Summary
Understanding gut microbiota alterations associated with HIV-infection and factors that drive these alterations may help explain gut-linked diseases prevalent with HIV. 16S rRNA sequencing of feces from HIV infected individuals revealed that HIV infection is associated with highly characteristic gut community changes and antiretroviral therapy does not consistently restore the microbiota to an HIV-negative state. Despite the chronic gut inflammation characteristic of HIV infection, the associated microbiota showed limited similarity with other inflammatory states and instead showed increased, rather than decreased, diversity. Metaanalysis revealed that the microbiota of HIV-infected individuals in the US was most similar to a Prevotella-rich community composition typically observed in healthy individuals in agrarian cultures of Malawi and Venezuela and related to that of US individuals with carbohydrate-rich/ protein- and fat-poor diets. By evaluating innate and adaptive immune responses to lysates from bacteria that differ with HIV, we explore the functional drivers of these compositional differences.
Introduction
HIV-1 infection induces rapid and substantial damage to gut-associated lymphoid tissues (GALT), with massive depletion of Th17 cells, a subset of CD4+ T cells that control intestinal bacteria (Brenchley et al., 2004). Loss of this CD4+ T cell subset may facilitate disease progression by allowing translocation of bacterial products such as lipopolysaccharide to blood causing systemic T-cell activation (Brenchley et al., 2006).
In healthy individuals, intestinal bacteria help control harmful pathogens, educate the immune system and aid in digestion (Lozupone et al., 2012), but distinctive compositions of this complex community has been associated with pathologies that are of increased prevalence with HIV-infection including metabolic disorders (Madge et al., 1999; Vijay-Kumar et al., 2010), chronic inflammation (Bjarnason et al., 1996; Willing et al., 2010), wasting/malnutrition (Smith et al., 2013; Wanke et al., 2000), atherosclerosis (Hsue et al., 2009; Koeth et al., 2013), and susceptibility to opportunistic infections (Chang et al., 2008). Considering the important role that both innate and adaptive immunity play in shaping gut microbiota composition (Carvalho et al., 2012; Hepworth et al., 2013), understanding how composition differs with HIV-infection, immunologic driving factors of these differences, and their implication for gut-linked diseases that increase in prevalence with HIV is of paramount importance.
Using 16S rRNA sequencing of feces, we show that HIV infection is associated with highly characteristic changes in gut community structure and that antiretroviral therapy (ART) does not consistently restore the microbiota to an HIV-negative like state. Using meta-analysis, we demonstrate that although the HIV-associated microbiota bears some resemblance to that of other chronic-inflammatory states, the differences between HIV-associated and healthy gut microbiota are also related to compositional changes previously associated with diet (Wu et al., 2011) and Western compared to agrarian cultures (De Filippo et al., 2010; Yatsunenko et al., 2012), suggesting an interplay between diet and the immune system in shaping gut microbiota composition. By comparing T-cell proliferation and cytokine responses to lysates from bacteria that differ with HIV infection in blood from HIV-positive and negative individuals, we begin to explore the relationship between immune dysfunction and HIV-associated microbiota change.
Results
Stool microbiota in chronic untreated HIV infection is distinctive
We sequenced the V4 region of 16S rRNA genes from fecal samples of individuals with chronic HIV infection (n=22), recent infection (n=3), and HIV negative controls (n=13)(Table 1, S1). Eleven individuals with chronic infection had not recently undergone ART and 8 had been on ART for ≥12 months with viral suppression for ≥6 months (see methods for further cohort details). Individuals were excluded during recruitment if they were pregnant, weighed <110 pounds, or had received antibiotics within the prior 30 days, resulting in inclusion of only relatively healthy HIV-positive subjects; No chronically HIV-infected individual had a peripheral CD4+ T cell count below 204 cells/µl (Table 1).
Table 1.
Description of cohorts
| Cohort | N | Age (years) |
Sex M:Male F:Female |
Race | BMI | Duration ART (months) |
CD4 (cells/uL) | Viral Load (Copies/mL) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| W | Wh | AA | AI | ? | ||||||||
| 1: Recent HIV infection | 3 | 31 ± 4.36 | M:2 F:1 | 3 | 0 | 0 | 0 | 0 | 32.5 ± 8.30 | 0 | 614 ± 521.8 (107–1342) | 164,880 ± 210,225 (418–449,000) |
| 2: Chronic HIV untreated | 11 | 30 ± 5.61 | M:11 F:0 | 6 | 2 | 2 | 1 | 0 | 25.0 ± 4.83 | 0 | 551 ± 218.2 (270–1095) | 139,109 ± 255,929 (1350–869,000) |
| 2*:Chronic HIV short-term ART | 6* | 34 ± 10.27 | M:5 F:1 | 3 | 2 | 1 | 0 | 0 | 27.1 ± 3.67 | 7.68 ± 0.78 | 609 ± 50.6 (547–681) | 83 ±105.0 (39–247) |
| 3: Chronic HIV long-term ART | 8 | 45 ± 8.30 | M:7 F:1 | 5 | 2 | 1 | 0 | 0 | 24.7 ± 6.45 | 33.6 ± 17.97 | 483 ± 258.0 (204–876) | 124 ± 280.8 (<20–819) |
| 4: HIV- seronegative | 13 | 37 ± 12.26 | M:8 F:5 | 10 | 1 | 1 | 0 | 1 | 25.6 ± 3.03 | 0 | NA | 0 |
Race abbreviations are W (White-not Hispanic), Wh (White-hispanic), AA (African American), AI (American Indian), ? (unknown)
3 of the 6 individuals in Cohort 2* are also in Cohort 2 because they were sampled both before and after short-term ART treatment. See also Table S1. Values are given as mean ± st. dev. with ranges in parentheses.
Principal Coordinates Analysis (PCoA) of fecal samples based on 16S rRNA sequences using both unweighted (Fig. 1) and weighted (Fig. S1) UniFrac showed a strong difference in microbiota of individuals with chronic untreated HIV infection compared to HIV-negative controls. UniFrac evaluates microbiota similarity based on shared evolutionary history of bacterial taxa, and the weighted metric differs from unweighted because it accounts for change in relative abundance in addition to presence/absence (Lozupone and Knight, 2005; Lozupone et al., 2007). Fecal microbiota with chronic untreated HIV infection also had greater overall diversity, with higher evenness as measured by the Shannon diversity index (p=0.013) and richness as measured by phylogenetic diversity (PD) (p=0.0003)(Table 2).
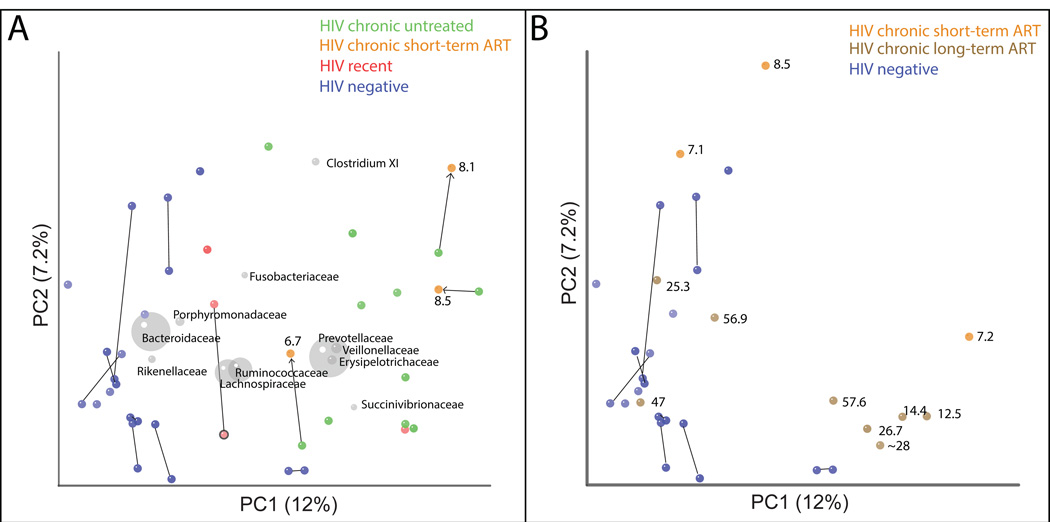
Fig. 1. Unweighted UniFrac clustering of HIV dataset.
Each point represents a single fecal sample. Panels A and B represent the same PCoA analysis but for clarity, Panel A only shows HIV positive individuals not on ART at the time of initial sample collection and HIV-negative controls, and Panel B shows only HIV positive individuals on ART. Points are labeled with ART duration at the time of sample collection in months. Samples from the same individual at two timepoints are joined with a line. Bacterial families are plotted as a weighted average of the coordinates of all samples, where the weights are the relative abundances of the taxon in the samples (grey circles). The size of the sphere representing a taxon is proportional to the mean relative abundance of the taxon across samples. See Fig. S1 for results with Weighted UniFrac.
Table 2.
Comparison of alpha diversity in HIV positive and negative individuals with and without ART treatment.
| observed species |
shannon | PD | |
|---|---|---|---|
| HIV negative | 508 ± 21.6 | 5.6 ± 0.19 | 27.9 ± 1.46 |
| Chronic HIV, ART untreated | 563 ± 30.5 | 6.3 ± 0.15 | 36.4 ± 1.31 |
| Chronic HIV on long-term ART | 469 ± 39.6 | 5.68 ± 0.16 | 28.69 ± 1.19 |
| Acute | 517 ± 64.4 | 5.76 ± 0.49 | 31.7 ± 3.06 |
| P-value (ART untreated versus negative) T test | 0.144 | 0.013 | 0.0003 |
| P-value (ART untreated versus ART treated) T test | 0.070 | 0.016 | 0.0006 |
Values are given as mean ± st dev.
To further determine the degree to which gut microbiota with chronic untreated infection differed from HIV-negative controls, we used a supervised learning technique called Random Forests (Knights et al., 2011a). The purpose of the Random Forests classifier is to learn a function that maps a set of predictive features to a discrete state, in this case healthy versus chronically infected with HIV and without ART. We used as features the relative abundance in each sample of 97% identity (ID) Operational Taxonomic Units (OTUs; clusters in which sequences have >=97% identity over their aligned 16S rRNA genes, which approximates assignment to the same species (Stackebrandt and Goebal, 1994)). The measure of the method’s success is its ability to classify new samples as coming from an HIV positive or negative individual. We only used one sample per individual when multiple timepoints were collected. The model could classify unknown samples according to HIV status with a 4.33% +/− 0.821 error rate, which is 10.6 times higher than the baseline error rate for random guessing of 45.83%, indicating that HIV-associated microbiota exhibit highly characteristic differences.
To investigate whether disease severity impacted gut microbiota in HIV infection, we determined whether microbiota diversity correlated with peripheral CD4+ T cell count or plasma HIV-1 RNA viral load in 11 individuals with chronic infection who were not on ART. As an estimate of divergence from healthy, we calculated the average pairwise weighted and unweighted UniFrac distance between each HIV microbiota sample and HIV-negative control samples. The magnitude of divergence from HIV-negative microbiota samples was not correlated with CD4+ T cell count or plasma HIV-1 RNA viral load (all Spearman correlation p-values > 0.73).
Individuals recently infected with HIV have relatively subtle microbiota differences
The microbiota of recently infected untreated individuals differed little from uninfected controls. Samples from 2 of 3 recently infected individuals clustered in an intermediate position between healthy and untreated chronic infection in an unweighted UniFrac PCoA plot (Fig. 1A). However, these individuals clustered with chronically infected subjects when using weighted UniFrac (Fig. S1), suggesting that changes in relative abundance of existing taxa may occur faster than gain/loss of taxa.
Effects of ART treatment
No chronically infected individual sampled pre and post 6.7–8.5 months of ART exhibited a strong shift towards the HIV-negative individuals (Fig. 1A), indicating that short-term ART was insufficient to restore the microbiota. However, the microbiota of two individuals treated for 7.1–8.5 months for whom we did not collect pre-ART samples showed a closer resemblance to HIV-negative individuals than to subjects with untreated chronic HIV-1 infection (Fig. 1B). Of the 8 HIV-chronic individuals on long-term ART (12.1–57.6 months), 5 clustered with untreated chronically infected individuals and 3 clustered with HIV-negative individuals (Fig. 1B). Individuals on ART for as long as 57.6 months could still cluster with HIV-positive subjects with chronic infection, indicating that ART duration alone does not predict restoration to a HIV-negative phenotype. Untreated HIV infected individuals had significantly higher alpha diversity (phylogenetic diversity and evenness) compared to ART-treated and HIV negative individuals, which were not significantly different from each other (Table 2).
Taxa that discriminate between HIV-negative individuals and chronic HIV infection
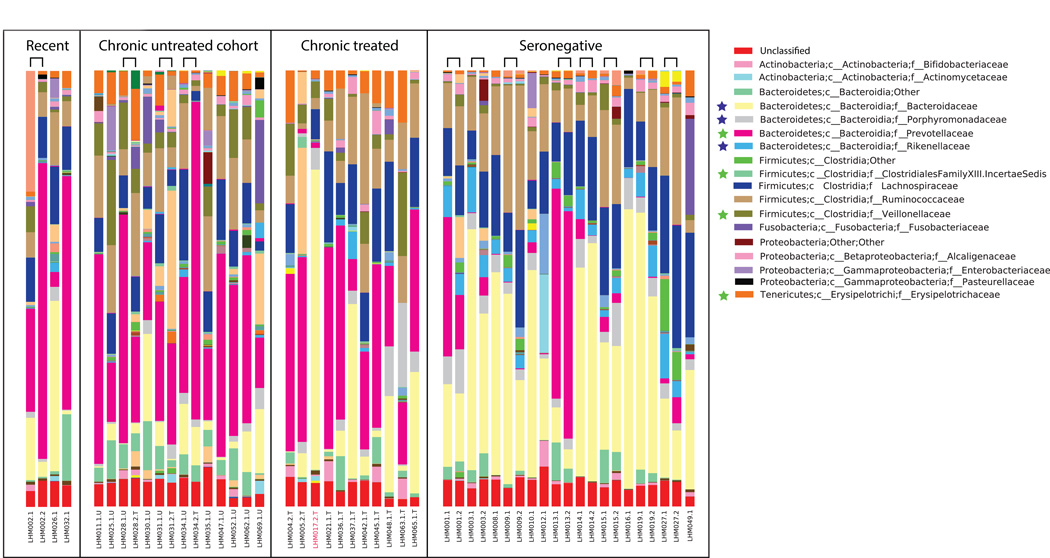
The fecal microbiota of untreated individuals with chronic infection exhibited significantly higher relative abundances of Prevotellaceae (Prevotella), Erysipelotrichaceae (Catenibacterium and Bulleidia), Veillonellaceae (Dialister and Mitsuokella), Clostridium cluster XIII and the genus Desulfovibrio compared to HIV negative (Fig. 1, 2, Table S2). HIV negative individuals had increased Bacteroidaceae (Bacteroides), Rikenellaceae (Alistipes), and Porphyromonadaceae (Parabacteroides)(Fig. 1,2, Table S2).
Fig. 2. Taxonomy bar charts showing dominance of Prevotellaceae in HIV infected subjects.
Families significantly different between ART naïve individuals chronically infected with HIV and HIV-negative controls are marked with a star (p<0.05 ANOVA with FDR correction, only the first time point was used for individuals who had multiple time points). Those enriched in HIV-negative have a blue star and in HIV (Chronic) in green. Samples from the same individual at two timepoints are adjacent and joined with a bridge. See Tables S2A, S2B, and S2C for lists of discriminatory genera, families, and 97% OTUs respectively and Fig. S2 for a phylogenetic tree relating discriminatory 97% OTUs.
To identify 97% ID OTUs that differentiate individuals with chronic HIV infection from HIV negative controls, we used the discriminative OTU features from the Random Forest classifier (see methods). Random Forests assigns an importance score to each OTU by estimating the increase in error caused by removing that OTU from the set of predictors. Discriminative OTUs depleted in HIV infection subjects were highly related (>97% ID) to type species in the families Bacteroidaceae (Bacteroides uniformis/fluxus, Bacteroides xylanisolvens, Bacteroides finegoldii, Bacteroides thetaiotaomicron, and Bacteroides massiliensis), Rikenellaceae (Alistipes putridinis), and Porphyromonadaceae (Parabacteroides distasonis) (Table S2). Species highly related to OTUs that increased in relative abundance with HIV infection included species in the families Erysipelotrichaceae (Eubacterium biforme, Catenibacterium mitsuoki), Prevotellaceae (Prevotella copri and Prevotella stercorea), Veillonellaceae (Dialister propionicifaciens, Mitsuokella jalaludinii, Allisonella histaminiformans, Megasphaera elsdenii, and Acidaminococcus fermentans), Ruminococcaceae (Ruminococcus callidus), and Desulfovibrionaceae (Desulfovibrio piger)(Table S2).
The identification of discriminatory 97% ID OTUs closely related to readily obtainable type species allows for investigation of immunologic driving factors of HIV-associated microbiota changes. However, since differences in biological attributes can occur even at the strain level (e.g. consider enterotoxic versus commensal Escherichia coli), we determined whether OTUs that change with HIV infection are related in phylogenetic lineages. Type species in lineages with a broad-conservation of HIV-associated change are more likely to share the traits that drove HIV-association. A positive association with HIV was broadly conserved across the Prevotellaceae but particularly in a lineage containing P. copri and P. stercorea (Fig. S2A). An HIV-negative association was conserved across the Bacteroides genus (Fig. S2A).
Evaluating the HIV-associated community type in a broader context
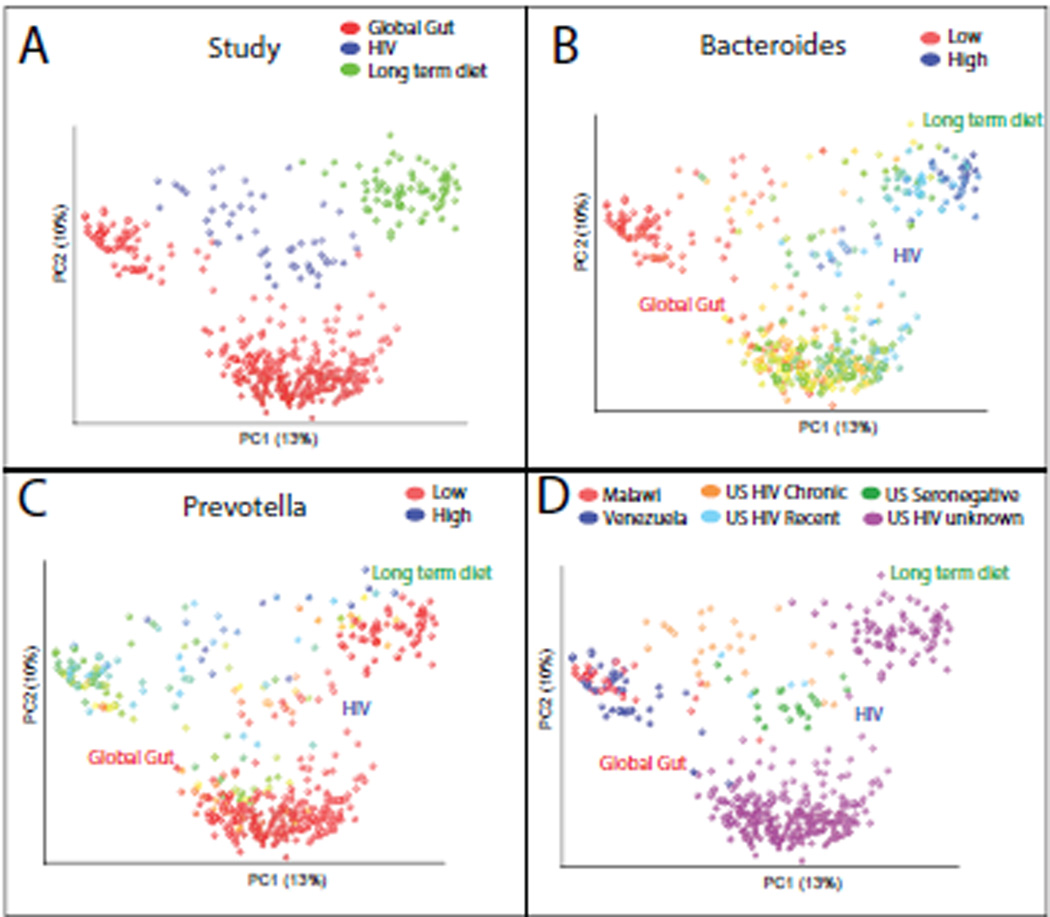
Since HIV-positive individuals suffer from increased incidence of diarrhea in the absence of obvious enteric pathogens and increased intestinal inflammation (Bjarnason et al., 1996; Brenchley and Douek, 2008), we had expected an expansion of bacteria that increase with other chronic intestinal inflammatory diseases. We were thus surprised that the most obvious compositional difference was an expansion of Prevotella and decrease in Bacteroides. To our knowledge, Prevotella had never before been described to be particularly pro-inflammatory, but Prevotella-rich/Bacteroides-poor gut microbiota had been observed in healthy individuals living in agrarian societies in Burkina Faso (De Filippo et al., 2010), Malawi and Venezuela (Yatsunenko et al., 2012), and in US adults who consumed diets poor in animal protein and saturated fats and rich in carbohydrates (as assessed over the prior year with a food frequency questionnaire) (Wu et al., 2011). Since Prevotella and Bacteroides are only two genera in the very complex gut community, we wanted to determine whether Prevotella-enrichment in HIV infection is an indicator for the same complex combinations of co-occurring taxa of Prevotella-rich microbiota in health. We thus combined this study’s data with data from 1) a survey of fecal microbiota in individuals from Malawi, the Amazonas State of Venezuela, and the US (Yatsunenko et al., 2012) and 2) a study linking long-term dietary patterns in healthy US adults with Bacteroides- versus Prevotella-rich enterotypes (Wu et al., 2011).
PCoA clustering with unweighted UniFrac was largely driven by technical variation between studies (Fig. 3A), as might be expected since they varied methodologically (Table S3)(Lozupone et al., 2013). However, by coloring the points by the relative abundance of Bacteroides (Fig. 3B) and Prevotella (Fig. 3C), it is clear that the major diversity differences within all three datasets are associated with a tradeoff between these genera along the same axis of variation. Samples from HIV positive individuals from the US clustered closer to individuals from Malawi and Venezuelan Amerindians than to HIV-negative individuals from the US (Fig. 3D). Additionally, the discriminative 97% ID OTUs that differentiated adults from Western and agrarian cultures using Random Forest analysis (Yatsunenko et al., 2012) were highly related and sometimes identical to those that discriminated between HIV negative and positive individuals (Fig. S2). This was particularly evident for the Bacteroidales order, where Western and HIV-negative associated OTUs clustered together in the Bacteroides and Alistipes genera and agrarian and HIV-positive associated OTUs clustered together in the Prevotella genus (Fig S2A). However, consistent with the notion that Bacteroides and Prevotella are indicator taxa for more complex community assemblages, parallels between agrarian cultures and HIV-positive individuals were also observed in other taxonomic groups (See Fig. S2 for more details).
Fig. 3.
Unweighted UniFrac PCoA plot comparing data from this study with a global survey of fecal microbial community composition conducted in individuals from Malawi, the Amazonas State of Venezuela, and the US (Yatsunenko et al., 2012) (Global Gut) and 2) a study linking long-term dietary patterns with gut microbial enterotypes (Wu et al., 2011) (Long term diet), using a reference mapping protocol. A) Points colored by study, B) Same plot but with points colored across a red-blue spectrum from low to high relative abundance of the genus Bacteroides, C) Same as for panel B but for the Prevotella genus, D) Points colored by country of residence and HIV status. See Fig. S3 for an additional meta-analysis exploring the relationship of HIV infection with Crohn’s disease.
Since HIV infection induces chronic gut inflammation (Bjarnason et al., 1996; Brenchley and Douek, 2008), we also used metaanalysis to determine whether there is a relationship with a chronic inflammatory disease of the gut using data from fecal samples collected from individuals with Crohn’s disease and healthy controls in (Willing et al., 2010). Samples from HIV infected individuals (including recent and chronic untreated and ART treated individuals) aligned with individuals with Ileal Crohn’s disease across a distinct axis of variation from that associated with diet/culture (See Fig. S3 and Supplemental Data).
Immune responses to selected bacteria modulated by HIV infection
Microbiota changes with HIV-infection can have several underlying causes including 1) a compromised ability of the innate and/or adaptive immune system to control commensal bacteria, 2) the indirect selection of inflammation-tolerant versus sensitive bacteria resulting from a chronic inflammatory state or 3) a loss of interaction with CD4+ T cells that produce regulatory responses that promote tolerance of beneficial microbes. Although the immunologic driving factors of microbiota changes in HIV-infection are likely to be complex, we began to explore drivers of compositional differences by examining cytokine and CD4+ T cell proliferative response to bacteria highly related to OTUs that differed with HIV infection status using Peripheral Blood Mononuclear Cells (PBMC) from a separate cohort of untreated chronic HIV-positive and HIV-negative individuals. We tested 1) two species from the genus Bacteroides, which had a conserved decrease with HIV infection (Bacteroides dorei and Bacteroides fluxus), 2) two species highly related to OTUs that increased with HIV infection, P. copri (Prevotellaceae) and E. biforme (Erysipelotrichaceae) (Table S2, Fig. S2) and 3) E. coli serotype O55:K59(B5), which did not change.
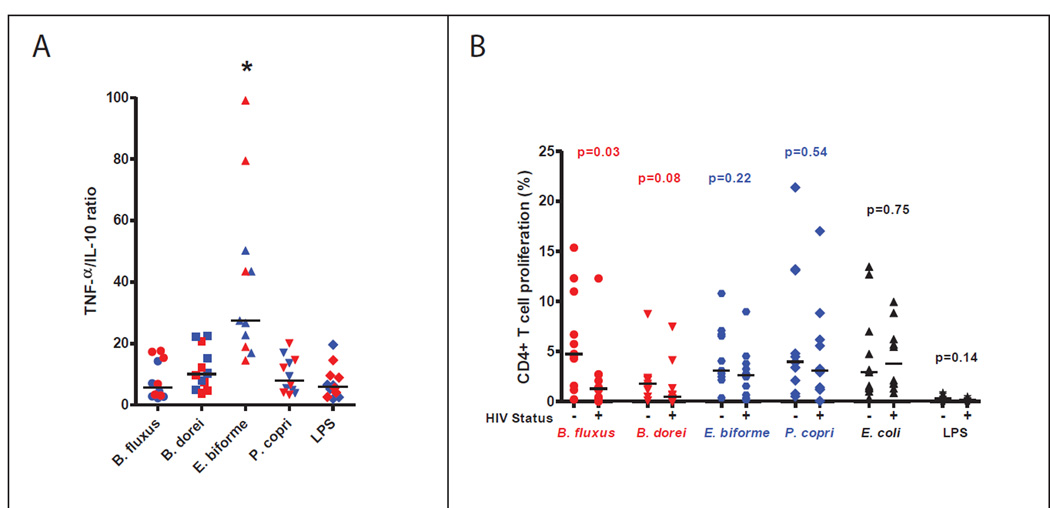
We first examined pro-inflammatory (TNF-α) and anti-inflammatory (IL-10) cytokines in 6 HIV-negative and 6 HIV-positive subjects after incubating PBMCs for 24 hours with bacterial lysate. HIV infected subjects tended to have elevated levels of both TNF-α and IL-10 to all bacteria in comparison with HIV-negative controls (Fig. S4). Previous studies have shown that both of these cytokines are elevated in plasma of HIV positive subjects (Dutertre et al., 2012; Said et al., 2010). However, E. biforme had a significantly higher TNF-α:IL-10 ratio (Fig. 4A) in both HIV-negative and HIV infected subjects, indicating that it is more pro-inflammatory than the other bacteria tested.
Fig. 4. Characterization of immune responses to selected bacteria in untreated HIV positive and HIV negative subjects.
A) TNF-α to IL-10 ratio (as measured using a bead array assay) after a 24 hour stimulation. An asterisk indicates that E. biforme had a significantly higher TNF-α/IL-10 ratio compared to the other bacterial isolates with the Kruskal-Wallis ANOVA and Dunns test. Red signifies HIV positive and blue HIV-negative subjects. B) Percentage of divided (Cell Trace low) CD4+ T cells enumerated by flow cytometry after a 6 day stimulation. Statistical significance was determined using Mann-Whitney T-tests. Bacteria that decreased in relative abundance with chronic, untreated HIV infection are red, those that increased are blue, and no difference is black. Points in the scatter plots depict each individual and the bar is the median. See also Fig. S4.
Next, we examined CD4+ T cell proliferative responses to these bacteria in 10 subjects with HIV infection and 11 HIV-negative controls. Because responses to gut bacteria in peripheral blood are often below the limit of detection directly ex vivo (Howe et al., 2009), we labeled PBMC with Cell Trace to track cell division and cultured the PBMC with the bacteria lysate for 6 days. This method amplifies bacteria specific CD4+ T cells to easily detected frequencies. All bacteria, but not LPS, induced CD4+ T cell proliferation in both HIV-infected and HIV negative subjects. However, responses to B. fluxus and B. dorei were reduced with HIV infection while responses to P. copri, E. biforme and E. coli were not significantly different (Fig. 4B).
Discussion
Because HIV targets central players of innate and adaptive immunity including CD4+ T cells, monocytes, and macrophages (McMichael et al., 2010; Mogensen et al., 2010), changes in gut microbiota with infection supports that the immune system plays an important role in shaping composition. That immune dysfunction can cause gut microbiota changes with concomitant health effects has been supported in mice lacking specific components of the immune system including TLR5, IL-22 or the inflammasome. Compositional shifts in gut microbiota in these models were associated with development of colitis (Carvalho et al., 2012; Elinav et al., 2011; Zenewicz et al., 2013) and metabolic syndrome (Vijay-Kumar et al., 2010), diseases that are increased in prevalence with HIV (Knox et al., 2000; Wanke, 1999).
Strong and characteristic change with HIV infection despite matching the control cohort for age, sex, race, and geographic location, is surprising since studies of the macaque microbiota in SIV infection failed to detect significant changes independent of colitis, and yet macaques experience a similar depletion of CD4+ T cells and inflammation in the gut compared to HIV infected humans (McKenna et al., 2008; Veazey et al., 1998). This difference may be related in part to SIV infection duration (e.g. macaques in one longitudinal study were only followed 8 weeks post infection (McKenna et al., 2008)). Microbiota changes in 2 of 3 recently infected HIV-positive humans in our study were subtle compared to changes with chronic infection, indicating that microbiota changes begin during early infection and continue over time.
The similarity between gut microbiota of HIV infected individuals in the US and HIV-negative individuals in agrarian cultures suggests that the relatively strong phenotype with HIV infection in humans may also be due to specific elements of our cohort’s lifestyle, since Western populations appear to have a stronger deviation of the pre- versus the post- HIV-infection microbiota. Indeed, similar to healthy humans in agrarian cultures, the healthy gut microbiota of macaques is already Prevotella-rich and Bacteroides-poor (McKenna et al., 2008). It will be of interest to see if HIV-associated changes in agrarian cultures are less pronounced than in the US or if they are more pronounced in macaques consuming a Western-type diet.
HIV-associated enteropathy is initiated during acute infection, continues through advanced disease and involves increased intestinal inflammation (Bjarnason et al., 1996; Brenchley and Douek, 2008). We thus had expected an expansion of bacteria that increase with other chronic intestinal inflammatory diseases and a decrease in diversity, which has been observed in several unhealthy or inflammatory states (Chang et al., 2008; Willing et al., 2010). Although a previous study that used 16S rRNA gene quantification of order level bacterial taxa showed a trend towards a greater proportion with HIV-infection of Enterobacteriales (Ellis et al., 2011), a taxon often associated with inflammatory conditions (Koren et al., 2012; Mukhopadhya et al., 2012), we did not detect significant changes in Enterobacteriales at either coarse or fine taxonomic levels. Furthermore, microbiota in untreated HIV-infection but not with ART had increased diversity compared to the control population. This is consistent with the resemblance to the microbiota in agrarian cultures, which also has increased diversity compared to that of healthy US adults (Yatsunenko et al., 2012), but not with other unhealthy states such as Inflammatory Bowel Disease (Willing et al., 2010) or recurrent Clostridium difficile infection (Chang et al., 2008). Since we excluded individuals with GI symptoms that warranted antibiotic treatment during the preceding 30 days, we cannot exclude the possibility that Enterobacteriales are enriched or diversity decreased in HIV-infected individuals with acute diarrhea or advanced AIDS.
However, our analysis of the pro/anti-inflammatory properties of cultured isolates related to OTUs that increased with chronic HIV infection indicated that E. biforme but not P. copri was significantly more pro-inflammatory than bacteria in the HIV negative-associated Bacteroides genus. E. biforme is enriched in individuals with Irritable Bowel Syndrome (Ponnusamy et al., 2011) and it increased in the active disease stage compared to remission in an individual with Ulcerative Colitis (Harrington et al., 2008). D. piger which increased with HIV infection, is significantly higher in IBD patients compared to healthy controls (Jia et al., 2012; Loubinoux et al., 2002). Enrichment of these taxa with HIV-infection is thus consistent with the relationship we observed between HIV and Ileal Crohn’s disease in both ART-treated and ART-untreated individuals at the community level.
Despite potential links between certain bacterial lineages and proinflammatory states in HIV infection, overall, the predominant pattern of microbiota change with HIV-infection was surprisingly along a Prevotella-Bacteroides gradient. Prevotella versus Bacteroides rich-communities correlate with overall-diversity differences observed both across cultures and within Western populations, although there is some controversy over whether they represent discrete “enterotypes” as initially proposed (Arumugam et al., 2011) or whether this variation occurs across a gradient (Koren et al., 2013; Wu et al., 2011; Yatsunenko et al., 2012). Diet has been implicated as a primary driving factor of this gradient since Prevotella were enriched in healthy US adults who consumed diets poor in animal protein and saturated fats and rich in carbohydrates and simple sugars (Wu et al., 2011), and in healthy individuals in agrarian cultures with diets dominated by maize-, cassava-, and other plant-derived polysaccharides (De Filippo et al., 2010; Yatsunenko et al., 2012). Although we do not have dietary information for our HIV positive cohort and thus do not know whether they uniformly consume a diet that is particularly carbohydrate-rich or animal fat/protein poor, that both HIV infection duration and ART treatment had an effect on the degree/consistency of Prevotella-richness with HIV infection further supports immunologic and not strictly dietary driving factors. This raises the intriguing possibility that the predominant type of gut microbiota diversity variation that occurs across cultures and within Western populations is driven by an interplay between diet and the immune system.
CD4+ T cells are profoundly depleted in GALT with HIV-infection (Brenchley and Douek, 2008). Interactions between CD4+ T cells and commensal gut bacteria and concomitant changes in microbiota composition are not well understood, although recent work has demonstrated that immunoglobulin A (IgA) is important for maintaining balance between gut bacteria and host (Kawamoto et al., 2012). B cell immunity and subsequent IgA production are dependent on CD4+ T follicular helper cells which are impaired in HIV infection (Cubas et al., 2013). Recently it was shown that innate lympoid cells regulate CD4+ T cell responses to intestinal bacteria (Hepworth et al., 2013). CD4+ T cell-microbe interactions are perhaps best understood for Bacteroides fragilis, which is in a genus that consistently and dramatically decreases with HIV infection. This species, and specifically a zwitterionic polysaccharide on its capsule called polysaccharide A (PSA), restores CD4+ T-cell development in germ-free mice by eliciting expansion of antigen-experienced (‘educated’) CD4+CD45Rblow T cells (Mazmanian et al., 2005). B. fragilis PSA can also mediate conversion of CD4+ T cells into anti-inflammatory Foxp3+ Treg cells (Round and Mazmanian, 2010) and can provide protection from inflammatory disease (Mazmanian et al., 2005; Mazmanian et al., 2008).When administered as a probiotic, another Bacteroides species, B. uniformis, can correct T-cell proliferation deficiencies displayed in “Western diet” fed mice (Gauffin Cano et al., 2012) and induces higher secretion of IL-10, a negative regulator of inflammatory response.
Given these interactions between Bacteroides species and CD4+ T cells, it is of interest that Bacteroides decrease in HIV-positive individuals when CD4+ T cell populations are compromised, suggesting that CD4+ T cell interactions may be essential for persistence of Bacteroides in the gut. Even within the overall compromised T-cell populations of HIV-positive subjects, we show that proliferative responses to Bacteroides species are preferentially depleted compared to other species tested. One potential explanation is that CD4+ T cell responses are diminished because of reduced levels of Bacteroides antigen. However, an increased proliferative response to P. copri and E. biforme was not observed despite their increase in relative abundance with HIV-infection, indicating that antigen concentration is not the only explaining factor. Another potential explanation given the known interaction of Bacteroides with CD4+T-cells via PSA, is that Bacteroides-specific T-cells are preferentially activated and thus killed, which is consistent with enhanced HIV infection of intestinal lamina propria CD4+ T cells generally upon exposure to commensal bacteria (Dillon et al., 2012). Conducting immunologic assays on samples for which we have matched gut microbiota compositional data will help to determine the degree to which T-cell proliferative response changes may be driven by changes in antigen presence/concentration.
Mouse experiments have supported that a Bacteroides-poor microbiota in the context of a Western diet may have negative health effects (Gauffin Cano et al., 2012). Oral administration of B. uniformis to mice consuming a high-fat diet resulted in decreased inflammation, body weight gain, liver steatosis, liver cholesterol and triglyceride concentrations and increased small adipocyte numbers. In this light, it is interesting that lipodystrophy, a disorder characterized by some of these same features has been increasingly observed in HIV positive individuals (Carr et al., 1998; Wanke, 1999). Although lipodystrophy and dyslipidemia is linked to ART, particularly the protease inhibitors (Carr et al., 1998), it also can occur in the absence of ART, leading some to suggest that it is in part due to HIV infection itself (Madge et al., 1999; Nicholaou et al., 2013).
Prevotella-rich microbiota have been linked with high plasma levels of trimethylamine-N-oxide (TMAO), a proatherogenic cardiovascular risk-associated compound that can be produced from dietary L-carnitine in red meat by intestinal microbiota (Koeth et al., 2013). Given high rates and early onset of atherosclerosis and Coronary Heart Disease in HIV infected individuals (Boccara et al., 2013; Hsue et al., 2009), it will be of interest to see if a Prevotella-rich microbiota in HIV-infected individuals is associated with high TMAO levels in plasma.
The failure of ART to consistently restore the gut microbiota to a state resembling HIV-negative individuals is consistent with persistence of gastrointestinal diseases with ART. HIV positive subjects on successful ART remain at greater risk of multiple inflammatory diseases, including atherosclerosis (Hsue et al., 2009). Diseases of nutrition ranging from wasting to lipodystrophy persist, or for lipodystrophy can even be exacerbated by ART (Carr et al., 1998; Wanke, 1999; Wanke et al., 2000). Although ART can improve chronic diarrhea in patients with advanced HIV infection and lead to disappearance of parasites such as Cryptosporidium sp. from stool (Foudraine et al., 1998), other parasites can persist (Boyles et al., 2012). A persistence of an HIV-associated microbiota in some individuals on long-term ART is also consistent with the observation that ART treatment generally does not completely restore CD4+ T cells in GALT (Estes et al., 2008). Discordance between CD4+ T cell counts in GALT and the periphery may explain why we did not observe a correlation between peripheral CD4+ T cell count and the degree of divergence in fecal microbiota composition from HIV-negative.
It will be of importance to further study the effects of HIV infection on the gut microbiota in the context of different disease states, cultures, and diets so that the complex interactions between lifestyle, diets and the immune system in shaping microbiota composition can be determined. Further studies of the immunomodulatory properties and inflammation-tolerance of bacterial isolates related to OTUs that change during HIV infection will provide insight into driving factors of gut microbiota changes and their potential impact on health.
Experimental Procedures
Subject Recruitment
Four cohorts were prospectively recruited from the University of Colorado Hospital Infectious Disease Group Practice: 1) Recent HIV-1 infection: Individuals likely infected within the prior 6 months. 2) Chronic HIV-1 infection untreated: Individuals infected for >6 months and ART drug-naïve or off treatment for >6 months. 3) Chronic HIV-1 infection on long-term ART: ART treatment for ≥12 months with a minimum of three ART drugs prior to study entry and viral suppressed for >6 months 4) Healthy controls: HIV negative individuals matched to HIV-infected participants for sex, age, geographic location and smoking status (See Supplemental Methods for further cohort characteristics). Individuals from Cohort 2 sometimes initiated ART treatment immediately following the initial visit and donated an additional fecal sample 6.7–8.5 months later. Informed consent was obtained from each subject and the study protocol was approved by the Colorado Multiple Institution Review Board.
DNA Extraction and Sequencing
Stool samples were collected on a sterile swab by the patient either during or 12 hours prior to the visit and stored at −80°C. DNA was extracted using the standard Power Soil Kit protocol (MoBio). Extracted DNA was PCR amplified with barcoded primers targeting the V4 region of 16S rRNA as detailed in (Yatsunenko et al., 2012). Control water samples that had undergone the same DNA extraction and PCR amplification procedures were also sequenced. Each PCR product was quantified using PicoGreen (Invitrogen) and equal amounts (ng) of DNA from each sample was pooled and cleaned using the UltraClean® PCR Clean-Up Kit (MoBio). Sequences were generated on two runs on a MiSeq personal sequencer (Illumina, San Diego, CA).
Sequence Data Analysis
Raw sequences were quality filtered and assigned to samples based on their barcodes using the default parameters of QIIME version 1.5.0 (Caporaso et al., 2010) (See Supplemental Methods). Sequences were assigned to 97% ID OTUs by comparing them to a non-redundant reference database of near-full length sequences (February 4, 2011 Greengenes database) (DeSantis et al., 2006) and unassigned sequences were clustered into de novo OTUs using UCLUSTref (Edgar, 2010). Since samples contained between 4,694 and 72,828 sequences, analyses were standardized at 4,600 sequences per sample to avoid biases. The sequence data has been deposited in the QIIME database (http://www.microbio.me/qiime/) and at EBI (http://www.ebi.ac.uk/ena/; accession ERP003611).
UniFrac PCoA analyses were conducted using QIIME as detailed in the Supplement. Random Forests classification on the HIV data was carried out using SourceTracker software (http://sourceforge.net/projects/sourcetracker/)(Knights et al., 2011b) and custom code as detailed in the Supplement. Alpha diversity was calculated using three different measures 1) observed species: the number of 97% ID OTUs observed in 4600 sequences; 2) Shannon Index (Shannon, 1948) and 3) Phylogenetic Diversity (PD) (Faith, 1992). Only one sample was used for individuals sampled at 2 timepoints.
Bacterial families and genera in each sample were determined using the RDP classifier retrained on the greengenes taxonomy (McDonald et al., 2012) and statistically compared as detailed in the Supplement. Discriminating OTUs were related to each other and to bacterial type strains using the ARB software package (Ludwig et al., 2004) as detailed in the Supplement.
To compare sequences from our samples to other studies, we used the QIIME database (http://microbio.me/qiime), which assigns sequences to 97% ID OTUs by comparing them to a reference database of near-full length sequences (February 4, 2011 Greengenes database) (DeSantis et al., 2006) using UCLUSTref (Edgar, 2010) as described in (Lozupone et al., 2013). Unassigned sequences were dropped from the analysis. We removed samples from individuals not between the ages of 10 and 66 and standardized at 4600 sequences/sample.
Immunologic assays
Bacterial lysates for B. dorei (DSM 17855), B. fluxus (DSM 22534), P. copri (DSM18205), E. biforme (DSM 3989) and E. coli serotype O55:K59(B5) were prepared and quantified as detailed in the Supplement. PBMCs were isolated from blood of 10 untreated HIV positive subjects with chronic infection (median CD4+ T cells 508 cells/ul; range 248–742 and VL 55495 copies of HIV RNA/ml; range 554–123000) and 11 HIV negative subjects as described in the Supplement. For the T cell proliferation assays, PBMCs were labeled with 2 µM Cell Trace to track cell division (Invitrogen), plated at 1 × 106 cells/ml in a 48 well plate, and cultured with 1 µg/ml of bacterial lysates, LPS, 2.5 µg/ml PHA, or medium alone at 37°C in a humidified 5% CO2 atmosphere. After 6 days, cells were washed with PBS containing 1% BSA and surfaced stained with anti-CD4 (PE-Cy5-5; BD Biosciences), anti-CD3 (Qdot 605; Invitrogen), anti-CD19 (Alexa Fluor 405; Caltag), CD45RA, and CD27 for 30 min at 4°C, then washed and fixed with 1% formaldehyde. Cells were analyzed using a LSR-II flow cytometer (BD Immunocytometry Systems) as described in the Supplement.
For the cytokine analysis, PBMCs (1 × 106 cells/ml) were cultured in 96 well plates with 1 µg/ml of bacterial lysates, LPS or medium alone. Cultures were incubated at 37°C in a humidified 5% CO2 atmosphere for 24 hours and culture supernatants were harvested and centrifuged to remove remaining cells and stored at −80°C. IL-10 and TNF-α levels were assayed using a 7-plex Inflammatory Cytokine kit (Meso Scale Development, Gaithersberg MD). Assays were performed per manufacturer’s instructions and analyzed using a Sector Imager 2400 (Meso Scale Discovery).
Supplementary Material
Highlights.
US adults with chronic HIV infection have highly characteristic gut microbiota
Antiretroviral therapy does not always restore gut microbiota to a healthy state
Microbiota in HIV-infection and with animal fat/protein poor diets are similar
CD4+ T cell response to Bacteroides species preferentially depleted with infection
Acknowledgements
This work was supported by a LHMP sponsored project (UO1HL098996 to AF, TC, SF and RK) and by the Colorado Clinical and Translational Sciences Institute (UL1TR000005). CL was supported by K01DK090285 and RK was supported by the Howard Hughes Medical Institute. We would like to thank Beverly Putnam and Christine Grismer for subject recruitment, Sam MaWhinney and Amanda Allshouse for statistical support, Dan Knights for contributing custom code for Random Forests analysis, and Maria del Carmen Portillo for help with anaerobic culture. We would particularly like to thank the study participants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnason I, Sharpstone DR, Francis N, Marker A, Taylor C, Barrett M, Macpherson A, Baldwin C, Menzies IS, Crane RC, et al. Intestinal inflammation, ileal structure and function in HIV. AIDS. 1996;10:1385–1391. doi: 10.1097/00002030-199610000-00011. [DOI] [PubMed] [Google Scholar]
- Boccara F, Lang S, Meuleman C, Ederhy S, Mary-Krause M, Costagliola D, Capeau J, Cohen A. HIV and coronary heart disease: time for a better understanding. Journal of the American College of Cardiology. 2013;61:511–523. doi: 10.1016/j.jacc.2012.06.063. [DOI] [PubMed] [Google Scholar]
- Boyles TH, Black J, Meintjes G, Mendelson M. Failure to eradicate Isospora belli diarrhoea despite immune reconstitution in adults with HIV--a case series. PloS one. 2012;7:e42844. doi: 10.1371/journal.pone.0042844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal immunology. 2008;1:23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nature medicine. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. The Journal of experimental medicine. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A, Samaras K, Burton S, Law M, Freund J, Chisholm DJ, Cooper DA. A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance in patients receiving HIV protease inhibitors. AIDS. 1998;12:F51–F58. doi: 10.1097/00002030-199807000-00003. [DOI] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell host & microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. The Journal of infectious diseases. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- Cubas RA, Mudd JC, Savoye AL, Perreau M, van Grevenynghe J, Metcalf T, Connick E, Meditz A, Freeman GJ, Abesada-Terk G, Jr., et al. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nature medicine. 2013;19:494–499. doi: 10.1038/nm.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SM, Manuzak JA, Leone AK, Lee EJ, Rogers LM, McCarter MD, Wilson CC. HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli. J Immunol. 2012;189:885–896. doi: 10.4049/jimmunol.1200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre CA, Amraoui S, DeRosa A, Jourdain JP, Vimeux L, Goguet M, Degrelle S, Feuillet V, Liovat AS, Muller-Trutwin M, et al. Pivotal role of M-DC8(+) monocytes from viremic HIV-infected patients in TNFalpha overproduction in response to microbial products. Blood. 2012;120:2259–2268. doi: 10.1182/blood-2012-03-418681. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CL, Ma ZM, Mann SK, Li CS, Wu J, Knight TH, Yotter T, Hayes TL, Maniar AH, Troia-Cancio PV, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–370. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes J, Baker JV, Brenchley JM, Khoruts A, Barthold JL, Bantle A, Reilly CS, Beilman GJ, George ME, Douek DC, et al. Collagen deposition limits immune reconstitution in the gut. The Journal of infectious diseases. 2008;198:456–464. doi: 10.1086/590112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- Foudraine NA, Weverling GJ, van Gool T, Roos MT, de Wolf F, Koopmans PP, van den Broek PJ, Meenhorst PL, van Leeuwen R, Lange JM, et al. Improvement of chronic diarrhoea in patients with advanced HIV-1 infection during potent antiretroviral therapy. AIDS. 1998;12:35–41. doi: 10.1097/00002030-199801000-00005. [DOI] [PubMed] [Google Scholar]
- Gauffin Cano P, Santacruz A, Moya A, Sanz Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PloS one. 2012;7:e41079. doi: 10.1371/journal.pone.0041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington CR, Lucchini S, Ridgway KP, Wegmann U, Eaton TJ, Hinton JC, Gasson MJ, Narbad A. A short-oligonucleotide microarray that allows improved detection of gastrointestinal tract microbial communities. BMC microbiology. 2008;8:195. doi: 10.1186/1471-2180-8-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe R, Dillon S, Rogers L, McCarter M, Kelly C, Gonzalez R, Madinger N, Wilson CC. Evidence for dendritic cell-dependent CD4(+) T helper-1 type responses to commensal bacteria in normal human intestinal lamina propria. Clin Immunol. 2009;131:317–332. doi: 10.1016/j.clim.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue PY, Hunt PW, Schnell A, Kalapus SC, Hoh R, Ganz P, Martin JN, Deeks SG. Role of viral replication, antiretroviral therapy, and immunodeficiency in HIV-associated atherosclerosis. AIDS. 2009;23:1059–1067. doi: 10.1097/QAD.0b013e32832b514b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Whitehead RN, Griffiths L, Dawson C, Bai H, Waring RH, Ramsden DB, Hunter JO, Cauchi M, Bessant C, et al. Diversity and distribution of sulphate-reducing bacteria in human faeces from healthy subjects and patients with inflammatory bowel disease. FEMS immunology and medical microbiology. 2012;65:55–68. doi: 10.1111/j.1574-695X.2012.00935.x. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, Kato LM, Fagarasan S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS microbiology reviews. 2011a;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Koren O, Ley RE, Field D, Knight R, DeSantis TZ, Kelley ST. Supervised classification of microbiota mitigates mislabeling errors. The ISME journal. 2011b;5:570–573. doi: 10.1038/ismej.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox TA, Spiegelman D, Skinner SC, Gorbach S. Diarrhea and abnormalities of gastrointestinal function in a cohort of men and women with HIV infection. The American journal of gastroenterology. 2000;95:3482–3489. doi: 10.1111/j.1572-0241.2000.03365.x. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine. 2013 doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS computational biology. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubinoux J, Bronowicki JP, Pereira IA, Mougenel JL, Faou AE. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS microbiology ecology. 2002;40:107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Applied and environmental microbiology. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, Achermann G, Wendel D, Vazquez-Baeza Y, Jansson JK, Gordon JI, Knight R. Meta-analysis of studies of the human microbiota. Genome research. 2013 doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madge S, Kinloch-de-Loes D, Mercy D, Johnson MA, Weller IVD. Lipodystrophy in patients naive to HIV protease inhibitors. AIDS. 1999;13:735. doi: 10.1097/00002030-199904160-00020. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme Journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, Lozupone CA, Hamady M, Knight R, Bushman FD. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS pathogens. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nature reviews Immunology. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH, Melchjorsen J, Larsen CS, Paludan SR. Innate immune recognition and activation during HIV infection. Retrovirology. 2010;7:54. doi: 10.1186/1742-4690-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhya I, Hansen R, El-Omer EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- Nicholaou MJ, Martinson JJ, Abraham AG, Brown TT, Hussain SK, Wolinsky SM, Kingsley LA. HAART-Associated Dyslipidemia Varies by Biogeographical Ancestry in the Multicenter AIDS Cohort Study. AIDS research and human retroviruses. 2013;29:871–879. doi: 10.1089/aid.2012.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. Journal of medical microbiology. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said EA, Dupuy FP, Trautmann L, Zhang Y, Shi Y, El-Far M, Hill BJ, Noto A, Ancuta P, Peretz Y, et al. Programmed death-1-induced interleukin-10 production by monocytes impairs CD4+ T cell activation during HIV infection. Nature medicine. 2010;16:452–459. doi: 10.1038/nm.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Goebal BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke CA. Epidemiological and clinical aspects of the metabolic complications of HIV infection the fat redistribution syndrome. AIDS. 1999;13:1287–1293. doi: 10.1097/00002030-199907300-00004. [DOI] [PubMed] [Google Scholar]
- Wanke CA, Silva M, Knox TA, Forrester J, Speigelman D, Gorbach SL. Weight loss and wasting remain common complications in individuals infected with human immunodeficiency virus in the era of highly active antiretroviral therapy. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2000;31:803–805. doi: 10.1086/314027. [DOI] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. e1841. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA. IL-22 Deficiency Alters Colonic Microbiota To Be Transmissible and Colitogenic. J Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.