins P, Tucker SC, Endress PK, editors. Aspects of floral development. Berlin: J. Cramer; 1988. pp. 45–56. [Google Scholar]
- Kirchoff BK. Homeosis in the flowers of the Zingiberales. American Journal of Botany. 1991;78:833–837. [Google Scholar]
- Kirchoff BK. A model of floral development and evolution in the Zingiberales. Journal of Cellular Biochemistry. 1993:4. Suppl. 17B. [Google Scholar]
- Kirchoff BK, Lagomarsino LP, Newman WH, Bartlett ME, Specht CD. Early floral development of Heliconia latispatha (Heliconiaceae), a key taxon for understanding the evolution of flower development in the Zingiberales. American Journal of Botany. 2009;96:580–593. doi: 10.3732/ajb.0800305. [DOI] [PubMed] [Google Scholar]
- Kress WJ. The phylogeny and classification of the Zingiberales. American Journal of Botany. 1986;73:744–745. [Google Scholar]
- Kress WJ. The phylogeny and classification of the Zingiberales. Annals of the Missouri Botanical Garden. 1990;77:698–721. [Google Scholar]
- Kress WJ, Specht CD. The evolutionary and biogeographic origin and diversification of the tropical monocot order Zingiberales. In: Columbus JT, Friar EA, Porter JM, Prince LM, Simpson MG, editors. Monocots: comparative biology and evolution (excluding Poales). Claremont: Rancho Santa Ana Botanic Garden; 2006. pp. 619–630. [Google Scholar]
- Kress WJ, Prince LM, Hahn WJ, Zimmer EA. Unraveling the evolutionary radiation of the families of the Zingiberales using morphological and molecular evidence. Systematic Biology. 2001;50:926–944. doi: 10.1080/106351501753462885. [DOI] [PubMed] [Google Scholar]
- Kress WJ, Prince LM, Williams KJ. The phylogeny and a new classification of the gingers (Zingiberaceae): evidence from molecular data. American Journal of Botany. 2002;89:1682–1696. doi: 10.3732/ajb.89.10.1682. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Moriarty EC. The importance of proper model assumption in Bayesian phylogenetics. Systematic Biology. 2004;53:265–277. doi: 10.1080/10635150490423520. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler Transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2007 Version 2.0, http://mesquiteproject.org . [Google Scholar]
- Magallon S, Castillo A. Angiosperm diversification through time. American Journal of Botany. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE) 2010:1–8. 14 November 2010, New Orleans. [Google Scholar]
- Milne I, Bayer M, Cardle L, et al. Tablet – next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences, USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proceedings of the National Academy of Sciences, USA. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai T. Notulae ad plantas Asiae Orientalis (XVI) Japanese Journal of Botany. 1941;17:189–203. [Google Scholar]
- Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Systematic Biology. 2004;53:571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A. Separating the wheat from the chaff: mitigating the effects of noise in a plastome phylogenomic data set from Pinus L. (Pinaceae) BMC Evolutionary Biology. 2012;12:100. doi: 10.1186/1471-2148-12-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OG. Musaceae, Zingiberaceae, Cannaceae, Marantaceae. In: Engler A, Prantl K, editors. Die natürlichen Pflanzenfamilien. Leipzig: W. Engelmann; 1889. pp. 1–43. 1887. [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biology. 2011;9 doi: 10.1371/journal.pbio.1000602. e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B. Identifiability of parameters in MCMC Bayesian inference of phylogeny. Systematic Biology. 2002;51:754–760. doi: 10.1080/10635150290102429. [DOI] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022594. e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratan A. Assembly algorithms for next-generation sequence data. Pennsylvania State University: USA; 2009. PhD Thesis. [Google Scholar]
- Rodriguez-de la Rosa RA, Cevallos-Ferriz SRS. Upper Cretaceous Zingiberalean fruits with in situ seeds from southeastern Coahuila, Mexico. International Journal of Plant Sciences. 1994;155:786–805. [Google Scholar]
- Rudall PJ, Bateman RM. Evolution of zygomorphy in monocot flowers: iterative patterns and developmental constraints. New Phytologist. 2004;162:25–44. [Google Scholar]
- Seberg O, Petersen G, Davis JI, et al. Phylogeny of the Asparagales based on three plastid and two mitochondrial genes. American Journal of Botany. 2012;99:875–889. doi: 10.3732/ajb.1100468. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution. 1999;16:1114–1116. [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Simmons MP, Freudenstein JV. Spurious 99 % bootstrap and jackknife support for unsupported clades. Molecular Phylogenetics and Evolution. 2011;61:177–191. doi: 10.1016/j.ympev.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Smith JF, Kress WJ, Zimmer EA. Phylogenetic analysis of the Zingiberales based on rbcL sequences. Annals of the Missouri Botanical Garden. 1993;80:620–630. [Google Scholar]
- Soltis DE, Soltis PS, Chase MW, et al. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Botanical Journal of the Linnean Society. 2000;133:381–461. [Google Scholar]
- Soltis DE, Smith SA, Cellinese N, et al. Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The origin and diversification of angiosperms. American Journal of Botany. 2004;91:1614–1626. doi: 10.3732/ajb.91.10.1614. [DOI] [PubMed] [Google Scholar]
- Specht CD, Yockteng R, Almeida AM, Kirchoff BK, Kress WJ. Homoplasy, pollination, and emerging complexity during the evolution of floral development in the tropical gingers (Zingiberales) Botanical Review. 2012;78:440–462. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006a;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. Phylogenetic models of rate heterogeneity: a high performance computing perspective. Proceedings of 20th IEEE/ACM International Parallel and Distributed Processing Symposium (IPDPS2006), High Performance Computational Biology Workshop. 2006b Rhodos, April 2006. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Steele PR, Hertweck KL, Mayfield D, McKain MR, Leebens-Mack J, Pires JC. Quality and quantity of data recovered from massively parallel sequencing: examples in Asparagales and Poaceae. American Journal of Botany. 2012;99:330–348. doi: 10.3732/ajb.1100491. [DOI] [PubMed] [Google Scholar]
- Stull GW, Moore MJ, Mandala VS, et al. A targeted enrichment strategy for massively parallel sequencing of angiosperm plastid genomes. Applications in Plant Sciences. 2013;1:1200497. doi: 10.3732/apps.1200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. Maximum-likelihood estimation of phylogeny from DNA sequence data. Methods in Enzymology. 2005;395:757–779. doi: 10.1016/S0076-6879(05)95039-8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson PB. Phylogeny of the Scitamineae – morphological and anatomical considerations. Evolution. 1962;16:192–213. [Google Scholar]
- Tomlinson PB. Anatomy of the monocotyledons. Oxford: Clarendon Press; 1969. III. Commelinales-Zingiberales. [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Lockhart PJ. Deciphering ancient rapid radiations. Trends in Ecology and Evolution. 2007;22:258–265. doi: 10.1016/j.tree.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Winkler H. Musaceae. In: Engler A, editor. Die natürlichen Pflanzenfamilien. Leipzig: W. Engelmann; 1930. pp. 505–541. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Xi Z, Ruhfel BR, Schaefer H, Amorim AM, et al. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proceedings of the National Academy of Sciences, USA. 2012;109:17519–17524. doi: 10.1073/pnas.1205818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XH, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. Journal of Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Xia XH, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 2003;26:1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomlefer WB. Guide to flowering plant families. Chapel Hill, NC: University of North Carolina Press; 1994. [Google Scholar]
- Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Systematic Biology. 2002;51:588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.