Bacteria swim by means of long flagella extending from the cell surface. These are assembled from thousands of protein subunits translocated across the cell membrane by an export machinery at the base of each flagellum. Unfolded subunits1,2,3 then transit through a narrow channel at the core of the growing flagellum to the tip, where they crystallize into the nascent structure. As the flagellum lengthens outside the cell the rate of flagellum growth does not change4. How is subunit transit maintained at a constant rate without a discernable energy source in the channel of the external flagellum5? We present evidence for a simple physical mechanism for flagellum growth that harnesses the entropic force of the unfolded subunits themselves. We show that a subunit docked at the export machinery can be captured by a free subunit through head-to-tail linkage of juxstaposed N- and C-terminal helices. We propose that sequential rounds of linkage would generate a multi-subunit chain that pulls successive subunits into and through the channel to the flagellum tip, and by isolating filaments growing on bacterial cells we reveal the predicted chain of head-to-tail linked subunits in the transit channel of flagella. Thermodynamic analysis confirms that links in the subunit chain can withstand the pulling force generated by rounds of subunit crystallization at the flagellum tip, and polymer theory predicts that as the N-terminus of each unfolded subunit crystallizes, the entropic force at the subunit C-terminus would increase rapidly overcoming the threshold required to pull the next subunit from the export machinery. This pulling force would adjust automatically over the increasing length of the growing flagellum, maintaining a constant rate of subunit delivery to the tip.
The structural subunits of the sequential rod, hook and flagellin filament of the bacterial flagellum (Extended Data Fig. 1) are unfolded and translocated across the cell membrane by a type III export machinery2,6, energised by ATP hydrolysis1,7 and the proton motive force (PMF)5,8,9. They pass into a central channel that extends the length of the growing flagellum outside the cell10,11,12. The channel has a c.20Å diameter so subunits must remain unfolded as they transit to the flagellum tip up to 15-20μm i.e. 10 cell lengths away2, where they crystallize into the growing structure13. Theoretical models describing the physical challenges presented by assembling flagella outside the living cell are set out in Supplementary Information 1, but how this feat is achieved remains a mystery. Models based on pushing or pumping mechanisms14,15 operating in the cell at the membrane entrance to the flagellum channel, e.g. powered by PMF, appear incompatible with constant rate transit, as lengthening of the pushed subunit column would engender a proportional increase in resistance and slowing of growth. Furthermore, the intrinsic flexibility of unfolded subunits would preclude transmission of compressive forces16. An alternative hypothesis17 involving diffusion of flagellin, relies on subunits adopting an unprecedented extended alpha-helical fold, and does not take into account interactions between subunits in the channel. In contrast to passive diffusion, the notion of active transport of subunits through the channel by a Brownian ratchet18 is compromised by the absence of ATP in the channel and the channel being too narrow to accommodate hypothetical motors.
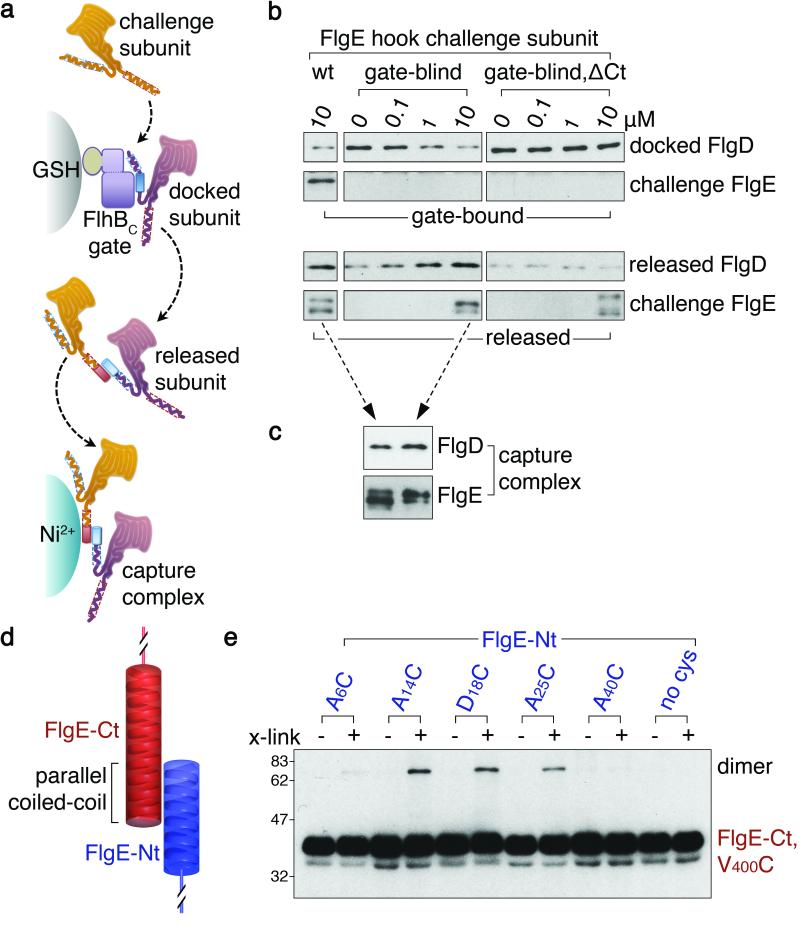
We set out to examine experimentally a fundamentally distinct hypothesis, in which we propose the energy for subunit transit though the channel resides in the unfolded subunits themselves as they move from the export machinery at the base of the flagellum2,19,20. We reasoned that such movement might be achieved by capture of each docked subunit by the preceding subunit already in the channel. Direct support for this possibility is provided by challenge experiments (Fig. 1) in which free flagellar hook subunit FlgE effected concentration-dependent release of another subunit docked at the export machinery gate component (i.e. in preformed subunit-gate complexes, FlgD-GSTFlhBC). Subunits docked at the gate component were captured, not displaced, by free subunit as the challenge subunit FlgE was engineered such that it was unable to bind the export gate (gate-blind; Extended Data Figs. 2 and 3), and heteromeric FlgD-FlgE subunit-subunit capture complexes were confirmed by affinity chromatography (Fig. 1c). Further capture assays established that gate-docked FlgD can be captured by other hook subunits, but not by filament cap subunit that is assembled later21,22 (Extended Data Fig. 4). We supposed that in the subunit docked at the export machinery, the extreme N-terminus would be available to the free C-terminus of the preceding subunit already in the channel, and that linkage of these juxtaposed termini could effect capture. Evidence for this was obtained by showing that C-terminally truncated gate-blind FlgE challenge subunit was unable to release gate-docked subunit in a concentration-dependent manner from the FlgD-FlhBC subunit-gate complex (Fig. 1b). We reasoned that links between N- and C- termini of unfolded adjacent subunits might adopt a parallel coiled-coil conformation (Fig 1d) and using unique cysteine-cysteine cross-links (FlgECt V400C with FlgE-Nt A14C, D18C or A25C) we trapped a subunit dimer compatible with just such a head-to-tail arrangement (Fig. 1e). Weaker cross-linking was detected between FlgE-Ct V400C and FlgE-Nt A6C, which are at opposite ends of the predicted coiled-coil, and no complexes were detected for FlgE-Nt A40C, which lies outwith the subunit extreme N-terminal helix (Fig. 1e). This conformation is different from that observed in assembled flagella, where subunit termini23 fold as anti-parallel coiled-coils that line the channel10,12 (Extended Data Fig. 5). The 8Å cysteine-cysteine cross-links could not form between subunits in the assembled hook, as cysteines are too far apart12,24 (Extended Data Table 1). The cross-linking data show that subunit capture is achieved by subunits linking head-to-tail via their terminal helices, predicted to form a parallel coiled-coil, and indicate that each subunit terminus contributes 14-25 residues to each coiled-coil link.
Figure 1. Free subunits capture subunits docked at the export gate via head-to-tail linkage of terminal helices.
a, Capture of FlgD subunits docked at glutathione (GSH) bead-immobilised GSTFlhBC export gate component by free challenge subunits. Released capture complexes were collected by Ni2+ affinity chromatography.
b, Docked FlgD subunit was challenged with increasing concentrations of free FlgE hook subunit, either wildtype (wt), unable to bind the export gate (gate-blind; Extended Data Fig. 3) or gate-blind and lacking C-terminal residues 359-403 (gate-blind, ΔCt). FlgE subunit lacking the C-terminus was attenuated for export (Extended Data Fig. 4e).
c, Released capture complexes generated in b, (dashed arrows) were isolated confirming that captured FlgD is linked to challenge subunit.
d, Juxtaposed carboxyl (FlgE-Ct) and amino (FlgE-Nt) terminal helices of sequential subunits adopt a parallel coiled-coil.
e, Subunit pairs linked head-to-tail (dimer) using in vitro site-specific cysteine-cysteine cross-links. FlgE and variants containing unique cysteines (A6C, A14C, D18C, A25C or A40C; Extended Data Table 2) within and adjacent to the N-terminal helix predicted to generate the coiled-coil were incubated, with (+) or without (−) BMOE cross-linker (x-link) and a FlgE variant (V400C) containing a unique cysteine within the C-terminal helix. FlgE derivatives lacked either C- or N-termini (FlgE-Nt, FlgE-Ct) to preclude self-interaction. No dimers were detected for subunits without engineered cysteines (Extended Data Fig. 5e).
All experiments were carried out at least three times and were biological replicates.
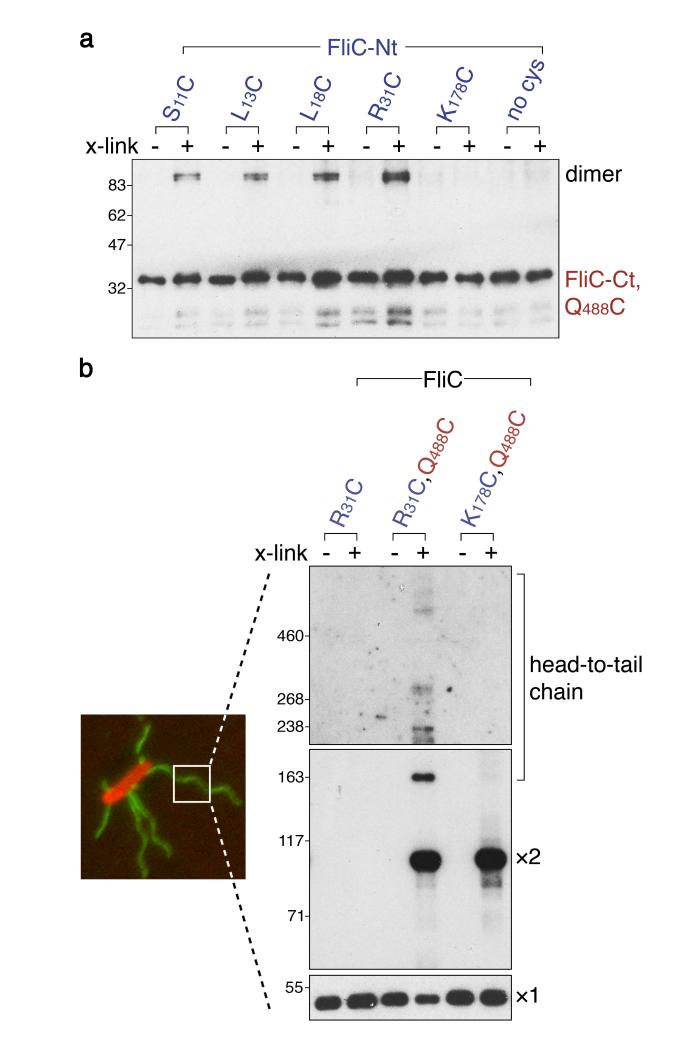
We suggest that successive rounds of such linkage could generate a subunit chain that extends through the flagellar channel. However, while a single unfolded hook subunit could span the channel in the rod/hook structure (~90nm), crystallizing into the tip of the hook as it simultaneously recruits the next subunit from the export machinery, the flagellar filament grows up to 20μm 2. Our proposed chain mechanism would therefore require multiple flagellin (FliC) subunits to link in the same way to span the transit channel of the flagellar filament. We therefore set out to visualize the linked flagellin subunits in vivo by isolating part of the predicted flagellin chain from within the flagellar filament growing on the surface of living cells. First, using again in vitro cysteine-cysteine cross-linking, we confirmed the same type of head-to-tail linkage between flagellin subunits that we had shown between hook subunits (Fig. 2a, FliC-CtQ488C or N489C with FliC-Nt S11C, L13C, L18C or R31C, but not with FliC-Nt K178C that lies outwith the N-terminal helix; Extended Data Table 1). The cross-linking distances indicate each flagellin terminus could contribute 21-32 residues to the coiled-coil link. We then set out to trap the predicted unfolded subunit chain during transit through the channel as the flagellum grows on bacteria. Using in vivo site-specific cross-linking of engineered flagellin containing cysteines shown to link in vitro (R31C, Q488C; Fig. 2a), we isolated higher order flagellin complexes and visualized, as a ladder, oligomeric chains containing three or more subunits (Fig. 2b). These oligomers could only form by head-to-tail linkage as a chain in the channel, and no chain was detected with flagellin containing R31C alone, or K178C and Q488C, which cannot engender head-to-tail links. Our data show that there are multiple linked subunits in the channel, incompatible with the recent theoretical model of filament growth17 in which transiting subunit do not interact with each other in the channel.
Figure 2. Head-to-tail linkage of flagellin subunits assembles a chain in the flagellum growing on the bacterial cell surface.
a, Flagellin pairs linked head-to-tail (dimer) using in vitro site-specific cysteine-cysteine cross-links. Flagellin (FliC-Nt, FliC-Ct) and its variants containing unique cysteines (S11C, L13C, L18C, R31C or K178C) within and adjacent to the N-terminal helix predicted to generate a coiled-coil were incubated, with (+) or without (−) BMOE cross-linker (x-link) and a FliC variant containing a unique cysteine (Q488C) within the C-terminal helix. No complexes were detected between subunits without engineered cysteines (Extended Data Fig. 6a)
b, Trapping of flagellin linked head-to-tail in chains within flagella (labelled green) growing on Salmonella cells (labelled red) using in vivo site-specific cysteine-cysteine cross-linking. Cells expressing recombinant full-length flagellin containing engineered cysteines (right panels; left lane, negative control R31C; centre lane, R31C and Q488C predicted to trap chain; right lane K178C and Q488C negative control) were incubated with (+) or without (−) BMOE cross-linker (x-link). Flagellar filaments were then isolated, depolymerised and resolved (immunoblot, panel exposure times decrease from top to bottom) to reveal monomer (x1), dimer (x2) and higher order head-to-tail chains of flagellin. We estimate between 6-10 linked subunits in the largest species. Dimers are predicted to form additionally by cross-linking between assembled flagellin C-terminal cysteines (Q488C) that line the channel and subunits in transit, so dimers are also seen in the controls (Extended Data Fig. 6b, c).
All experiments were carried out at least three times and were biological replicates.
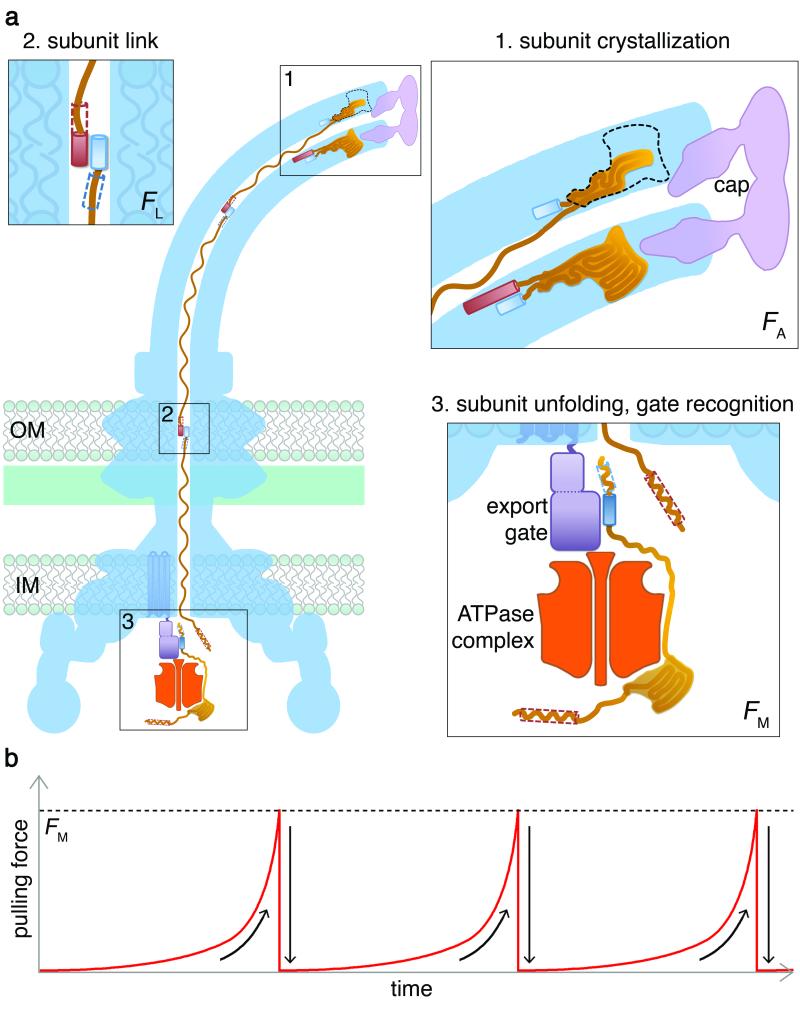
Having provided direct evidence of subunit capture, linkage and the resulting chain, we propose a physical mechanism for subunit transit energised by the thermal motion of the chain of linked unfolded subunits anchored at the flagellum tip. In this mechanism, depicted in Fig. 3 and supported with calculations in Supplementary Information 2, subunit folding at the tip would gradually decrease the number of free residues in the channel and intrinsically provide an increasing pulling force on the next linked subunit docked at the export machinery. The proposed mechanism requires strong anchoring of subunit at the tip25, and breaking this anchor would require an estimated a force FA of ~11nN. Interestingly, strong anchoring (FA of ~6-6.7nN) could also be achieved in the absence of subunit assembly into the crystallized structure, explaining how some flagellar proteins, e.g. filament cap and hook-filament junction proteins, are exported even when no longer required for flagellum assembly26 (similar export without assembly is observed for effectors secreted through related virulence needles; Supplementary Information 2a and 3). Secondly, the mechanism requires parallel coiled-coil linking of sequential subunit termini and, as each hook subunit contributes 14-25 residues to the coiled-coil, breaking this link would require an estimated force FL of 400-700pN. Breaking the links between the more abundant flagellin subunits would require a force of the same order of magnitude. The chain mechanism requires that subunit anchoring at the flagellum distal tip must be stronger than subunit-subunit linking in the channel, which in turn must be stronger than the force FM required to pull subunits from the export machinery into the channel. Affinity of subunit for the export gate component, measured by ITC (Extended Data Fig. 2a), estimates FM at ~30pN, fulfilling the requirement FM<FL<FA.
Figure 3. An entropic chain mechanism for flagellum growth outside the cell.
a, 1. Subunit crystallization beneath the cap foldase13 provides a strong anchor (force to break anchor, FA; Supplementary Information 2). 2. Sequential subunits are linked (force to break link, FL) head-to-tail in a chain by juxtaposed terminal helices forming parallel coiled-coils. 3. Subunits docked at the export ATPase6 are unfolded1 and docked by the export machinery. The N-terminal helix of the docked subunit is then captured (force to break docking FM) into the subunit chain by the free C-terminal helix of an exiting subunit in the flagellar channel.
b, Successive rounds of subunit capture from the export machinery. As an unfolded subunit crystallizes, the pulling force at its free end increases to reach the FM threshold (dashed line) whereupon the next subunit is captured from the export machinery. The pulling force then drops rapidly as the new unfolded subunit enters the channel. This process repeats for each subunit captured into the chain.
Polymer theory describes the entropic pulling force at the C-terminus of the unfolded subunit chain anchored at its N-terminus by crystallization into the flagellum, and explains how this force adjusts as channel length increases27,28 (Supplementary Information 2c). In an anchored chain of N residues each of length a, when a distance R separates the C-terminus from the N-terminal anchor point the force depends on R/Na (Extended Data Fig. 6). As a subunit crystallizes, fewer residues are left free in the channel (Na decreases), the pulling force F(R,Na) at the export machinery increases, eventually reaching the critical value FM and the next subunit is pulled from the export machinery into the channel. Once this occurs, Na increases, the entropic force drops rapidly and the “slack” of residues is available for a new round of subunit crystallization at the tip. This process repeats for each subunit pulled into the channel (Fig. 3b). The same principle applies even if the subunit chain breaks inside the channel, either spontaneously or by shearing of the flagellum. The most cell-distal subunit in the remaining chain could diffuse to the tip4 and start crystallizing, thus providing an increasing anchoring that would gradually re-start the pulling mechanism. Crucially, the entropic force F(R,Na) automatically adjusts with the length of the multi-subunit chain (Supplementary Information 3 and Extended Data Table 2) engendering a constant rate of subunit transit independent of channel length. The chain mechanism we propose is therefore supported by both experimentation and thermodynamic analysis, and we suggest that it explains the published observations on how flagella grow outside the bacterial cell.
Online Only Methods
Bacterial Strains, Recombinant Plasmids and Antibodies
Wildtype S. typhimurium SJW1103 is motile29, derived mutants carry stable lesions in flagellar genes flhDC (SJW136830), ΔflgD::kmR and ΔfliC::kmR (this study) in which flagellar genes were replaced by a kanamycin resistance cassette, using the λ Red recombinase system31. Recombinant genes were expressed in E. coli C4132 or S. typhimurium SJW1368. Bacteria were cultured at 30-37°C in Luria-Bertani (LB) broth, containing (where appropriate) ampicillin (100μg.ml−1), tetracycline (3μg.ml−1), chloramphenicol (20μg.ml−1) and/or amino acid p-benzoyl-L-phenylalanine (pBpa, 1mM) and harvested by centrifugation (8,000×g for 15min). Recombinant proteins were expressed from IPTG (0.8mM) inducible plasmids pET15b, pET20b33, pGEX-4T-334, pDULE35 or pACT736 or from L-arabinose (0.0002-0.2%w/v) inducible pBAD1837. To construct recombinant plasmids encoding: wildtype, derivative and fusion genes (a full list and description of the constructs used in this study can be found in Extended Data Table 3). S. typhimurium flhB, flgG, flgD, flgE, fliK, fliC, fliD and sptP22 were each amplified from chromosomal DNA by PCR or overlap-extension PCR using Pfu Ultra DNA polymerase. PCR products were inserted XbaI-HindIII/SalI into pBAD18 with or without 3×FLAG or C-terminal 6His tags. For GST fusion constructs, gene products were inserted BamHI-XhoI into pGEX-4T-3. For C-terminal His-tagged, 3×FLAG tagged and pACT7 constructs, gene products were inserted NcoI-BamHI into pET15b or NdeI-BamHI into pET20b and pACT7. Inserts were verified by DNA sequencing. Analysis by immunoblotting was carried out using the following antibodies: FLAG antibody (Sigma, suitable for immunoblotting); His antibody (Qiagen, suitable for immunoblotting); FliC antibody (non-commercial, suitable for immunoblotting); FlgD antibody (non-commercial, suitable for immunoblotting); FliD antibody (non-commercial, suitable for immunoblotting).
Purification of Proteins
Proteins were purified as described19,38,39, recombinant proteins engineered to possess affinity tags are listed in Extended Data Table 3. Briefly, recombinant cells were resuspended in buffer, GSTFlhB derivatives in buffer A (50 mM Tris-HCl [pH 7.4], 200mM NaCl), C-terminally His6 tagged derivatives in buffer B (50mM Tris-HCl [pH 7.4], 400mM NaCl, 5-10mM imidazole), derivatives to be purified by ion exchange in 50mM Na-phosphate [pH 7.0-7.8], 5-50mM NaCl and lysed by cell disruption (Constant systems). Membranes, unlysed cells, and insoluble proteins were removed (40,000×g, 1h), and the cleared cell lysate was incubated with affinity resin (glutathione sepharose [GSH-resin; GE Healthcare], Ni2+-Nitrilotriacetic acid agarose [Ni2+-affinity; Qiagen] or anion exchange resin (Q HP [GE Healthcare]). After washing (>10 column volumes buffer), complexes were eluted using buffer containing either 10mM glutathione (GSH-resin), 600mM imidazole (Ni2+-affinity) or an increasing NaCl gradient (Q HP), dialyzed against an appropriate buffer.
Isothermal titration calorimetry
ITC was performed at 20°C using a VP-ITC system (Microcal Inc). Purified proteins were dialyzed into 50mM Tris-HCl (pH 7.4), 100mM NaCl. The calorimetry cell was filled with 1.4ml of GSTFlhBC at 58μM before making sequential injections of between 0.5μl and 4.0μl of 1,600μM FlgD wildtype at 200-s intervals from the syringe to a final volume of 116.5μl. Controls contained titrant or titrand against buffer alone. The heat due to the binding reaction is the difference between the heat of reaction and the heat of dilution. The heat evolved following each injection was obtained from the integral of the calorimetric signal using Origin software (Microcal).
Affinity chromatography copurification
Copurification of protein complexes was achieved with either glutathione Sepharose 4B or Ni2+-affinity20. Purified proteins or cleared cell lysates (input) were incubated together for 1h with affinity resin. After extensive washing with buffer A (GSH-resin) or buffer B containing 10-50mM imidazole (Ni2+-affinity) proteins were eluted in buffer containing either 20mM glutathione (GSH-resin) or 600mM imidazole (Ni2+-affinity) or in SDS loading buffer. Fractions were analysed by immunoblotting.
Flagellar subunit export
S. typhimurium harboring plasmids encoding flagellar subunits were cultured at 37°C in LB broth containing L-arabinose (0.0002-0.2% w/v) to mid-log phase (0.6-0.8A600). To remove accumulated exported subunits from the initial cultures, cells were centrifuged (16,000×g, 6min) and resuspended in new media, for a further 40mins cultures were incubated at 37°C. The cells were pelleted by centrifugation (16,000×g, 3min) and the clarified culture supernatant was passed through a 0.2μm nitrocellulose filter. Exported flagellar subunits were precipitated by 10% (v/v) trichloroacetic acid (TCA) on ice for 1h. Whole cell samples and exported fractions were resuspended in SDS-PAGE loading buffer (volume calibrated to cell density [A600]) and visualized by immunoblotting.
Photo-cross-linking assay with unnatural amino acids
To allow a greater shift in the SDS-PAGE migration of cross-linked complexes an autocleavage defective FlhBC variant (FlhBCN269A40) was used. Binding of FlgD subunit to GSTFlhBC and GSTFlhBCN269A was comparable (Extended Data Fig. 7b). Copurified protein complexes of GSTFlhBC N269A and FlgD wildtype or unnatural amino acid (pBpa35) derivatives were isolated as described above and eluted in buffer A containing 20mM glutathione. Samples were UV irradiated (350nm) for 8×5minutes at 4°C. Irradiated (+) and non-irradiated (−) fractions were analysed by immunoblotting.
Subunit capture
Preformed GSTFlhBC-FlgDFLAG complex bound to glutathione sepharose washed with 5 volumes of Buffer C, then divided and subsequently challenged with increasing concentrations [0, 0.1, 1 or 10μM] of individual, purified, subunit derivatives for 20mins at room temperature. After incubation, the unbound/released fraction was collected and a sample taken for analysis. Bound fractions were washed extensively with Buffer C, and eluted in SDS loading buffer. 10μM subunit challenged unbound/released fractions with were adjusted to contain 5mM imidazole, 0.5% w/v BSA and then Ni2+-affinity purified, samples were washed extensively with buffer (50mM Na-phosphate [pH 7.4], 250mM NaCl, 10mM imidazole, 0.5%w/v BSA), copurified captured samples were eluted in SDS loading buffer. Released, bound and captured samples were analysed by immunoblotting.
Rationale for choice of residues in cross-linking experiments
To distinguish subunit anti-parallel coiled-coil termini in the assembled structure from those compatible with subunits linked by alternative parallel coiled-coils we employed short range (8Å) cross-linking of engineered cysteine pairs in juxtaposed subunits. Residue pairs (for replacement with cysteines) were chosen that would not form cross-links between/within assembled subunits, i.e. cysteines would be more than 8Å away from each other. This was determined by measuring lateral and axial distances between cysteines in adjacent subunits in the assembled structure. For in vivo assays where two cysteines were introduced into each subunit, intra-subunit distances were also measured. This was achieved by analysing atomic models of the assembled hook and filament structures, generated by fitting the atomic resolution data of hook and filament subunits into their respective electron cryomicrographs10,11,12 Extended Data Fig. 5 and Extended Data Table 1. This cross-linking strategy was also designed to examine interactions with all faces of the helical termini.
In vitro site-specific cysteine cross-linking
Purified N- and C-terminally truncated hook subunits (either FlgE or FliC, see Extended Data Table 3 for full descriptions) containing engineered cysteines in their remaining termini were dialysed into buffer C (50mM Na-phosphate [pH7.0], 200mM NaCl, 1mM β-mercaptoethanol) and mixed (1:1 molar-ratio), with or without 150μM Bismaleimidoethane (BMOE)41, then incubated at room temperature for 40mins before the reaction was quenched using 4mM N-ethylmaleimide. Samples were then Ni2+-affinity purified and washed extensively with buffer C containing 10mM imidazole, isolated protein complexes were eluted in SDS loading buffer and analysed by immunoblotting.
In vivo site-specific cysteine cross-linking
Flagellin subunits subjected to in vivo cystiene cross-linking were isolated by adapting the method of Erlander et al42. S. typhimurium ΔfliC::kmR harboring pBAD18 plasmids encoding flagellin subunit derivatives with engineered cysteines (see Extended Data Table 3) were cultured at 37°C in LB broth containing L-arabinose (0.002% w/v) and 1mM dithiothreitol to late-log phase (~1.0 A600). Cultures were centrifuged (16,000×g, 3min), the cell pellet was resuspended in M9 media containing L-arabinose (0.2% w/v), with or without BMOE (0.1mM), and cultures were incubated at 37°C for a further 30mins. To remove free exported subunit monomers, cultures were centrifuged (16,000×g, 3min), the supernatant discarded and the cell pellet resuspended in 50mM Na-phosphate (pH 7.0), 200mM NaCl with or without BMOE (0.1mM) then shaken vigorously for 45min at 4°C. The cross-linking reaction was then quenched using 4mM N-ethylmaleimide. To remove cells, samples were centrifuged (1200×g, 4min) and the supernatant containing sheared flagellar filaments carefully decanted, this step was repeated. To pellet flagellar filaments the supernatant was centrifuged (108,000×g, 40min) and the resulting pellet was resuspended in 50mM Na-phosphate (pH 7.0), 200mM NaCl and incubated for 20min at 65°C to depolymerise filaments. Subunit concentration was assessed and the samples were precipitated by 10% (v/v) TCA on ice for 1h. Cross-linked subunit samples were resuspended in LDS sample buffer (volume calibrated to protein concentration) and visualized by electrophoresis on 3-8% Tris-Acetate PAGE (Invitrogen) followed by immunoblotting.
Fluorescence microscopy of Salmonella cells and flagella
S. typhimurium were cultured at 37°C in LB broth to late-log phase (~1.0 A600). Cultures were centrifuged (16,000×g, 3min), the cell pellet was resuspended, without vigorous mixing, in buffer A and incubated for 20min with 2% (v/v) paraformaldehyde (in 20mM PIPES, pH 7.4). Cells were centrifuged (16,000×g, 3min). The resulting cell pellet resuspended in buffer A and spread on a lysine coated glass cover slide, incubated at 25°C for 5min. Cells fixed on cover sides were probed43 using flagellin anti-sera followed by AlexaFluor-488/594-conjugated secondary antibody, cell membranes were stained for 10min with SynaptoRed, and visualized using a fluorescence microscope (Leica DM IRBE). Images were captured by a CCD digital camera (Hamamatsu) and processed using OpenLab (Improvision).
Supplementary Material
Acknowledgements
We thank Philip Hinchliffe for help with analysis of atomic structures, Koji Yonekura and Keiichi Namba for providing information on filament structure. Howard Berg and Nick Greene for helpful discussions, Allister Crow for advice on ITC, and Paraminder Dhillon for technical assistance. The work was supported by a Wellcome Trust Programme Grant (C.H. and G.M.F.).
Footnotes
Supplementary Information is also linked to the online version of the paper at www.nature.com/nature.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Full methods and associated references are also available in the online version of the paper.
References
- 1.Akeda Y, Galán JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 2.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nature Rev. Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrusci P, et al. Architecture of the major component of the type III secretion system export apparatus. Nature Struct. Mol. Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner L, Stern AS, Berg HC. Growth of flagellar filaments of Escherichia coli is independent of filament length. J. Bacteriol. 2012;194:2437–2442. doi: 10.1128/JB.06735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galán JE. Energizing type III secretion machines: what is the fuel? Nature Struct. Mol. Biol. 2008;15:127–128. doi: 10.1038/nsmb0208-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas J, Stafford GP, Hughes C. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. USA. 2004;101:3945–3950. doi: 10.1073/pnas.0307223101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibuki T, et al. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nature Struct. Mol. Biol. 2011;18:277–282. doi: 10.1038/nsmb.1977. [DOI] [PubMed] [Google Scholar]
- 8.Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 9.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 10.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 11.Samatey FA, et al. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature. 2004;431:1062–1068. doi: 10.1038/nature02997. [DOI] [PubMed] [Google Scholar]
- 12.Fujii T, Kato T, Namba K. Specific arrangement of alpha-helical coiled coils in the core domain of the bacterial flagellar hook for the universal joint function. Structure. 2009;17:1485–1493. doi: 10.1016/j.str.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Yonekura K, et al. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- 14.Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 2008;4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- 15.Tanner DE, Ma W, Chen Z, Schulten K. Theoretical and computational investigation of flagellin translocation and bacterial flagellum growth. Biophys. J. 2011;100:2548–2556. doi: 10.1016/j.bpj.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blundell JR, Terentjev EM. Buckling of semiflexible filaments under compression. Soft Matter. 2009;5:4015–4020. [Google Scholar]
- 17.Stern A, Berg H. Single-file diffusion of flagellin in flagellar filaments. Biophys. J. 105:182–184. doi: 10.1016/j.bpj.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julicher F, Ajdari A, Prost J. Modeling molecular motors. Rev. Mod. Phys. 1997;69:1269–1281. [Google Scholar]
- 19.Minamino T, Macnab RM. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 2000;182:4906–4914. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans LD, Stafford GP, Ahmed S, Fraser GM, Hughes C. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc. Natl. Acad. Sci. USA. 2006;103:17474–17479. doi: 10.1073/pnas.0605197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamino T, Doi H, Kutsukake K. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci. Biotechnol. Biochem. 1999;63:1301–1303. doi: 10.1271/bbb.63.1301. [DOI] [PubMed] [Google Scholar]
- 22.Stafford GP, et al. Sorting of early and late flagellar subunits after docking at the membrane ATPase of the Type III export pathway. J. Mol. Biol. 2007;374:877–882. doi: 10.1016/j.jmb.2007.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vonderviszt F, Ishima R, Akasaka K, Aizawa S. Terminal disorder: a common structural feature of the axial proteins of bacterial flagellum? J. Mol. Biol. 1992;226:575–579. doi: 10.1016/0022-2836(92)90616-r. [DOI] [PubMed] [Google Scholar]
- 24.Shaikh TR, et al. A partial atomic structure for the flagellar hook of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA. 2005;102:1023–1028. doi: 10.1073/pnas.0409020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vonderviszt F, Závodszky P, Ishimura M, Uedaira H, Namba K. Structural organization and assembly of flagellar hook protein from Salmonella typhimurium. J. Mol Biol. 1995;251:520–532. doi: 10.1006/jmbi.1995.0453. [DOI] [PubMed] [Google Scholar]
- 26.Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J. Bacteriol. 1984;159:1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sotta P, Lesne A, Victor JM. Monte Carlo simulation of a grafted polymer chain confined in a tube. J. Chem. Phys. 2000;112:1565–1573. [Google Scholar]
- 28.Blundell JR, Terentjev EM. Semiflexible filaments subject to arbitrary interactions: a Metropolis Monte Carlo approach. Soft Matter. 2011;7:3967–3974. [Google Scholar]
- 29.Yamaguchi S, et al. Genetic analysis of three additional fla genes in Salmonella typhimurium. J. Gen. Microbiol. 1984;130:3339–3342. doi: 10.1099/00221287-130-12-3339. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi K, Ohto Y, Aizawa S, Macnab RM, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 33.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Kaelin WG, et al. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 35.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nature Methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 36.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E.coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vonderviszt F, Závodszky P, Ishimura M, Uedaira H, Namba K. Structural organization and assembly of flagellar hook protein from Salmonella typhimurium. J. Mol. Biol. 1995;251:520–532. doi: 10.1006/jmbi.1995.0453. [DOI] [PubMed] [Google Scholar]
- 39.Evans LD, Hughes C. Selective binding of virulence type III export chaperones by FliJ escort orthologues InvI and YscO. FEMS Microbiol. Lett. 2009;293:292–297. doi: 10.1111/j.1574-6968.2009.01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraser GM, et al. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 2003;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen LL, Rosa JJ, Turner S, Pepinsky RB. Production of multimeric forms of CD4 through a sugar-based cross-linking strategy. J. Biol. Chem. 1991;266:18237–18243. [PubMed] [Google Scholar]
- 42.Erlander SR, Koffler H, Foster TF. Physical properties of flagellin from Proteus vulgaris, a study involving the application of the archibald sedimentation principle. Arch. Biochem. Biophys. 1960;90:139–153. doi: 10.1016/0003-9861(60)90625-1. [DOI] [PubMed] [Google Scholar]
- 43.Stafford GP, Hughes C. Salmonella typhimurium flhE, a conserved flagellar regulon gene required for swarming. Microbiol. 2007;153:541–547. doi: 10.1099/mic.0.2006/002576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.