Summary
Hierarchical models (HM) have been used extensively in multisite time series studies of air pollution and health to estimate health effects of a single pollutant adjusted for other pollutants and other time-varying factors. Recently, Environmental Protection Agency (EPA) has called for research quantifying health effects of simultaneous exposure to many air pollutants. However, straightforward application of HM in this context is challenged by the need to specify a random-effect distribution on a high-dimensional vector of nuisance parameters. Here we introduce reduced HM as a general statistical approach for analyzing correlated data with many nuisance parameters. For reduced HM we first calculate the integrated likelihood of the parameter of interest (e.g. excess number of deaths attributed to simultaneous exposure to high levels of many pollutants), and we then specify a flexible random-effect distribution directly on this parameter. Simulation studies show that the reduced HM performs comparably to the full HM in many scenarios, and even performs better in some cases, particularly when the multivariate random-effect distribution of the full HM is misspecified. Methods are applied to estimate relative risks of cardiovascular hospital admissions associated with simultaneous exposure to elevated levels of particulate matter and ozone in 51 US counties during 1999–2005.
Keywords: Air pollution, Multilevel models, Multisite time series data, Nuisance parameters, Random effects
1 Introduction
The US Environmental Protection Agency (EPA) estimated that thousands of premature deaths and hundred of thousands cases of illness may be avoided by reducing pollution (EPA, 2011). Most epidemiological studies of air pollution and health have estimated the health effects associated with ambient exposure to individual pollutants adjusted for exposure to other pollutants and confounders. However, National Research Council (NRC) has recently questioned whether the current approach of setting separate National Ambient Air Quality Standards (NAAQS) for each of the six criteria pollutants adequately protects population health, as this approach may greatly under-estimate risk (NRC, 2004). To meet the challenges of the NRC recommendations, new statistical methods are needed to account for multiple exposures and their interactions.
Previous multisite time series studies of the health effects of air pollution have estimated risks associated with exposure to a single pollutant. Dominici et al. (2000) developed a two-stage hierarchical model to combine information across locations on the association between daily changes of a given pollutant and daily changes in the health outcome, adjusted for other pollutants and confounders. This approach has been applied to several national US studies for estimating independent associations of various pollutants of epidemiologic interest with different health outcomes, including mortality and cardiovascular and respiratory emergency hospital admissions (Dominici et al., 2006; Bell et al., 2004; Peng et al., 2008, 2009). Two-level random-effect models have also been used to estimate health effects of exposure to individual pollutants and to identify factors that explain heterogeneity in the health risks across European cities (Katsouyanni et al., 2001). Addressing the potential for biased estimates due to measurement error of correlated exposures in multipollutant models, Zeka and Schwartz (2004) applied methodology developed by Schwartz and Coull (2003) to estimate independent effects of individual pollutants that minimizes the impact of measurement error.
To estimate the health effects of simultaneous exposure to multiple pollutants, we specify a hierarchical model (HM) that, at the first stage, flexibly specifies the air pollution-health outcome risk surface by incorporating interactions among pollutants and allowing for smooth nonlinear functions of pollutant concentrations. In the full HM, we define βi to be the random effects describing the association between the health outcome and the multiple exposure variables included in the regression model (e.g. nonlinear functions of main effects and interactions of pollution variables and potential confounders) for the ith location. The parameter of primary scientific interest (θi) is the increased health risk when daily ambient levels of the pollutants considered are simultaneously above their national standards compared to when daily levels are below their national standards. Our goals are to obtain more precise estimates of θi by borrowing strength across locations, to estimate overall regional or national risks θ*, and to identify site-specific factors (e.g. population demographics, traffic patterns, long-term averages of other pollutants) that modify the association between simultaneous exposure to multiple pollutants and adverse health outcomes.
More generally, the hierarchical modeling approaches we consider apply to problems where the parameter of interest θi can be defined as a known function of βi where dim(βi) ≫ dim(θi). Many difficulties may arise upon implementation of standard Generalized Linear Mixed Models (GLMM) or full HM in the presence of a high-dimensional vector of random effects (βi). First, one must specify a multivariate random-effect distribution on the full vector βi, which might not be of primary scientific interest. There is an extensive literature on the consequences of misspecification of random-effect distributions in GLMM (Verbeke and Lesaffre, 1997; Heagerty and Kurland, 2001; Litière et al., 2008; Agresti et al., 2004). Though small to moderate misspecification of the random-effect distribution may not have a large impact in the estimation of fixed effects, there are situations for which misspecification can result in efficiency loss and biased estimates of the random effects (Neuhaus et al., 1992; Heagerty and Kurland, 2001; Agresti et al., 2004; Litière et al., 2010; McCulloch and Neuhaus, 2011). Several approaches have been proposed for specifying flexible semi- or non-parametric distributions for the random effects (Laird, 1978; Magder and Zeger, 1996; Komárek and Lesaffre, 2008; Gallant and Nychka, 1987; Chen et al., 2002). However, most of these approaches cannot be implemented in the context of a high-dimensional vector of random effects, and the validity of the assumption on the random-effect distribution is sometimes difficult to verify (Agresti et al., 2004; Litière et al., 2008). Second, if one is interested in estimating effect modification, at the second stage the full HM presents the additional challenge of specifying a high-dimensional multivariate regression model. Third, implementing diagnostic methods for misspecification of a multivariate random-effect distribution can be very challenging. Fourth, it may be computationally intensive and/or challenging to implement an MCMC sampler that mixes well and converges quickly to the stationary distribution as the number of random effects increases.
In this paper, we introduce reduced hierarchical models as a general statistical approach for eliminating nuisance parameters in hierarchical models with a large number of random effects. The reduced HM combines information across clusters (e.g. locations) directly on the parameter of interest θi. At the first stage, we calculate an integrated likelihood for θi, and at the second stage, we specify a flexible random-effect distribution directly on the θi. Reduced HM overcome many of the practical challenges in the specification and implementation of full HM in the context of a high-dimensional vector of nuisance parameters. Though developed to study health effects of simultaneous exposure to multiple pollutants, reduced HM are widely applicable for other studies of multiple exposures, and in general to clustered datasets with a large number of nuisance parameters. Accordingly, much of the methods section is presented in a general context while maintaining a close connection to the scientific motivation for this work.
Previous studies have used likelihoods of the parameter of interest at the first stage of a hierarchical model for conducting a meta analysis of randomized trials of a treatment for stomach ulcers (Efron, 1996; Liao, 1999). Specifically, Efron (1996) used a conditional likelihood for the clinical trial-specific log odds ratio (θi) and developed empirical Bayes methods for combining the likelihoods in order to conduct inference (interval estimation) on the θi. Liao (1999) also eliminated nuisance parameters at the first stage using conditional likelihoods, but he modeled the θi using a Bayesian approach, assuming a normal random-effect distribution for the θi. In these two studies, the vector of cluster-specific parameters βi is just two-dimensional, and a conditional likelihood for θi is available in closed form. While not explicitly defining a likelihood function to eliminate nuisance parameters at the first stage of the HM, Warn et al. (2002), building upon the work by Smith et al. (1995), reparameterized the cluster-specific parameters βi as (λi, θi), where θi is the parameter of interest, and then proposed to use noninformative priors for the nuisance parameter λi, which were assumed to be independent across clusters. However, it may not always be possible to define such a reparametrization (e.g., if θi is a complex function of βi), and this approach still requires sampling the nuisance parameter λi at each iteration of the MCMC, which can become computationally expensive when the dimension of λi is large. In this paper we generalize parameter reduction for HM to very general situations where (1) no conditional or marginal likelihood is available; (2) an integrated likelihood is not available in closed form; (3) there does not exist a reparametrization (λi; θi) of the within-cluster parameter space; (4) the second-level model includes cluster-specific covariates; and (5) flexible specifications of the random-effect distribution are desired. This generalization is referred to as reduced HM. Additionally, while there are several practical advantages of the reduced HM arising from the elimination of nuisance parameters at the first stage, even in the specific context where this approach has been applied previously (2-dimensional setting with conditional likelihood available in closed form), there is a lack of evidence supporting the reduced HM as performing competitively with the full HM across a range of scenarios. To address this gap, we will provide a critical evaluation of the reduced HM as an alternative to fitting the full HM in a series of simulation studies.
In Section 2, we describe the multisite time series data used to estimate the health risks associated with simultaneous exposure to multiple pollutants. In Section 3, we describe the level-one model of an HM aimed at estimating the association between joint exposure to ozone and fine particulate matter and hospital admissions. In Section 4, we introduce the reduced HM in a general setting where an integrated likelihood is estimated for each cluster and a flexible random-effect distribution is specified directly on the cluster-specific parameter of interest. Section 5 describes our simulation study. In Section 6, we present our results from the data analysis. We provide discussion and concluding remarks in Section 7.
2 Data
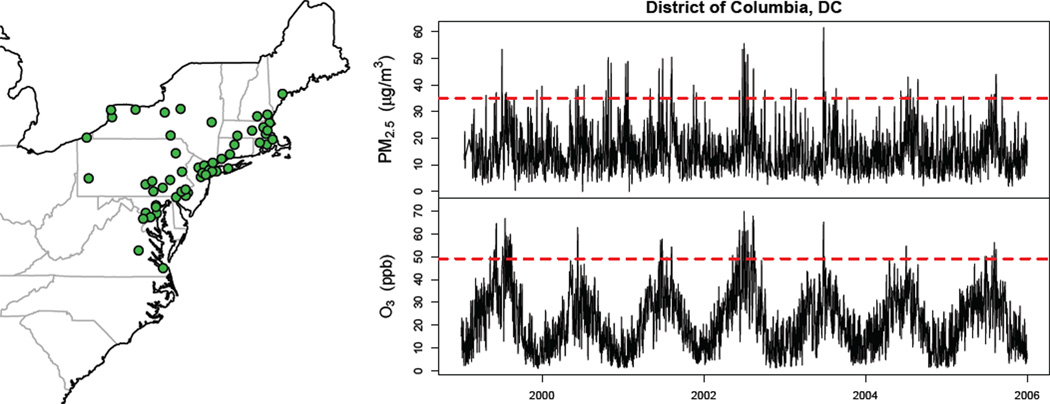
We used data from a national database consisting of parallel time series from 60 counties in the northeastern United States during the period 1999–2005. Daily counts of emergency hospital admissions for cardiovascular diseases (CVD), which comprise heart failure (ICD-9 code 428), heart rhythm disturbances (426–427), cerebrovascular events (430–438), ischemic heart disease (410–414, 429), and peripheral vascular disease (440–448) were obtained from billing claims of US Medicare enrollees. CVD admissions were stratified by two age categories, 65–74 and ≥ 75. Concentrations of fine particulate matter (PM2.5; units µg/m3) and ozone (O3; units parts per billion), which for many counties are measured on either a 1-in-3 or 1-in-6 day schedule, were obtained from the US EPA’s Air Quality System. Daily temperature and dewpoint temperature were obtained from the National Climatic Data Center. Among the 60 northeastern US counties with available data, we considered the 51 counties having at least 100 days where PM2.5 and O3 were measured concurrently, as well as at least one day when both pollutants were above their national standard (defined below). Figure 1 shows a map of the locations, as well as example time series of PM2.5 and O3 for Washington, DC.
Figure 1.
Left panel shows a map of the 51 northeastern US counties used for a multi-site time series study of the association between joint exposure to PM2.5 and O3 and hospitalization for cardiovascular diseases. Right panel shows daily times series of PM2.5 (in µg/m3) and O3 (in parts per billion; ppb) for the District of Columbia for the period 1999–2005. Horizontal lines correspond to the daily national standards for each pollutant.
3 Poisson regression model for multiple pollutants
In this section we describe the first level of an hierarchical model for estimating health effects associated with simultaneous exposure to fine particulate matter (PM2.5) and ozone (O3). We assume for county i on day j for age group k, the number of CVD admissions yijk has a Poisson distribution with mean model
| (1) |
where nijk is the number of individuals of the kth age group at risk, and ns(·) denotes natural cubic splines with the specified degrees of freedom (df) and bij (j = 1, 2) and γij (j = 3,…, 7) representing the spline coefficients. The product of the cubic spline bases for PM2.5 and O3, which includes both main effects and interaction terms, provides a flexible specification of the unknown joint pollutant-hospital admissions exposure-response surface. Here age denotes an indicator for being in the ≥ 75 age category (versus 65–74); dow is a vector of indicator variables for day of week; is the current day’s (average of the previous three days’) average temperature; and is the current day’s (average of the previous three days’) average dew point temperature. The smooth function of calendar time ns(j; 7 df/year, γi7) accounts for seasonality and longer-term, time-varying trends in hospital admissions.
This within-county model extends those developed to study PM2.5 and O3 individually (Dominici et al., 2006; Bell et al., 2004) by allowing for nonlinear associations of each of the pollutants and their interaction. In particular, the choice of covariates and df in the smooth functions are based on those used by Dominici et al. (2006). Previous studies have assessed the sensitivity of health effect estimates from single-pollutant models to adjustment for temperature and the smooth function of calendar time, finding that results were robust across specifications of the confounder model (Peng et al., 2006; Welty and Zeger, 2005).
To place model (1) within the more general context of HM for two-level clustered data, we introduce some notation. Let bi = (bi1, bi2) be the vector of random effects for the exposure-response surface characterizing the relation between joint exposure to ozone and fine particulate matter and the health outcome. Let γi = (γi0, γi1,…, γi7) be the vector of random effects describing the association between the confounders and the health outcome, and define βi = (bi, γi). Note that these random effects are introduced in order to model variation across counties, not as a random-effects parameterization of penalized splines (the number of df in the spline terms is fixed). Let xij denote the full vector of covariate data for day j in county (cluster) i, and let denote the 15-dimensional subvector of xij that is the concatenation of the basis terms for the main effects and interactions of the spline bases for ozone and fine particulate matter, ns(PM2.5ij; 3 df, bi1) · ns(O3ij; 3 df, bi2).
We next define a variable that identifies whether the daily levels of either PM2.5 and/or O3 are above or below their corresponding 24-hour National Ambient Air Quality Standards (NAAQS),
The values 35 µg/m3 and 0.049 ppm were derived from the NAAQS, which are defined in Appendix A of the Supplementary Materials.
We define θi to be the log of the expected number of CVD admissions on days when both PM2.5 and O3 are above their respective national standards divided by the expected number of CVD admissions on days when both pollutants are lower than their national standards, adjusted for the potential confounding variables:
| (2) |
Here NiA (NiD) are the number of days when both pollutants are above (below) their respective national standards in county i during the study period 1999–2005. Derivation of the formulation for the parameter of interest is in Appendix B of the Supplementary Materials. Other definitions of θi that may be of interest, such as the log of the expected number of CVD admissions on days when only PM2.5 (or when only O3) is above its national standard divided by the expected number of CVD admissions on days when both pollutants are lower than their national standards could be defined similarly and the same methods (described below) could be straightforwardly applied.
4 Reduced hierarchical model
Rather than specify a full HM on the large number of random effects βi, we define a reduced HM directly on the parameter of interest θi:
| (3) |
Here Li(θi) denotes a likelihood function (detailed below) and RE(θi|α) denotes an arbitrary random-effect distribution. Note that the likelihood function in general depends on the vector of outcome data from the ith cluster yi and on the set of covariate data xi, though we suppress this dependency in our notation. To conduct inference in the Bayesian framework, a prior distribution is placed on α.
The reduced HM may be further generalized by allowing the random-effect distribution RE(θi|α) to depend on cluster-level covariates zi, in order to study potential effect modification. In particular, for the second-stage model we assume and place the random-effect distribution on the α0i. The second level model may also be extended to allow the θi to be spatially correlated across clusters.
4.1 Integrated Likelihood
In the general setting where the parameter of interest θi is a complicated function of the level 1 parameters βi as in (2), we propose to use an integrated likelihood for Li(θi). For notational simplicity the cluster-specific subscript i is suppressed in what follows. An integrated likelihood for the ith cluster may be expressed as
| (4) |
where πθ(θ) is the prior distribution for θ and fθ|Y is the corresponding posterior distribution of θ based on the data from only that cluster. Note that in the special case where the cluster-specific parameters β can be reparametrized as (θ,λ), this expression can be rewritten as fy|θ(y|θ) = ∫ fy|θ,λ (y|θ, λ) πλ|θ(λ|θ)dλ, where fy|θ,λ is the joint likelihood, and πλ|θ is the prior density of λ given θ (Berger et al., 1999).
When such a reparametrization of β is not available or when fy|θ(y |θ) is not available in closed form, we propose a simulation approach to approximate (4) as follows:
Assign a prior distribution to the vector β of level-1 parameters, such that the induced prior distribution πθ(θ) on θ = h(β; x) is diffusely spread out over the range of plausible values for θ. Simulate R prior samples from πθ(θ).
Fit a within-cluster model to generate R samples β(r) from the posterior fβ|y(β|y).
Obtain the posterior samples θ(r) = h(β(r); x).
Select a grid of points {θk} covering the range of θ and apply a Gaussian kernel smoother to estimate both fθ|y (θ|y) and πθ(θ) on this grid.
We repeat this process for each cluster i to obtain approximations f̂yi|θi (yi | θi), i = 1,…, I. Note that the choice of prior distribution for β in Step 1 will depend on the form of the function h. Also note that while this procedure requires drawing from the posterior fβi|yi (βi | yi), since this is done within each cluster independently, the sampling is greatly simplified as compared to fitting the full HM where the βi are correlated across clusters (i.e. sampling from fβ1,…,βI |y1,…,yI (β1,…, βI |y1,…, yI)). In addition, since this step is performed a single time prior to fitting the reduced HM, estimating the parameters of the reduced HM remains fast. Further details of our implementation are in Appendix C of the Supplementary Materials.
4.2 Dirichlet process mixture model for RE(θi|α)
To allow for flexible specification of the random-effect distribution we propose to use a Dirichlet process mixture model for RE(θi|α). The Dirichlet process mixture model (Ferguson, 1973; Neal, 2000) can be expressed as the limit as the number of components K goes to infinity of the mixture model
where Discrete(ci | p1,…, pK) corresponds to the p.m.f. ℙ(ci = k) = pk (k = 1, …, K) and δ/K is the concentration parameter written so that it approaches 0 as K goes to infinity. Here we consider a normal mixture so that F(· | ϕc) = N(· | µc, τc), and we select the conjugate prior so that G0 = NormalGamma(λ, γ, a, b), i.e. τc ~ Gamma(τ | a, b) and µc | τc ~ N(λ, γτc).
4.3 Computational details
The reduced HM (3) may be fit using Markov Chain Monte Carlo (MCMC) methods (Metropolis et al., 1953; Gilks et al., 1995) to generate samples from the posterior distribution of the unknown parameters
where π(α) denotes the prior distribution on the vector of parameters of the random-effect distribution. At each iteration of the MCMC algorithm, a sample is drawn from the full conditional
| (5) |
for each cluster i. When the integrated likelihood has been estimated using the approach from Section 4.1, we replace Li(θi) in equation (5) by f̂ yi|θi (yi | θi). Since fc(θi) is not a known distribution, we sample from it by applying a Metropolis-Hastings step. In the Metropolis-Hastings step, we need to evaluate the likelihood f̂yi|θi at an arbitrary point θ. We do this by selecting the grid point θk that is closest to θ and evaluating the likelihood at that grid point.
For generating posterior samples of α when RE(θi | α) is the Dirichlet process mixture model defined in Section 4.2, we adapt an MCMC sampling algorithm described by Neal (2000). Details are in Appendix C of the Supplementary Materials.
5 Simulation study
There are instances for which the reduced HM may be preferred to the full HM due to practical considerations such as its simplified implementation and the ease with which prior information may be incorporated directly on the parameter of interest. However, a more thorough understanding of situations when the reduced HM works well is needed. In this section we conduct simulation studies to compare performance of the reduced HM to the full HM across a range of scenarios.
We base our studies on data from a meta-analysis of 41 randomized trials of a treatment for stomach ulcers, provided by Efron (1996). Rather than use the multipollutant case study as a basis for simulation studies, a meta-analysis example is used to highlight the broad utility of the reduced HM methodology across diverse applications. In addition, even in the simpler context of this application (two-dimensional vector of random effects βi) for which a full HM may be straightforwardly implemented, the relative performance of the reduced HM to the full HM is not well understood and, as we shall see, the full HM may not always be the optimal choice even in the low-dimensional case.
The data from the ith trial is {yi = (yi0, yi1), xi = (ni0, ni1)}, where yi0, yi1 are the number of occurrences of ulcers for the control and treatment groups, and ni0, ni1 are the number of subjects in the control and treatment groups, respectively. Let pi = (pi0, pi1) be the vector of probabilities of the occurrence of ulcers in the control and treatment groups. The distribution of the data from experiment (cluster) i is assumed to be , and the parameter of interest is the log odds ratio
| (6) |
In this example, a full HM would require the specification of a random-effect distribution for pi = (pi1, pi0). Alternatively, a commonly used specification first defines a one-to-one transformation of the pi into ℝ2 through the logit link and assumes a bivariate normal distribution for the random effects:
| (7) |
For a reduced HM, we first summarize the information contained in experiment i about the log odds ratio θi through a likelihood function, and we then specify a random-effect distribution directly on the θi. For this problem, a conditional likelihood for θi is available in closed form. By conditioning on the margins of the two-by-two table for each experiment, the conditional likelihood may be expressed as
| (8) |
We may then use for the likelihood function in the reduced HM (3). Computing integrated likelihoods for each of the randomized trials in the ulcer data set (Efron, 1996), we found them to be generally quite similar to the corresponding conditional likelihoods, and so only the conditional likelihoods were considered in the simulation study.
We simulated data under four data generating mechanisms, and we estimated model parameters under four HM formulations. We next describe each of the hierarchical modeling approaches used to fit the data, after which we detail the four data generating models.
5.1 Hierarchical models
We fit each simulated data set using four approaches: a full HM assuming the logistic model (7) with a normal random-effect distribution on the βi (FHM); a reduced HM using the conditional likelihood from equation (8) with a normal random-effect distribution on the θi (RHM-L-N); a reduced HM using the conditional likelihood from (8) with a flexible random-effect distribution on the θi (RHM-L-DP); and a reduced HM using a normal approximation to the likelihood with a normal random-effect distribution on the θi (RHM-N-N). For the flexible random-effect distribution, we considered the Dirichlet Process normal mixture model described in Section 4.2. For each approach, we estimated the cluster-specific log odds ratios θi as well as the overall log odds ratio θ* = 𝔼(θi), where the expectation is taken over all of the clinical trials included in the analysis. Additionally, we obtained 95% posterior intervals for the overall (θ*) and cluster-specific (θi) parameters. Details of estimation for each of the four models are in Appendix D of the Supplementary Materials.
5.2 Data generating models
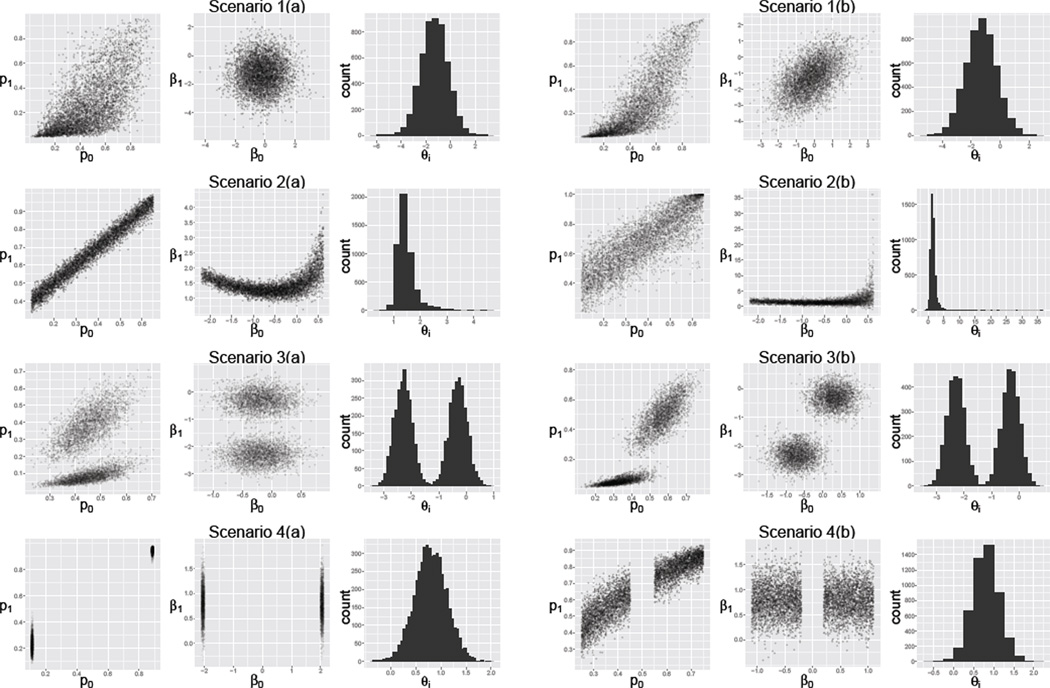
We considered four data generating models. We always assumed yi0 ~ Binom(ni0, pi0) and yi1 ~ Binom(ni1, pi1), and we selected different models for generating pi0 and pi1 (i = 1,…, I). Note that each model for generating pi0 and pi1 induces a distribution on the log odds ratio θi through (6). Thus, each time we generated a dataset, we obtained I values of the cluster-specific, true log odds ratios θi (one for each cluster i). The models were selected in order to distinguish among scenarios where the full HM is expected to outperform the reduced HM and vice versa. Figure 2 shows, for each of the four data generating models, the distribution of the (pi0, pi1), along with the corresponding distributions of the and the log odds ratios θi.
Figure 2.
Plots of simulated data under each scenario from four data-generating models. First row displays data from model 1, scenarios (a)–(b); second row shows data from model 2, scenarios (a)–(b); third row corresponds to model 3, scenarios (a)–(b); and fourth row to model 4, scenarios (a)–(b). For each scenario 5000 data points (pi0, pi1) are plotted, as well as the corresponding points (βi0, βi1) under the transformation logit(pik) = βi0 + βi1 I(k = 1), and histograms of the corresponding log odds ratios θi.
In each case, we set ni0 = ni1 = n, and we considered n = 100 for I = 100, 50, and 25. These parameter values were selected to correspond to a large within-cluster sample size for either a large, moderate, or small number of clusters.
Model 1 - Bivariate Normal
We generated data from
where , and we considered two different values for Σ,
(see scenarios 1(a) and 1(b) in Figure 2). These parameter values were selected to be the same order of magnitude of those from the ulcer data set. Since this model fully specifies a normal random-effect distribution on the βi, particularly in scenario 1(b) where a moderate correlation between the random effects is assumed, we expected it to favor the full HM (7).
Model 2 - Uniform/Beta
We generated pi0 ~ Uniform(0.1, 0.6) and pi1 | pi0 ~ Beta(m = pi0 + 0.3, ϕ), where the beta distribution is parametrized by its mean m and variance ϕ. We considered two values for ϕ, namely ϕa = 0.001 and ϕb = 0.01. Since this model is not based on either the full or reduced HM a priori we didn’t expect it to favor either of these two approaches (see scenarios 2(a) and 2(b) in Figure 2).
Model 3 - Normal Mixture
We generated (pi0, pi1) by
where we fixed β* = (−0.2; 1.3), α = 0.5, and Σ = diag{(0.1, 0.1)}. We considered two values for ν, namely . This data generating model was selected because the random-effect distribution will be misspecified for both the full and the reduced HM (since θi = βi1), when a normal random-effect distribution is assumed; thus, we expected neither approach to perform particularly well (see scenarios 3(a) and 3(b) in Figure 2).
Model 4 - Normal-θi
Finally, we generated data by first simulating values for the log odds ratios θi and for the log odds , which induces a distribution on the . In particular, we simulated θi ~ N(µ, σ2) and λi ~ 0.5U(−u2, −u1) + 0.5U(u1, u2), where we fixed µ = 0.8, σ2 = 10. We considered two scenarios for u1 and u2, namely (u1a, u2a) = (2, 2.1) and (u1b, u2b) = (0.2, 1.1). This model was chosen because it was expected to favor the reduced HM over the full HM, since the normal random-effect distribution on the (βi0, βi1)′ for the full HM will be misspecified, while the random-effect distribution for θi in the reduced HM will be correctly specified (see scenarios 4(a) and 4(b) in Figure 2).
5.3 Results
We evaluated the relative performance of the four modeling approaches (FHM, RHM-L-N, RHML-DP, and RHM-N-N) in estimating both the cluster-specific (θi) and overall (θ*) log odds ratios. Because disparity in performance across methods was attenuated for the smaller values for the numbers of clusters, in this section we focus our discussion on results for I = 100 (Table 1). Results for cases I = 25 and I = 50 are in Tables S1 and S2 of the Supplementary Materials.
Table 1.
Simulation results for the cluster-specific log odds ratios θi: squared error loss for the posterior mean estimates θ̃i (sq. error), and coverage of 95% posterior intervals. Results for the mean log odds ratio θ*: bias, standard deviation, and rMSE of the posterior mean estimates θ̃* and coverage of 95% posterior intervals. Methods compared are the full hierarchical model (FHM), reduced HM with conditional likelihood and normal random-effect distribution (RHM-L-N), reduced HM with normal approximation to the likelihood and normal random-effect distribution (RHM-N-N), and reduced HM with conditional likelihood and Dirichlet-Process normal mixture for the random-effect distribution (RHM-L-DP).
| Simulation | Cluster θi | Overall θ* | |||||
|---|---|---|---|---|---|---|---|
| Sq. Error | Coverage | Bias | SD | rMSE | Coverage | ||
|
Model 1: Bivariate Normal |
1(a)* | θ*=−1.3 | |||||
| (i) FHM | 14.4 | 0.95 | 0.00 | 0.11 | 0.11 | 0.94 | |
| (ii) RHM-L-N | 14.8 | 0.95 | 0.02 | 0.11 | 0.11 | 0.95 | |
| (iii) RHM-L-DP | 14.8 | 0.95 | 0.03 | 0.11 | 0.11 | 0.94 | |
| (iv) RHM-N-N | 18.0 | 0.94 | 0.09 | 0.10 | 0.14 | 0.89 | |
| 1(b)* | θ*=−1.3 | ||||||
| (i) FHM | 14.9 | 0.95 | −0.01 | 0.12 | 0.12 | 0.94 | |
| (ii) RHM-L-N | 18.9 | 0.94 | 0.04 | 0.11 | 0.12 | 0.93 | |
| (iii) RHM-L-DP | 18.9 | 0.94 | 0.06 | 0.11 | 0.12 | 0.92 | |
| (iv) RHM-N-N | 27.5 | 0.92 | 0.14 | 0.10 | 0.17 | 0.74 | |
|
Model 2: Uniform/ Beta |
2(a) | θ* = 1.46 | |||||
| (i) FHM | 7.5 | 0.88 | −0.03 | 0.04 | 0.05 | 0.87 | |
| (ii) RHM-L-N | 8.0 | 0.91 | −0.03 | 0.04 | 0.05 | 0.90 | |
| (iii) RHM-L-DP | 7.5 | 0.95 | −0.04 | 0.04 | 0.06 | 0.90 | |
| (iv) RHM-N-N | 9.6 | 0.89 | −0.07 | 0.04 | 0.08 | 0.66 | |
| 2(b) | θ*= 1.67 | ||||||
| (i) FHM | 99.5 | 0.90 | −0.14 | 0.08 | 0.16 | 0.55 | |
| (ii) RHM-L-N | 104.2 | 0.91 | −0.13 | 0.08 | 0.16 | 0.56 | |
| (iii) RHM-L-DP | 96.2 | 0.92 | −0.14 | 0.09 | 0.17 | 0.57 | |
| (iv) RHM-N-N | 137.6 | 0.89 | −0.23 | 0.07 | 0.24 | 0.11 | |
|
Model 3: Normal Mixture |
3(a) | θ*= −1.3 | |||||
| (i) FHM | 10.6 | 0.95 | −0.01 | 0.11 | 0.11 | 0.97 | |
| (ii) RHM-L-N | 11.6 | 0.95 | 0.00 | 0.11 | 0.11 | 0.97 | |
| (iii) RHM-L-DP | 9.8 | 0.96 | 0.02 | 0.10 | 0.11 | 1.00 | |
| (iv) RHM-N-N | 11.6 | 0.95 | 0.06 | 0.10 | 0.12 | 0.94 | |
| 3(b) | θ* = −1.3 | ||||||
| (i) FHM | 10.1 | 0.95 | −0.02 | 0.12 | 0.13 | 0.93 | |
| (ii) RHM-L-N | 13.5 | 0.95 | 0.02 | 0.12 | 0.12 | 0.94 | |
| (iii) RHM-L-DP | 12.1 | 0.96 | 0.03 | 0.12 | 0.12 | 0.99 | |
| (iv) RHM-N-N | 15.0 | 0.94 | 0.12 | 0.11 | 0.17 | 0.80 | |
|
Model 4: Normal-θi |
4(a) | θ*= 0.8 | |||||
| (i) FHM | 7.9 | 0.84 | 0.00 | 0.06 | 0.06 | 0.85 | |
| (ii) RHM-L-N | 7.2 | 0.93 | 0.01 | 0.06 | 0.06 | 0.94 | |
| (iii) RHM-L-DP | 7.3 | 0.97 | −0.01 | 0.06 | 0.06 | 0.96 | |
| (iv) RHM-N-N | 7.5 | 0.90 | −0.05 | 0.05 | 0.07 | 0.86 | |
| 4(b) | θ*= 0.8 | ||||||
| (i) FHM | 5.1 | 0.93 | 0.00 | 0.05 | 0.05 | 0.93 | |
| (ii) RHM-L-N | 5.2 | 0.94 | 0.00 | 0.05 | 0.05 | 0.94 | |
| (iii) RHM-L-DP | 5.2 | 0.96 | −0.01 | 0.04 | 0.05 | 0.94 | |
| (iv) RHM-N-N | 5.2 | 0.94 | −0.02 | 0.04 | 0.05 | 0.93 | |
For scenarios 1(a) and 1(b), the summary statistics for RHM-L-DP are based on 999 and 998 simulation repetitions, respectively. The other repetitions were excluded because the MCMC didn’t converge within the maximum number of iterations.
The main disparity in performance across the reduced HM (RHM-L-N and RHM-L-DP) and full HM approaches occurred for estimation of the cluster-specific parameters θi; methods (except RHM-N-N) performed comparably for estimating the overall θ*. The two situations where FHM yielded similar or slightly better cluster-specific estimates than the reduced HM were those for which the data generating model implied considerable correlation between β0i and β1i, which could be captured to varying degrees by the bivariate normal random-effect distribution on the βi. This occurred for data generating models 1(b) and 3(b), which had correlation of ≈ 0.5 and 0.8, respectively (see Figure 2). Because nuisance parameters are eliminated before pooling, the reduced HM do not take advantage of this correlation structure. For the other scenarios, the reduced HM generally performed comparably to or better than the FHM. Comparing the reduced HM with different random-effect distributions, we found that RHM-L-DP performed just as well or only slightly worse than RHM-L-N when the true distribution was normal (models 1(a)–(b) and 4(a)–(b)), but performed moderately better when the true random-effect distribution was non-normal (models 2(a)–(b) and 3(a)–(b)).
Across simulation scenarios we generally found that the model using the normal approximation to the likelihood (RHM-N-N), although most efficient computationally, was not competitive with the other approaches. For estimating θi, the RHM-N-N either performed comparably (scenarios 2(a), 3(a) and 4(a)–(b)), or moderately worse (scenarios 1(a)–(b), 2(b), and 3(b)) than the other approaches. For estimating the overall θ*, the RHM-N-N generally had larger rMSE and coverage markedly lower than the nominal rate (exceptions are scenarios 3(a) and 4(c)). One reason for the poor performance of RHM-N-N is that the normal approximation to the likelihood does not provide a good approximation in this application, particularly when yi1 or yi0 is equal to zero or n (which occurs most frequently under models 1(a)–(b) and 3(a), scenarios where RHM-N-N performs worst). In addition, we note that under scenario 2(b), none of the approaches performed particularly well for estimating the mean (θ*) of the highly skewed random-effect distribution for θi.
5.4 Conclusions
Our simulation studies were designed to assess the relative performance of the reduced HM to the full HM across different scenarios of misspecification of the random-effect distribution. We found that large correlation in the random effects βi generally led to slightly improved estimation of the cluster-specific θi by the full HM as compared to the reduced HM. However, in other scenarios, namely those for which the random-effect distribution for the full HM was misspecified, the reduced HM achieved superior performance. In addition, for estimating the overall θ* we found performance to be very similar across methods. Overall, in our simulation studies the reduced HM performed nearly as well as the full HM, and even performed better in some cases.
6 Application
We applied the reduced HM to our multisite time series study of 51 urban counties in the north-eastern US for the period 1999–2005. Our goal was to estimate the county-specific and overall log relative risks of emergency cardiovascular hospital admissions associated with levels of PM2.5 and O3 above their national standards.
We considered three types of reduced HM. The first uses a normal approximation to the likelihood at the first stage and a normal random-effect distribution at the second stage (RHM-N-N). The second uses an integrated likelihood at the first stage and a normal random-effect distribution at the second stage (RHM-L-N). The third uses an integrated likelihood at the first stage and a Dirichlet process normal mixture for the random-effect distribution (RHM-L-DP). The parameter of interest θi, defined in (2), is the log relative risk of cardiovascular admissions when PM2.5 and O3 are both above their national standards compared to when both are below their standards. For each reduced HM we assumed little prior information, by incorporating diffuse priors on the overall θ* We first fit each reduced HM without including any second-level covariates. We subsequently considered inclusion, at the second stage, of a county-specific measure of the average level of NO2 during the study period to demonstrate how reduced HM may be used to identify effect modification. Long-term average NO2 may be an important effect modifier because it a proxy for traffic exposure. This was done by assuming, at the second level that θi = α0i + α1zi, where zi is the long-term average NO2 for the ith county, and placing each of the normal and flexible (Section 4.2) random-effect distributions on the α0i. Details of the implementations for each reduced HM are in Appendix C of the Supplementary Materials.
Prior to fitting the reduced HM using the integrated likelihood (RHM-L-N and RHM-L-DP), we evaluated the performance of the integrated likelihood in the air pollution context through simulation study (detailed in Appendix E). Brifley, we considered a model based on our air pollution and health outcome data for which the integrated likelihood may be written in closed form. We simulated data under this model, applied our approach to estimate the integrated likelihood (described in Section 4.1), and compared our estimated integrated likelihood to the true integrated likelihood, finding that the estimate closely matched the truth.
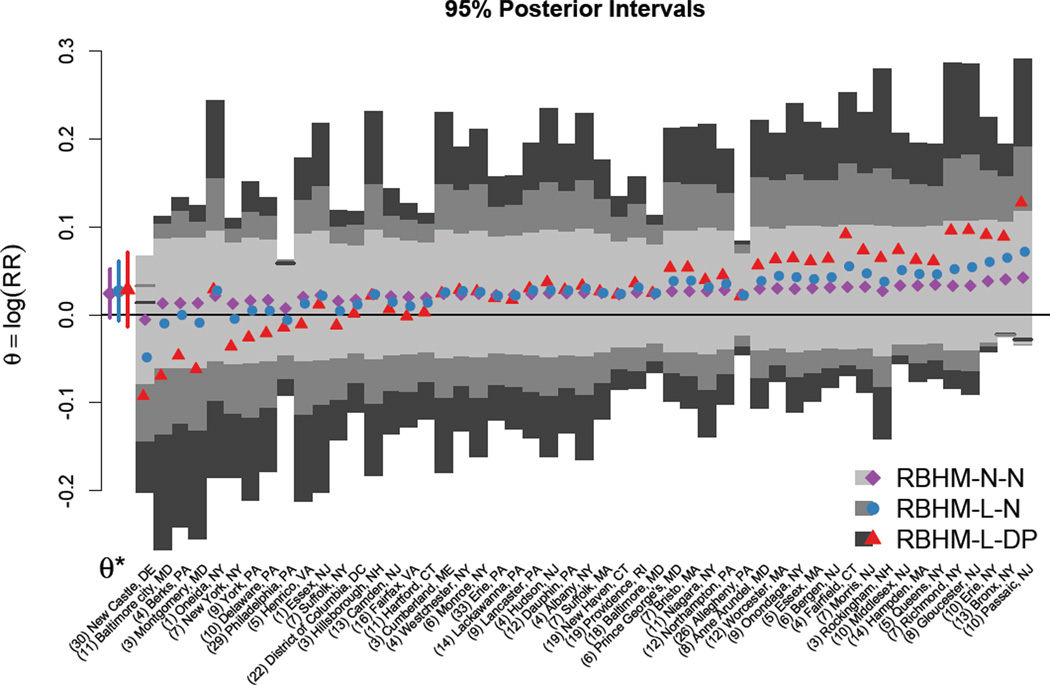
Figure 3 shows the posterior mean estimates and 95% posterior intervals for the overall θ* and for the cluster-specific θi obtained under each reduced HM. We found that on average, across all counties, there was an increase in CVD admissions on days when both ozone and fine particulate matter were above their national standards compared to days when both pollutants were below their national standards. In particular, we estimated that the overall log relative risk of CVD admissions associated with levels of O3 and PM2.5 both above their national standards (θ*) was 0.024 (95% posterior interval −0.004 to 0.053) for RHM-N-N, 0.027 (−0.007 to 0.061) for RHM-L-N, and 0.029 (−0.014 to 0.071) for RHM-L-DP. A log relative risk of 0.024 corresponds (approximately) to a 2.4% increase in cardiovascular hospital admissions on days when both O3 and PM2.5 are above their standards compared to days when both pollutants are below their standards. We also found variability across counties in the estimate of the cluster-specific effects θi. For most counties, θi was estimated to be positive, though for each county the posterior interval covered zero. The random-effect estimates exhibited the largest shrinkage for RHM-N-N, followed by RHM-L-N, with the RHM-L-DP estimates remaining furthest from the overall regional estimate.
Figure 3.
Results of a multisite time series study of 51 northeastern US counties, 1999–2005. County-specific (θi) and overall (θ*) estimates, with 95% posterior intervals, of the log relative risk of cardiovascular admissions on days when both O3 and PM2.5 exceed their national standard compared to days when both pollutants are below their standards, across three reduced HMs: normal approximation to the likelihood with normal random-effect distribution (RHM-N-N) and integrated likelihood with normal (RHM-L-N) and flexible (RHM-L-DP) random-effect distributions. Counties are ordered from left to right by increasing values of where θ̂i is the MLE and is the estimated standard error. The number of days with both O3 and PM2.5 greater than their national standards is listed beside each city.
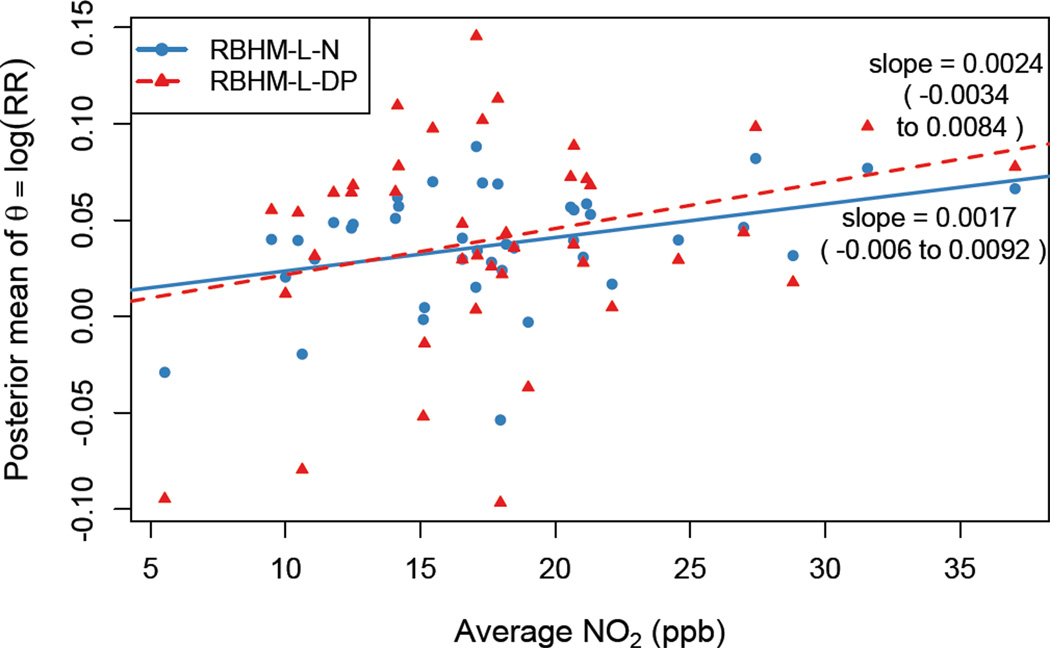
Figure 4 shows the posterior mean estimates of the location-specific θi from the reduced HM including average NO2 as a covariate at the second stage, plotted against the location’s long-term average NO2. The positive slopes (α1) suggest that the risk of cardiovascular admissions associated with daily levels of O3 and PM2.5 greater than their national standards is higher in locations with greater NO2 levels and lower in locations with lower NO2 levels, though the estimates were not statistically significant. More precisely, we estimated that an interquartile range increase in long-term average NO2 is associated with a percentage increase in the relative risk of cardiovascular hospital admissions associated with O3 and PM2.5 both above their national standards of 1.2% (−3.8% to 6.2%) under RHM-L-N, and 1.6% (−2.2% to 5.7%) under RHM-L-DP.
Figure 4.
For the 41 northeastern US counties with NO2 measurements, estimates of θi from the reduced HM incorporating long-term average NO2 as a covariate in the second stage model. Estimates of slopes α1 (95% posterior intervals) are shown beside the corresponding trend line. The parameter of interest θi is the log relative risk of cardiovascular admissions on days when both O3 and PM2.5 exceed their national standard compared to days when both are below their standards.
We performed several diagnostic assessments and sensitivity analyses to evaluate our model fit and demonstrate the robustness of our results to model specification (see Appendix F of the Supplementary Materials for details). Though the within-county model (1) does not account for the potential for autocorrelation in the hospitalization time series, exploratory data analysis revealed little evidence of residual autocorrelation in our data. In particular, when we fit model (1) separately for each county and inspected the autocorrelation function (ACF) of the deviance residuals, we did not find a consistent pattern in the ACF. We further investigated whether there was spatial correlation across counties by plotting a variogram of the estimated county-specific θi, as well as whether there was residual spatial correlation in the county-specific estimates after accounting for long-term average NO2 (Appendix F). We did not find evidence of spatial dependence across counties in the risk of cardiovascular admissions associated with O3 and PM2.5 both above their national standards. To assess the sensitivity of our results to the specification of the exposure-response surface, we refit the reduced HM where the joint association of ozone and PM2.5 with the health outcome in equation (1) was instead modeled as the product of cubic spline bases with just 2 df. We found that the resulting cluster-specific estimates θi were very similar and that the overall estimates θ* were nearly identical.
7 Discussion
While previous studies have estimated health effects of single pollutants, understanding how complex mixtures of pollutants affect health remains a challenging goal. Quantifying health risks resulting from exposure to a single pollutant is a useful analytical construct, but it is not representative of true exposure. It is therefore critical to develop models for estimating health effects of simultaneous exposure to multiple pollutants.
In this paper we developed methodology for estimating both county-specific and regional average risks of multipollutant exposure. This approach extends previous single pollutant models by allowing for nonlinear smooth functions of multiple pollutants and their interactions at the first stage and for effect modification at the second stage. Because flexible associations of several exposures are modeled concurrently, the inclusion of interactions of spline terms leads to a high-dimensional vector of random effects. As a result, several challenges to the application of the usual full HM framework are introduced. To address these challenges, we have proposed the reduced HM as a general statistical approach for combining information across locations directly on the parameter of interest, in the context of many nuisance parameters. In this approach, information about the parameter of interest is summarized through a likelihood function (e.g. integrated likelihood) in the first stage. At the second stage, a flexible random-effect distribution (e.g. Dirichlet process normal mixture) is specified directly on the parameter of interest. We conducted simulation studies to compare performance of the reduced HM to the full HM, and we applied the reduced HM to a multisite time series study of 51 northeastern US counties during the period 1999–2005.
In comparison with the reduced HM, on first inspection the full HM is the seemingly optimal approach, as it uses all of the available data in a single model to combine information across clusters. However, many practical difficulties may arise upon implementation. First, for the full HM one must specify a random-effect distribution on the vector βi parametrizing the within-cluster model. This may be difficult when the βi are high-dimensional or when they do not have meaningful interpretations (e.g. regression spline coefficients as in equation (1)). Additionally, for conducting Bayesian inference, prior distributions must be selected for the parameters of the random-effect distribution (e.g. mean vector β* and variance-covariance matrix Σ), which may also be complicated if these parameters do not have meaningful interpretations. If there does not exist a reparametrization of βi such that βi = (θi, λi) for λi a (q−1)-dimensional nuisance parameter, then prior information about the quantity of interest θi = h(βi; xi) cannot be easily translated into prior information about the model parameters βi. Moreover, if one is interested in effect modification of cluster-specific covariates zi at the second level, then a potentially high-dimensional multivariate regression model for βi | β*, zi must be specified. Finally, fitting the model (e.g. implementing the MCMC sampler) will become increasingly challenging and computationally intensive as the dimension of βi (number of random effects) increases.
For the reduced HM, on the other hand, rather than specify a high-dimensional random-effect distribution on parameters that are not of primary scientific interest, one only needs to specify a random-effect distribution for a one-dimensional parameter that has a meaningful interpretation. Additionally, it is frequently much easier to incorporate prior information about the parameter of interest θi than about a large vector of nuisance parameters βi that may be hard to interpret (e.g. spline coefficients). Furthermore, reducing a hierarchical model on a high-dimensional vector of parameters to a hierarchical model on a much lower dimensional space yields simpler implementation and greater computational efficiency, and makes model diagnostics and sensitivity analyses more wieldy.
Although the reduced HM overcomes many difficulties in the specification and implementation of the full HM, it also introduces new challenges. At the first stage, one must eliminate nuisance parameters to obtain the likelihood function Li(θi). While the literature on likelihood-based methods for eliminating nuisance parameters is vast (Pawitan, 2001; Edwards, 1992), in this paper we restricted our attention to those likelihoods that correspond to true probability distributions, including the integrated and conditional likelihood. In the case of large within-cluster sample sizes, the choice of which likelihood function to use should make little difference compared with the impact of the selection of the random-effect distribution. For smaller sample sizes, an integrated likelihood, though more computationally intensive than a normal approximation, allows greater flexibility for capturing the true form of the likelihood. Second, while the reduced HM avoids the need to specify a high-dimensional random-effect distribution on the complex βi, use of the integrated likelihood for θi still necessitates specifying priors for βi (Section 4.1). However, because we seek an objective likelihood function in the sense that it should summarize the information contained in the data about the parameter of interest such that the prior has as little influence as possible, any prior distribution for βi that induces a vague prior for θi will suffice. For the applications we have considered, assuming diffuse normal priors for each component of βi leads to a prior for θi that is at over a large range of reasonable values for θi, and we have found this approach to work well. Alternative approaches for approximating the likelihood function one could also be considered, such as the data cloning method of Lele et al. (2007). Third, while one gains simplicity by eliminating nuisance parameters at the outset, it is possible that some information may be lost before combining information across clusters.
We conducted a series of simulation studies to evaluate the relative performance of the reduced HM as compared to the full HM across a range of potential scenarios (Section 5). For the full HM, because one must specify random-effect distributions for a larger number of parameters, which may also be hard to interpret, there is more potential for model misspecification than for the reduced HM where a random-effect distribution is placed on the lower-dimensional parameter of interest. On the other hand, if the parameter of interest θi is correlated with nuisance parameters within a cluster, then information may be lost by reducing the parameter space to a single parameter and pooling the θi. We based the simulation study on an application for which a conditional likelihood for θi was available in closed form so as to focus on the impact on inference of misspecifying the random-effect distribution, rather than of misspecifying the likelihood function. In addition, though prior studies have considered the special case of reduced HM where a conditional likelihood is available (Efron, 1996; Liao, 1999), the relative performance of this approach as compared to the full HM had not been previously studied. When we refit the reduced HM using an integrated likelihood for a subset of the simulations (I = 100 and n = 100), we found that the performance for estimating the cluster-specific and overall parameters were either identical or just slightly worse than using the conditional likelihood. Across simulation scenarios, we found that the reduced HM generally achieved comparable performance to the full HM, and even had superior performance in some cases. We also performed a separate simulation study to evaluate the performance of our approach for estimating the integrated likelihood (Section 4.1) in a scenario based on our multipollutant application, finding that the estimated integrated likelihood closely matched the true integrated likelihood (Appendix E of the Supplementary Materials). Taken together, our findings from these simulation studies highlight the utility of the reduced HM both specifically to the multipollutant application and more generally to the context of two-level clustered data.
Development of reduced HM was motivated by methodological needs for estimating health risks of joint exposure to multiple pollutants. We applied the reduced HM methodology to estimate the risk of emergency cardiovascular admissions associated with simultaneous exposure to fine particulate matter and ozone. For the overall effect θ*, we found marginal evidence of increased risk on days when both pollutants exceeded their national standards compared to when both were below their national standards. The reduced HM with normal random-effect distribution on the parameter of interest θi (RHM-L-N) led to more shrinkage of the county-specific random effects than the reduced HM with flexible random-effect distribution (RHM-L-DP). Further, the RHM-L-N had narrower credible intervals for the county-specific parameters θi than RHM-L-DP. If the normal random-effect distribution is misspecified (e.g. if the analysis is missing an important county-level effect modifier) then the RHM-L-N may understate statistical uncertainty in the θi. We illustrated how diagnostics on the reduced parameter space could be performed to assess modeling assumptions, by investigating spatial autocorrelation in the risk of simultaneous exposure to PM2.5 and O3. Though we did not find evidence of spatial autocorrelation in the θi in this application, it would be straightforward to model spatial dependence in the second stage of the reduced HM by specifying a spatial model for cov(θi, θj). We also demonstrated that the reduced HM can easily accommodate effect modifiers. Specifically, we examined the inclusion of long-term county-level NO2, a surrogate for traffic exposure. We found a larger relative risk of cardiovascular admissions associated with levels of PM2.5 and O3 higher than their national standards in locations with high average NO2 compared to locations with low average NO2, although the effect modification was not statistically significant. For our within-county model (1) and parameter of interest θi (defined in (2)) we only considered the association of current day’s exposure to PM2.5 and O3 with hospitalization on the same day, though previous days’ exposure (e.g., at different lags from the present day) may also be predictive of health outcome. This choice of lag was motivated by previous single pollutant studies, which have found that the strongest effects for PM2.5 and O3 occur at short (current or 1 day prior) lags (Dominici et al., 2006; Bell et al., 2004). Furthermore, to demonstrate our methodology, we considered just a single example of a policy-relevant parameter of interest. The US EPA is considering introducing joint national standards to better protect human health from the risks of exposure to complex mixtures, and so studies providing a scientific basis for joint standards are needed (Dominici et al., 2010). Depending on the scientific question, alternative parameters of interest may be specified and the same methodology applied. We could consider, for example, the gradient of the air pollution-hospitalization exposure-response surface at the national standards, or the relative risk of adverse health events when both PM2.5 and O3 exceed their national standard at different temporal lags compared to when just one of the pollutants exceeds its standard. In the future we will apply this approach to systematically conduct a national investigation of the health effects associated with simultaneous exposure to multiple pollutants. Methods can be extended to an arbitrarily large number of pollution variables and locations, and to consider joint pollutant exposure at different lags as well as multiple parameters of interest that summarize different salient features of the multivariate exposure-response surface.
There are several extensions to the reduced HM methodology we have proposed. First, we assumed a within-location model that had the same form across locations. However, this assumption could be relaxed. One could specify different within-cluster models for each cluster, as long as the interpretation of the parameter of interest remains constant across models. For example, for the within-cluster model (1) in the multipollutant application, the full HM would require a common spline basis (e.g. common knot locations) for the joint O3 and PM2.5 association across locations, while the reduced HM can allow for locally optimized spline bases. Thus the reduced HM approach can readily accommodate heterogeneity in the appropriate model to use across locations. In this manuscript we focused on two-level clustered datasets and a scalar parameter of interest. However, the reduced HM could be generalized to three- or higher-level models, and to situations where the parameter of interest θi = h(βi) is a multivariate parameter with dim(θi) < dim(βi).
We have described the reduced HM within the context of estimating health risks of exposure to many pollutants. However, this hierarchical modeling strategy is broadly applicable to clustered data in which the parameter of interest is a known function of the vector of parameters βi of the within-cluster model. The meta-analysis of stomach ulcer treatment that served as the basis for our simulation study is one example. Another example is the estimation of heat wave mortality risk in multisite time series studies (Bobb et al., 2011). One can build a location-specific model similar to (1) where the exposure-response function of interest is the temperature-mortality relation, adjusted for time-varying covariates. One can then define a heat wave day indicator variable as a function of temperature on current and previous days. The parameter of interest θi, defined as the log relative risk of mortality on heat wave days compared to non-heat wave days (see for example Peng et al. (2011)), can then be written as a known function of the temperature-mortality exposure-response function (parameterized by βi), and the reduced HM framework may be applied.
The reduced HM is especially useful in situations where βi is high-dimensional, where the components of βi are not easily interpretable, or where one wishes to incorporate prior information directly on the parameter of interest. For such applications, the reduced HM allows one to specify a random-effect distribution directly on the parameter of interest θi and to study effect modification by specifying an across-cluster regression model for θi. Further, the reduced parameter space leads to simpler implementation, which facilitates the specification of flexible random-effect distributions that do not require strong assumptions on the random effects. For problems that are very high-dimensional in the number of clusters, the number of observations within a cluster, and the number of parameters in the within-cluster model, it may not be computationally feasible to fit a full HM. In such cases, the reduced HM is a practical alternative.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences grant numbers T32ES012871, R01ES012054, R01ES019560; and the United States Environmental Protection Agency grant numbers RD-83479801, RD-83241701.
Footnotes
Supplementary Materials
The reader is referred to the online Supplementary Materials for technical appendices and additional simulation study results.
References
- Agresti A, Caffo B, Ohman-Strickland P. Examples in which misspecification of a random effects distribution reduces efficiency, and possible remedies. Computational Statistics & Data Analysis. 2004;47:639–653. [Google Scholar]
- Bell M, McDermott A, Zeger S, Samet J, Dominici F. Ozone and short-term mortality in 95 US urban communities 1987–2000. Journal of the American Medical Association. 2004;292(19):2372. doi: 10.1001/jama.292.19.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Liseo B, Wolpert R. Integrated likelihood methods for eliminating nuisance parameters. Statistical Science. 1999 [Google Scholar]
- Bobb JF, Dominici F, Peng RD. A Bayesian model averaging approach for estimating the relative risk of mortality associated with heat waves in 105 U.S. cities. Biometrics. 2011;67(4):1605–1616. doi: 10.1111/j.1541-0420.2011.01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang D, Davidian M. A Monte Carlo EM algorithm for generalized linear mixed models with flexible random effects distribution. Biostatistics. 2002;3:347–360. doi: 10.1093/biostatistics/3.3.347. [DOI] [PubMed] [Google Scholar]
- Dominici F, Peng R, Barr C, Bell M. Protecting human health from air pollution: shifting from a single-pollutant to a multipollutant approach. Epidemiology. 2010;21(2):187. doi: 10.1097/EDE.0b013e3181cc86e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici F, Peng RD, Bell ML, Pham L, McDermott A, Zeger SL, Samet JM. Fine particulate air pollution and hospital admission for cardiovascular and respiratory diseases. Journal of the American Medical Association. 2006;295(10):1127–1134. doi: 10.1001/jama.295.10.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici F, Samet JM, Zeger SL. Combining evidence on air pollution and daily mortality from the 20 largest US cities: a hierarchical modelling strategy. Journal of the Royal Statistical Society, Series A. 2000;163(3):263–302. [Google Scholar]
- Edwards A. Likelihood. Baltimore: Johns Hopkins University Press; 1992. [Google Scholar]
- Efron B. Empirical Bayes methods for combining likelihoods. Journal of the American Statistical Association. 1996;91(434):538–565. [Google Scholar]
- EPA. The Benefits and Costs of the Clean Air Act from 1990 to 2020. Washington, D.C.: Technical report U.S. Environmental Protection Agency Office of Air and Radiation; 2011. [Google Scholar]
- Ferguson T. A Bayesian analysis of some nonparametric problems. The Annals of Statistics. 1973;1(2):209–230. [Google Scholar]
- Gallant AR, Nychka DW. Semi-nonparametric maximum likelihood estimation. Econometrica. 1987;55(2):363–390. [Google Scholar]
- Gilks W, Richardson S, Spiegelhalter D. Markov Chain Monte Carlo in Practice: Interdisciplinary Statistics. Chapman and Hall/CRC; 1995. [Google Scholar]
- Heagerty P, Kurland B. Misspecified maximum likelihood estimates and generalised linear mixed models. Biometrika. 2001;88(4):973. [Google Scholar]
- Katsouyanni K, Touloumi G, Samoli E, Gryparis A, Tertre AL, Monopolis Y, Rossi G, Zmirou D, Ballester F, Boumghar A. Confounding and effect modification in the short-term effects of ambient particles on total mortality: results from 29 european cities within the aphea2 project. Epidemiology. 2001;12(5):521. doi: 10.1097/00001648-200109000-00011. [DOI] [PubMed] [Google Scholar]
- Komárek A, Lesaffre E. Generalized linear mixed model with a penalized Gaussian mixture as a random effects distribution. Computational Statistics & Data Analysis. 2008;52(7):3441–3458. [Google Scholar]
- Laird N. Nonparametric maximum likelihood estimation of a mixing distribution. Journal of the American Statistical Association. 1978;73(364):805–811. [Google Scholar]
- Lele SR, Dennis B, Lutscher F. Data cloning: easy maximum likelihood estimation for complex ecological models using bayesian markov chain monte carlo methods. Ecol Letters. 2007;10(7):551–563. doi: 10.1111/j.1461-0248.2007.01047.x. [DOI] [PubMed] [Google Scholar]
- Liao J. A hierarchical Bayesian model for combining multiple 2×2 tables using conditional likelihoods. Biometrics. 1999;55(1):268–272. doi: 10.1111/j.0006-341x.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- Litière S, Alonso A, Molenberghs G. The impact of a misspecified random-effects distribution on the estimation and the performance of inferential procedures in generalized linear mixed models. Statistics in Medicine. 2008;27:3125–3144. doi: 10.1002/sim.3157. [DOI] [PubMed] [Google Scholar]
- Litière S, Alonso A, Molenberghs G. Rejoinder to “A note on type II error under random effects misspecification in generalized linear mixed models”. Biometrics. 2010 doi: 10.1111/j.1541-0420.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- Magder LS, Zeger SL. A smooth nonparametric estimate of a mixing distribution using mixtures of Gaussians. Journal of the American Statistical Association. 1996;91(435):1141–1151. [Google Scholar]
- McCulloch CE, Neuhaus JM. Prediction of random effects in linear and generalized linear models under model misspecification. Biometrics. 2011;67(1):270–279. doi: 10.1111/j.1541-0420.2010.01435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metropolis N, Rosenbluth A, Rosenbluth MN, Teller AH, Teller E. Equation of state calculations by fast computing machines. Journal of Chemical Physics. 1953;21(6):1087–1092. [Google Scholar]
- Neal R. Markov chain sampling methods for Dirichlet process mixture models. Journal of Computational and Graphical Statistics. 2000;9(2):249–265. [Google Scholar]
- Neuhaus J, Hauck W, Kalbeisch J. The effects of mixture distribution misspecification when fitting mixed-effects logistic models. Biometrika. 1992;79(4):755–762. [Google Scholar]
- NRC. Research priorities for airborne particulate matter: IV. Continuing research progress. Washington, D.C.: Technical report, National Research Council of the National Academies; 2004. [Google Scholar]
- Pawitan Y. In All Likelihood: Statistical Modelling and Inference Using Likelihood. Oxford University Press; 2001. [Google Scholar]
- Peng R, Chang H, Bell ML, McDermott A, Zeger SL, Samet JM, Dominici F. Coarse particulate matter air pollution and hospital admissions for cardiovascular and respiratory diseases among Medicare patients. Journal of the American Medical Association. 2008;299(18):2172–2179. doi: 10.1001/jama.299.18.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Dominici F, Louis T. Model choice in time series studies of air pollution and mortality. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2006;169(2):179–203. [Google Scholar]
- Peng RD, Bell ML, Geyh AS, McDermott A, Zeger SL, Samet JM, Dominici F. Emergency admissions for cardiovascular and respiratory diseases and the chemical composition of fine particle air pollution. Environmental Health Perspectives. 2009;117(6):957–963. doi: 10.1289/ehp.0800185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng RD, Bobb JF, Tebaldi C, McDaniel L, Bell ML, Dominici F. Toward a quantitative estimate of future heat wave mortality under global climate change. Environ. Health Perspect. 2011;119:701–706. doi: 10.1289/ehp.1002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J, Coull BA. Control for confounding in the presence of measurement error in hierarchical models. Biostatistics. 2003;4(4):539–553. doi: 10.1093/biostatistics/4.4.539. [DOI] [PubMed] [Google Scholar]
- Smith T, Spiegelhalter D, Thomas A. Bayesian approaches to randomeffects meta-analysis: A comparative study. Statistics in Medicine. 1995;14(24):2685–2699. doi: 10.1002/sim.4780142408. [DOI] [PubMed] [Google Scholar]
- Verbeke G, Lesaère E. The effect of misspecifying the random-effects distribution in linear mixed models for longitudinal data. Computational Statistics & Data Analysis. 1997;23:541–556. [Google Scholar]
- Warn DE, Thompson SG, Spiegelhalter DJ. Bayesian random effects meta-analysis of trials with binary outcomes: methods for the absolute risk difference and relative risk scales. Statistics in Medicine. 2002;21(11):1601–1623. doi: 10.1002/sim.1189. [DOI] [PubMed] [Google Scholar]
- Welty LJ, Zeger SL. Are the acute effects of particulate matter on mortality in the national morbidity, mortality, and air pollution study the result of inadequate control for weather and season? A sensitivity analysis using flexible distributed lag models. American Journal of Epidemiology. 2005;162(1):80–88. doi: 10.1093/aje/kwi157. [DOI] [PubMed] [Google Scholar]
- Zeka A, Schwartz J. Estimating the independent effects of multiple pollutants in the presence of measurement error: an application of a measurement-error-resistant technique. Environmental Health Perspectives. 2004;112(17):1686–1690. doi: 10.1289/ehp.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.