Abstract
CAAX proteins play essential roles in multiple signalling pathways, controlling processes such as proliferation, differentiation and carcinogenesis 1. The ~120 mammalian CAAX proteins function at cellular membranes and include the Ras superfamily of small GTPases, nuclear lamins, the γ-subunit of heterotrimeric GTPases, and several protein kinases and phosphatases 2. Proper localization of CAAX proteins to cell membranes is orchestrated by a series of post-translational modifications of their C-terminal CAAX motifs 3 (where C is cysteine, A is an aliphatic amino acid and X is any amino acid). These reactions involve cysteine prenylation, -AAX tripeptide cleavage, and methylation of the carboxyl prenylated Cys residue. The major CAAX protease activity is mediated by the Ras and a-factor converting enzyme 1 (Rce1), an integral membrane protease of the endoplasmic reticulum 4,5. Information on the architecture and proteolytic mechanism of Rce1 has been lacking. Here, we report the crystal structure of a Methanococcus maripaludis homolog of Rce1, whose endopeptidase specificity for farnesylated peptides mimics that of eukaryotic Rce1. Its structure, comprising eight transmembrane α-helices, and catalytic site, are distinct from other intramembrane proteases (IMPs). Catalytic residues are located ~10 Å into the membrane and are exposed to the cytoplasm and membrane through a conical cavity that accommodates the prenylated CAAX substrate. The farnesyl lipid is proposed to bind to a site at the opening of two transmembrane α-helices, which then positions the scissile bond adjacent to a glutamate-activated nucleophilic water molecule. This study suggests that Rce1 is the founding member of a novel IMP family, the glutamate IMPs.
Rce1 is a type II CAAX prenyl endopeptidase first identified in Saccharomyces cerevisiae together with the type I CAAX-processing enzyme, ZMPSTE24/Ste24p or AFC1p 4,6. ZMPSTE24/Ste24p is a zinc metalloprotease with a specific role in the processing of prelamin A in all eukaryotes and a-factor in yeast. Rce1 in contrast, has a much wider specificity, processing all farnesylated and geranylgeranylated CAAX proteins. However, extensive sequence and biochemical analyses were unable to classify Rce1 within the three conventional IMP families. These are the rhomboids 7, the intramembrane metalloproteases - S2P 8, and the aspartyl proteases - presenilin 9 and SPP. The membrane proteases ZMPSTE24/Ste24p 10,11 and FlaK 12 have their catalytic sites at the membrane interface. Rce1 belongs to the ABI (Abortive Infection) family of putative integral membrane proteases with homologs in all three domains of life. The ABI family is defined by three conserved motifs 13,14 that constitute the catalytic site of the ABI proteases, and whose importance has been demonstrated by mutational analysis of yeast Rce1p 15,16. Rce1 inactivation resulted in the mislocalization of Ras proteins from the plasma membrane 17. The consequent disruption of Ras signalling inhibited Ras-induced transformation of fibroblasts 17, but accelerated progression of K-RAS-induced myeloproliferative disease 17,18. Rce1-deficient mice develop lethal dilated cardiomyopathy 19, and Rce1 is also essential for the survival of photoreceptor cells 20.
To understand the structure and catalytic mechanism of Rce1, we examined the expression and solubility properties of ~30 Rce1 homologues (including human, yeast and prokaryotes) using fluorescence size exclusion chromatography (FSEC) and differential scanning fluorimetry (data not shown). The archaeal Methanococcus maripaludis Rce1 (MmRce1) was identified as a suitable candidate for structural studies and the full-length protein (276 residues, 15% sequence identity to human Rce1) was crystallized in complex with a conformation-sensitive monoclonal antibody Fab fragment (Extended Data Fig. 1). The structure of MmRce1-Fab was determined by molecular replacement using the Fab fragment as a search model, and the complex was refined to 2.5 Å resolution (Extended Data Table 1).
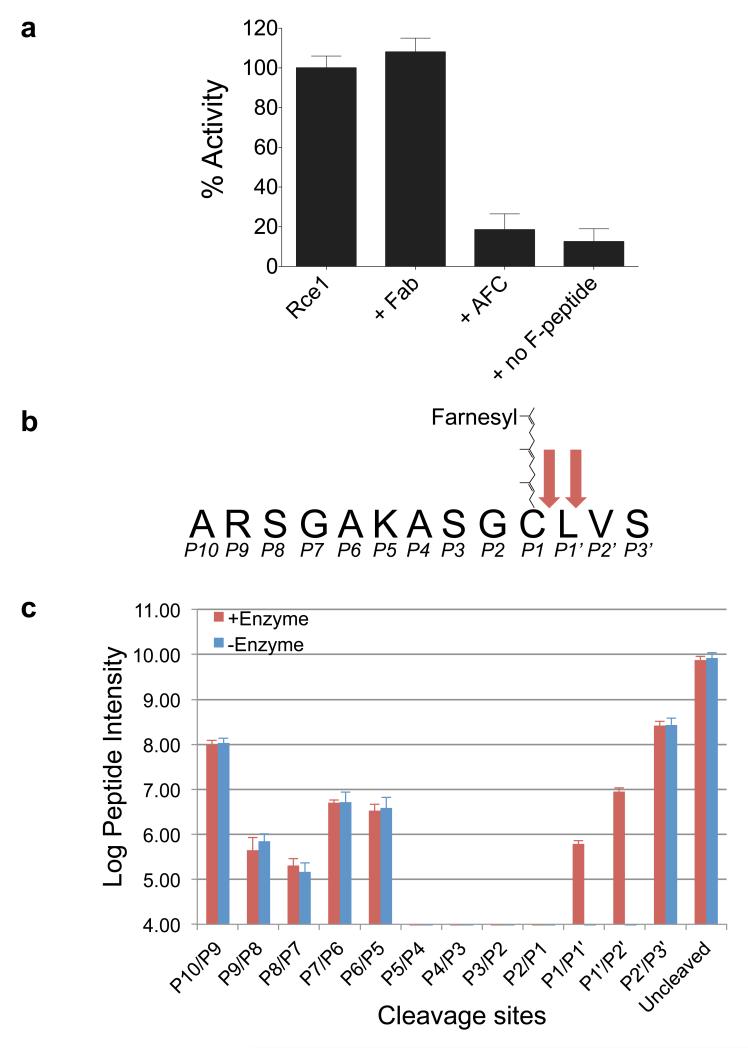
A fluorescence-based protease assay showed that MmRce1 hydrolyses a farnesylated peptide modelled on the C-terminus of human RhoA (Fig. 1a, b and Extended Data Fig. 2). Similar to its eukaryotic orthologs 21,22, peptide hydrolysis is dependent on a farnesylated cysteine, although in contrast to human Rce1 (ref. 21), MmRce1 did not proteolyze geranylgeranylated peptides (data not shown). Eukaryotic Rce1 is an endoprotease cleaving specifically C-terminal and adjacent to the farnesyl cysteine (P1) 4,23. Mass spectrometry analysis indicated that MmRce1 is also an endoprotease, although with slightly relaxed specificity, cleaving the CAAX motif C-terminal to both P1 and P1′ (Fig. 1b, c). MmRce1 is inhibited by N-acetyl-S-farnesyl-L-cysteine (AFC), the minimal analog of farnesylated peptides (Fig. 1a). The ability of MmRce1 to cleave farnesylated peptides specifically, validates it as a model for understanding the mechanism of CAAX processing by eukaryotic Rce1.
Figure 1. MmRce1 is an endoprotease specific for farnesylated peptides.
a, Proteolytic activity of wild type MmRce1 compared with the MmRce1-Fab complex (+ Fab), MmRce1 incubated with molar excess of AFC (+ AFC) and MmRce1 incubated with a non-farnesylated peptide (+ non F-peptide). Apparent Km: 19.7 μM ± 1.0 μM, apparent k: 0.175 ± 0.0027 sec−1. The mean and standard deviation of 3 experiments are shown. b, A schematic representation of the RhoA-derived farnesylated peptide. The two cleavage sites identified by mass spectrometry are marked with red arrows. c, Semi-quantitative mass spectrometry graph of the uncleaved and truncated MmRce1 farnesylated peptides. Many of the truncated forms were also present in the ‘- Enzyme’ control sample. These might be bi-products from peptide synthesis that are isobaric with truncations corresponding to positions (P2′/P3′ and P6/P5 to P10/P9). Only the P1′/P2′ and P1/P1′ truncations [being ARSGAKASGC(farnesyl)L and ARSGAKASGC(farnesyl), respectively] are found in the ‘+ Enzyme’ sample. The mean and two standard deviation of four experiments are shown.
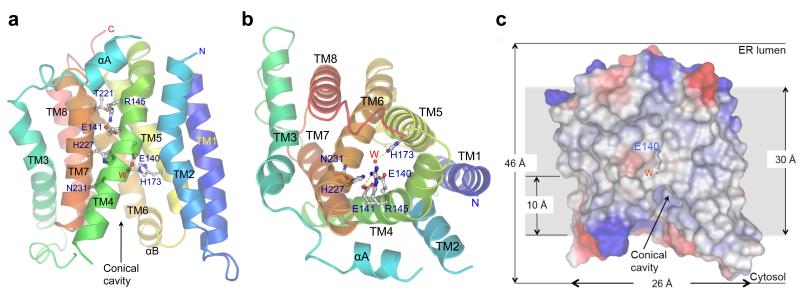
The structure of MmRce1 comprises eight conserved transmembrane α-helices (TM1-8), with two peripheral membrane α-helices (αA and αB) (Fig. 2a, b). Rce1 is topologically distinct from other IMPs, and to our knowledge, represents a novel protein fold. The molecule is approximately 35 Å in length, 26 Å in width and 46 Å in height, allowing it to be embedded in the lipid membrane (Fig. 2c). Except for αA and αB that interconnect TM2 with TM3 and TM7 with TM8, respectively, short loops link TM helices.
Figure 2. MmRce1 is an integral membrane protease with eight TM α-helices.
a, b, Two views showing ribbon-representations of MmRce1 (molecule C from the asymmetric unit). The side chains of the five invariant ABI domain residues (and conserved Arg145 and Thr210) are shown. The catalytic water (W) is a red sphere. a, View of the molecule parallel to the membrane. b, View of the molecule from the ER lumen side. c, The molecular surface of MmRce1, colour-coded by electrostatic potential. The lipid membrane (~30 Å) is indicated by the grey background, based on the distribution of non-polar residues and transmembrane helices. The Glu140 side chain and the catalytic water (W) are shown. The catalytic water (W) is located ~10 Å into the membrane.
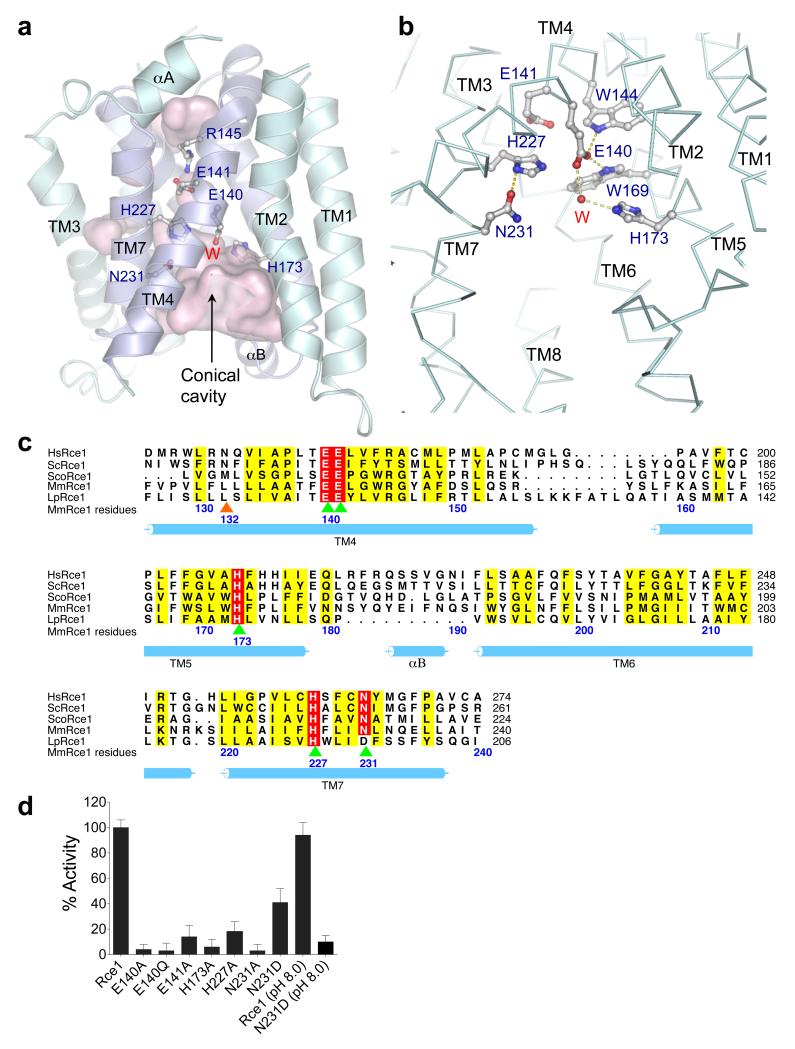
The ~100-residue ABI domain corresponds to TM4, TM5, TM6 and TM7 (Fig. 3a, c). These four TM helices form an anti-parallel helix bundle possessing approximate two-fold symmetry that is surrounded by the less conserved TM helices 1, 2, 3 and 8 (Extended Data Fig. 3). The three conserved motifs of the ABI domain are positioned on TM4, TM5 and TM7 (Fig. 3a, c). Seven TM helices delineate a large conical catalytic cavity (volume 1,400 Å3) (Fig. 2a, c and Fig. 3a). The catalytic residues Glu140, His173, His227 and Asn231 are located at the top of the cavity inside the membrane (Figs 2c and 3a). This conformation of TM helices allows unrestricted solvent access to the catalytic site from the cytoplasm at the base of the cavity. One side of the conical cavity is open to the membrane through a gap between TM2 and TM4. Access to the periplasm (ER lumen) is blocked by the conserved Arg145, which interacts with Thr210 and the invariant Glu141 (Fig. 2a, b).
Figure 3. The conserved catalytic ABI domain of Mm Rce1 is responsible for catalysis.
a, Outline of the MmRce1 cavities in pink surface representation. The side chains of the five invariant ABI domain residues (and conserved Arg145) and the catalytic water (W) are shown. TM4, 5, 6 and 7 constitute the ABI domain and are in dark blue. b, Detailed interactions of the catalytic water (red sphere) with the catalytic residues (Glu140 and His173). Hydrogen bonds are shown as dotted lines. His227 and Asn231 are the oxyanion hole residues. c, Multiple sequence alignment of the ABI domains from Rce1 homologues representing all three domains of life. HsRce1: Homo sapiens (UniProt: Q9Y256), ScRce1: Saccharomyces cerevisiae (UniProt: Q03530), ScoRce1: Streptomyces coelicolor (UniProt: Q9XAK4), MmRce1: Methanococcus maripaludis (UniProt: Q6LZY8), LpRce1: Lactobacillus plantarum (UniProt: C6VK86). Residues mutated in this study, and which disrupt activity, are indicated in green and orange arrows. Green arrows depict catalytic residues. The orange arrow indicates a putative farnesyl lipid-binding residue of TM4. d, Proteolytic activity of wild type MmRce1 and point mutants towards a farnesylated peptide. Mutation of any of the five conserved ABI domain residues impairs MmRce1 catalytic activity. The mean and standard deviation of 3 experiments were considered for data analysis.
To understand the relevance of the MmRce1 structure in the context of the MmRce1-Fab complex, we assayed MmRce1 in the presence of Fab. Formation of the MmRce1-Fab complex had no influence on MmRce1 catalytic activity even though MmRce1 and Fab form extensive contacts (1,100 Å2) that would prevent conformational changes of the seven TM helices delineating the catalytic cavity (Fig. 1a and Extended Data Fig. 1). Thus in the crystal structure of MmRce1, catalytic residues are correctly aligned for cleavage of a farnesylated peptide, although it is possible that conformational changes may be required to accommodate larger prenylated protein substrates. Both the bacterial rhomboid GlpG 24-27 and S2P 8 have been proposed to undergo conformational shifts to mediate substrate gating, and structural changes of presenilin 9 and FlaK 12 are necessary to align catalytic site residues.
Rce1 shares no sequence similarity with other proteases 13. It therefore represents a novel protease, although interestingly with no paralogs in eukaryotes. Both cysteine- and metallo-enzyme based catalytic mechanisms have been proposed for Rce1 (refs 13, 15). However, the absence of an evolutionarily conserved cysteine residue (Fig. 3c and Extended Data Fig. 3) precludes a thiol-based mechanism 13,16. Strong evidence also indicates that Rce1 is not a metalloenzyme. MmRce1 activity is unaffected by EDTA or Zn2+ (Extended Data Fig. 4a). Its concentration-dependent inactivation by 1,10 phenanthroline, a hydrophobic metal chelator, results from non-specific protein unfolding (Extended Data Fig. 4). Furthermore, we could not detect Zn2+ bound to MmRce1 by means of either PIXE (proton-induced X-ray emission) or TXRF (total X-ray reflection fluorescence) (data not shown), and no Zn2+ ions were identified in the MmRce1 crystal structure.
All five conserved residues of the ABI domain (Fig. 3c) have been implicated in catalysis 15,16. Three residues of S. cerevisiae Rce1p - Glu156, His194 and His248 (equivalent to Glu140, His173 and His227 of MmRce1) are critical for catalysis 15,16, whereas mutation of either Glu157 or Asn252 (Glu141 and Asn231 of MmRce1) impairs catalytic activity 16 (Fig. 3c). We confirmed that MmRce1 catalytic activity is strictly dependent on Glu140 and His173, whereas mutation of either Glu141 or His227 severely disrupts activity (Fig. 3d). Similar to its eukaryotic homologs 16, replacing Asn231 with Ala abolished MmRce1 catalytic activity (Fig. 3d). Interestingly, in a few prokaryotic Rce1 homologs Asp replaces Asn231 (Fig. 3c and refs 13,14). When substituted into MmRce1, Asp reduces protease activity to ~40% of wild type (Fig. 3d). However at higher pH, whereas the wild type MmRce1 protease activity is unchanged, the N231D mutant is essentially inactive, suggesting that deprotonation of Asp231 inactivates MmRce1(N231D) (Fig. 3d). The low activity of the N231A mutant and pH dependent-activity of MmRce1(N231D) indicates a catalytic role for a hydroxyl group at this position. Using thermal shift and CD assays, we detected no affect of the mutants on either MmRce1 stability or conformation (Extended Data Figs 5 and 6).
The more buried position of Glu141 (Fig. 3a, b) suggests it may play an indirect structural role, and that the critical catalytic residues of MmRce1 are Glu140, His173, His227 and Asn231. The side chains of these four residues project into the solvent-filled catalytic cavity. Glu140 and His173, the residues most critical for catalysis, face each other from opposite TM helices and coordinate a bridging water molecule (Fig. 3b). Two conserved aromatic residues (Trp144 and Trp169) contact the carboxylate side chain of Glu140, potentially raising its pKa (Fig. 3b). Thus, a likely catalytic mechanism would involve Glu140 and His173 general base-catalyzed deprotonation of a water molecule for nucleophilic attack on the scissile bond of the peptide substrate. The side chains of His227 and Asn231, positioned on successive helical turns of TM7 opposite the catalytic dyad of Glu140 and His173, are likely to donate hydrogen bonds to stabilize the oxyanion transition state. Protonation of the leaving amino group of the –AAX tripeptide could be catalyzed by either Glu140 or His173 (Extended Data Fig. 7).
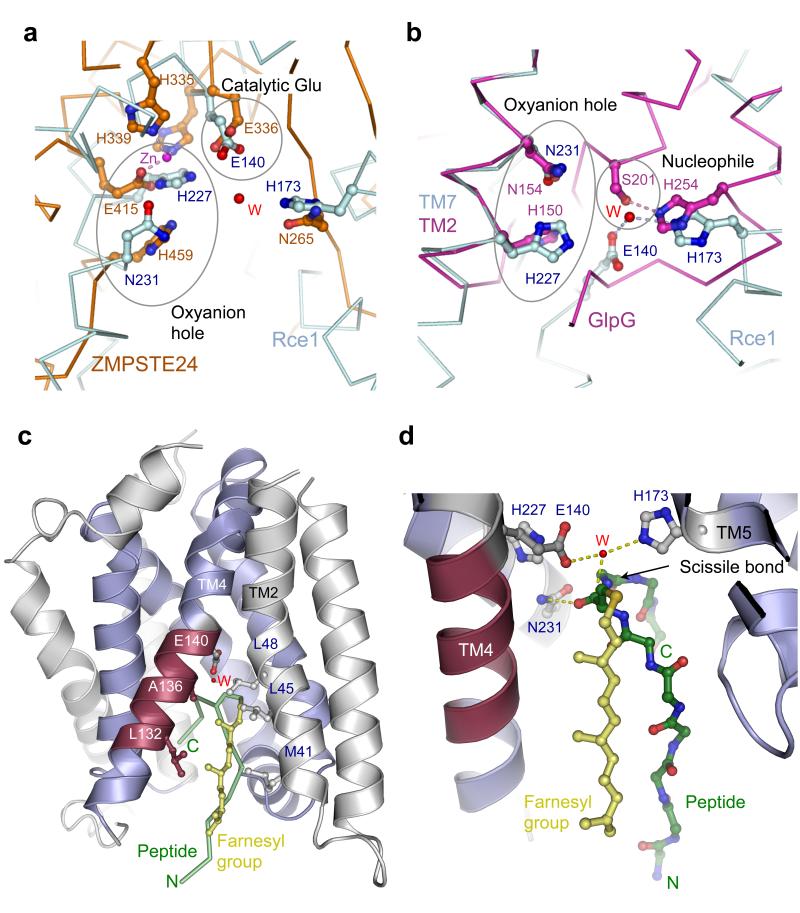
Although Rce1 belongs to a novel protease family, the proposed catalytic mechanism shares similarities with other proteases (Extended Data Fig. 7). The membrane-associated zinc-metalloproteases S2P 8 and ZMPSTE24 (refs 10,11) employ a glutamate-activated water molecule for cleavage of the scissile bond (Fig. 4a and Extended Data Fig. 7), as does the recently described fungal glutamic peptidase (SGP) 28. In S2P, an Asn residue is proposed to stabilize the oxyanion 8. Our inspection of the ZMPSTE24/Ste24p catalytic sites (refs 10,11) suggests a similar role for Asn and/or His residues (Fig. 4a and Extended Data Fig. 7). A striking similarity exists with the GlpG proteases where the proposed oxyanion hole is formed by His150 and Asn154 of its invariant H(X)3N motif, located on successive turns of a transmembrane helix 25-27,29, analogous to the proposed oxyanion hole of His227 and Asn231 in MmRce1. Superimposing His227 and Asn231 onto their equivalents of the H(X)3N motif of GlpG, reveals that the proposed Glu-activated nucleophilic water of MmRce1 exactly matches the position of the nucleophilic hydroxyl group of the Ser201 side chain of GlpG (Fig. 4b). In support of the notion that His173 of MmRce1 activates the nucleophilic water for attack onto the peptide substrate, its imidazole side chain superimposes onto His254 of the GlpG Ser-His catalytic dyad (Fig. 4b and Extended Data Fig. 7). Thus MmRce1, analogous to metalloproteases, aspartyl proteases and SGP, employs a carboxylate to activate a nucleophilic water molecule, however, both MmRce1 and SGP are unique by not polarizing the carbonyl group of the scissile peptide.
Figure 4. MmRce1 catalytic mechanism shares similarities with ZMPSTE24 and GlpG.
a, Comparison of the catalytic site conformations of MmRce1 (cyan) and ZMPSTE24 (orange, PDB code 4AW6) 11. Both proteins share similar cavities leading to the catalytic site. b, Comparison of the catalytic site conformations of MmRce1 (cyan) and GlpG (magenta, PDB code 2O7L) 27. The catalytic water (W) from MmRce1 is located in a similar position as the catalytic serine hydroxyl (Ser201) in GlpG. c, Cartoon representation of MmRce1 with a farnesylated peptide modelled at its catalytic site. ABI helices are shown in blue, and the conserved TM4 fragment (Leu132 – Thr138) is shown in red. TM2 and TM4 non-polar residues flanking the farnesyl lipid are labelled. The farnesylated peptide (GAKASGC(farnesyl)LVS) is depicted as green ribbon and the farnesyl lipid as yellow ball and stick model. d, The catalytic site of MmRce1 is shown with the modelled farnesylated peptide depicted as green ball and stick model and the farnesyl lipid as yellow ball and stick model. The catalytic residues (Glu140 and His173), the catalytic water (W) and the oxyanion residues (His227 and Asn231) are shown.
To understand how MmRce1 recognizes its substrates, we modelled a farnesylated peptide, based on the CAAX motif of RhoA, at the catalytic site of Rce1. The peptide adopts a β-hairpin conformation with the scissile bond positioned adjacent to the putative nucleophilic water molecule (Fig. 4c, d). The model is consistent with the proposed catalytic mechanism, and suggests that the farnesyl lipid would enter MmRce1’s catalytic site from the membrane, sealing the opening between the non-polar faces of TM2 and TM4. The interaction of the farnesyl lipid at this site would position the isoprenyl cysteine relative to the catalytic water, contributing to defining the CAAX motif cleavage site. There is sufficient room to accommodate the CAAX motif aliphatic residues adjacent to TM5, TM6 and TM7, whereas the six residues N-terminal to the CAAX motif, and its C-terminal ‘X’ residue would occupy the solvent-filled conical cavity leading into the cytoplasm. The large volume of the cavity, capable of accommodating diverse residues, is consistent with the sequence variety of Rce1 substrates N-terminal to the CAAX motif. Mutations of TM5, which might alter the size of the –AAX-motif binding pocket, modify the CAAX specificity of yeast Rce1p 30. Supporting the model that the opening between TM2 and TM4 creates the farnesyl lipid-binding site, substituting bulky Trp residues for either Leu45 of TM2 or Leu132 of TM4 to disrupt this site, substantially reduced MmRce1 activity (Extended Data Fig. 8), without affecting the protein structure (Extended Data Figs 5 and 6). To gain insight into the structure of eukaryotic Rce1 and its capacity to process both farnesylated and geranylgeranylated peptides, we generated a model of human Rce1 (Extended Data Fig. 8). The model predicts a longer TM4 helix that might provide a more extensive hydrophobic surface to promote favourable interactions with the C20 prenyl chain of a geranylgeranyl lipid.
This study establishes Rce1 as a founding member of a novel family of Glu-dependent IMPs. Insights into the structure and mechanism of Rce1 have implications for the development of antagonists of CAAX motif processing, which would have the potential to disrupt Ras signalling pathways.
Methods
Cloning
Homologs of human Rce1 were identified based on their membership of the Interpro CAAX amino terminal protease family, accession id IPR003675, containing the conserved ABI domain. 30 open reading frames were PCR amplified from bacterial and archaeal strains obtained from DSMZ (www.dsmz.de) and cloned using the In-Fusion method (Clontech) into a pTriEX-derived vector encoding a TEV-cleavable C-terminal green fluorescent protein-His7 tag (pOPIN-GFP).
Expression and homolog screening
Constructs were screened for expression levels varying four parameters: a) isopropyl-β-D-thiogalactopyranoside (IPTG) concentration – 100, 400 or 1000 μM; b) induction temperature – 20, 25 or 30 °C; c) induction duration – 4 or 16 hours; and d) Escherichia coli (E. coli) expression strain – BL21(DE3), C41(DE3) or C43(DE3). Expression levels were quantified by measuring in-cell fluorescence in a Carey fluorimeter fitted with a plate reader. Expression of full-length constructs was assayed for by running samples on SDS-PAGE gels and performing in-gel fluorescence imaging.
The best-expressed Rce1 homologs were further tested with a detergent screen for optimization of both solubilization efficiency and sample monodispersity. A range of detergents with differing properties was assayed (OM, DM, UDM, DDM, HG, OG, NG, OTG, FosCholine10, FosCholine12, Cymal6, Cymal7, LDAO, SDS, MEGA9, MEGA10, OGNG and MNG3 - Anatrace) and the solubilization efficiency was measured by quantitating the GFP signal of samples following solubilization and pelleting of insoluble material. Sample monodispersity was tested using fluorescence-detection size-exclusion chromatography (FSEC) on a Superdex 200 10/300 GL column coupled with an off-line fluorimeter to follow the elution profiles of the GFP-fused membrane proteins. Taken together these methods identified that the Methanococcus maripaludis S2 Rce1 (MmRce1) homolog was the best expressed protein and that it could be efficiently solubilized in 1% (w/v) n-Undecyl-β-D-maltopyranoside (UDM).
Expression and purification of MmRce1 and mutants
MmRce1-GFP-His7 protein was expressed in E. coli strain C41(DE3). Cultures were grown in L-Broth medium at 30 °C until the optical density (A600) reached 0.5, induced with 1 mM IPTG and left shaking overnight at 20 °C. Cells were collected by centrifugation at 4,000 g (30 min at 4 °C), and were stored at −80 °C. The harvested cell pellet was resuspended in Lysis buffer (15 mL buffer per 1 L of cell culture) containing 50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 50 μg/mL lysozyme, 20 μg/mL DNase I and 2 protease inhibitor cocktail tablets (Roche) per 60 mL lysate. The mixture was passed 3 times through an Emulsiflex homogenizer (Avestin) at 15,000 p.s.i. and centrifuged at 12,000 g for 15 min at 4 °C to remove cell debris. The resulting supernatant was centrifuged at 100,000 g for 1 hour at 4 °C to pellet the membrane fraction. The membrane was resuspended in Solubilization buffer containing 50 mM Tris-HCl [pH 8.0], 200 mM NaCl and 1% (w/v) UDM (Anatrace) adjusted to a final protein concentration of 3 mg/mL and was incubated with rotation for 90 min at 4 °C. The solution was centrifuged at 100,000 g for 50 min at 4 °C to remove the non-solubilized membrane fraction.
The protein was bound to Ni-nitrilotriacetic acid (NTA) Superflow resin (Qiagen) in Buffer A containing 50 mM Tris-HCl [pH 8.0], 150 mM NaCl and 0.029% (w/v) UDM, washed in Buffer A plus 50 mM imidazole and finally eluted from the column in Buffer A plus 200 mM imidazole. The C-terminal GFP-His7 was cleaved by tobacco etch virus (TEV) protease during overnight dialysis at 4 °C and GFP-His7 and His7-tagged TEV were removed by passing through a second Ni-NTA Superflow column. MmRce1 was further purified by size exclusion chromatography using a Superdex 200 10/300 GL size exclusion column (GE Healthcare) in Buffer B containing 20 mM MES [pH 6.5], 200 mM NaCl, 0.029% (w/v) UDM and 0.86% (w/v) n-Octyl-β-D-galactopyranoside (OG) (Anatrace). OG was added to Buffer B in order to reduce the total micelle size and improve protein monodispersity, as assessed using Multi Angle Light Scattering (data not shown). The MmRce1 fractions collected were analyzed by SDS-PAGE. The protein was stored at 4 °C for up to 2 days or flash-frozen and kept at −80 °C for longer periods of time.
Antibody generation
All animal experiments described here were approved by the Institutional Animal Care and Use Committee of Kyoto University Graduate School of Medicine.
To raise antibodies against conformational epitopes of MmRce1, 4-week old female BALB/c mice were immunized with 0.1 mg of reconstituted MmRce1-proteoliposome four times at 10-day intervals. Two days following the last injection, the mouse spleens were removed and the splenocytes were fused with mouse myeloma P3U1 cells using polyethylene glycol (PEG) 31.
To screen for antibodies that could specifically recognize native receptors and eliminate those that recognized flexible loops, N- and C-termini and unstructured regions of MmRce1, a liposome/denature ELISA method was used, as described 32. Candidate clones producing conformational antibodies against MmRce1 were screened by small-scale SEC and the established clones were isolated by limiting dilution to produce monoclonal hybridoma cell lines. The binding affinities of the established clones for MmRce1 were measured using a Biacore T100 (GE Healthcare) 32.
For large-scale antibody production, the hybridomas were transplanted into BALB/c mice. IgGs were collected from mouse ascites by 40% (w/v) ammonium sulfate precipitation and purified by Protein G Sepharose FF chromatography (GE Healthcare). The Fab645-2 fragment was obtained from IgG using a Fab preparation kit (Pierce).
Purification of the MmRce1-Fab complex
Purified MmRce1 and Fab645-2 were mixed at a molar ratio of 1:1.5 and were incubated on ice for 30 min prior to size exclusion chromatography. The complex co-eluted on a Superdex 200 10/300 GL size exclusion column (GE Healthcare) equilibrated with Buffer B and the fractions collected were analyzed by SDS-PAGE.
Crystallization of the MmRce1-Fab complex
The MmRce1-Fab645-2 complex was concentrated up to 8 mg/mL using Vivaspin 50 kDa cut-off centrifugal concentrators (Sartorius Stedim Biotech). The first crystallization trials were carried out by using the sitting-drop vapour-diffusion method with 96-well plates (Greiner) at 20 °C, screening against the commercial screens MemGold, MemStart, MemSys and MemPlus (Molecular Dimensions). Crystals were obtained in various conditions, but further crystallization screening and optimization identified one crystallization condition as the most promising, in terms of crystal quality and reproducibility (MemPlus F1: 12.5 mM 3-(N-morpholino) propanesulfonic acid (MOPS) [pH 7.0], 350 mM NaCl and 28% w/v PEG 1000). The final crystallization condition was 12.5 mM MOPS [pH 7.0], 350 mM NaCl and 30% v/v PEG 400. Well-diffracting crystals were obtained in 24-well plate hanging drops by vapour diffusion at 20 °C, grew to maximum dimensions in three weeks and were directly flash-frozen and stored in liquid nitrogen.
Data collection and processing
Diffraction data were collected from a single cryo-cooled crystal on beamline I04-1 at the Diamond Light Source, UK. Two identical datasets were collected on the same crystal with a single kappa angle oscillation difference of 45° between them. Two thousand images were collected for each dataset, with an oscillation range of 0.1° per image, to a maximum resolution of 2.5 Å. Both datasets were indexed with XDS 33. The data sets could be indexed equally well in P1 with two different unit cell parameters (a = 72.9 Å, b = 72.9 Å, c = 101.7 Å, α = 79.4°, β = 79.4°, γ= 76.2° and a = 113.0 Å, b = 90.0 Å, c = 99.3 Å, α = 89.0°, β = 102.3°, γ= 90.0°). Further data analysis with XTRIAGE 34 confirmed pseudotranslation with NCS vector 0.5, 0.5, 0.0. Therefore, the data were re-indexed with the larger unit cell and merged and scaled with SCALA 35 from the CCP4 program suite 36.
Structure determination and refinement
The structure was determined by molecular replacement using an antibody Fab fragment structure (PDB code, 3VG9) as a search model using PHASER 37. The Matthews coefficient calculated for the larger unit cell indicated the presence of four MmRce1- Fab645-2 complexes in the asymmetric unit (MmRce1 chains labelled C, F, I, L). PHASER successfully placed all four Fab645-2 molecules with a Z-score of 6.8. The electron density map obtained from PHASER showed α-helix-like features for the MmRce1. However, this map was not interpretable and had poorly-defined molecular boundaries for MmRce1. To improve the phases, solvent flattening and NCS-averaging were carried out with DM 38. For this purpose, a solvent mask for MmRce1 was calculated by placing dummy atoms at the putative MmRce1 positions. Density-modified maps clearly showed all the α-helices and some of the loops of MmRce1. Using this map, iterative manual model building was performed with COOT 39 and refined with PHENIX 34. Systematically, at every stage of model building, simulated annealed omit maps were calculated to check for model bias and also for phase improvement. Water molecules and detergent molecules were added towards the end of the refinement (Extended Data Fig. 9). Data collection and refinement statistics are shown in Extended Data Table 1. Ramachandran map definitions were defined using MOLPROBITY 40.
Site-directed mutagenesis
All MmRce1 point mutants were produced using the QuickChange™ site-directed mutagenesis kit from Agilent Technologies.
All point mutants were able to interact with the conformation-sensitive antibody Fab645-2 on a gel filtration column (Superdex 200 10/300 GL), suggesting that their overall fold did not differ from that of native MmRce1.
In vitro assay of MmRce1 enzymatic activity
A fluorescence resonance energy transfer (FRET) assay was used to demonstrate that purified MmRce1 could cleave the CAAX cleavage site of a peptide substrate designed based on the C-terminus of human RhoA. The sequence of the peptide was DABCYL-ARSGAKASGC(farnesyl)LVS-EDANS (where DABCYL is 4-{[4-(dimethyloamino)phenyl]azo}benzoic acid and EDANS is 5-[(2-aminoethyl)amino]naphthalene-1-sulfonic acid; Cambridge Peptides). The lyophilized peptide was dissolved in dimethyl sulfoxide (DMSO) and stored at a final concentration of 10 mM at −80 °C. In the intact substrate, DABCYL quenches the fluorescence of EDANS. Proteolytic cleavage at the C-terminal side of the farnesylated Cys separates the fluorophore and quencher, thus resulting in an increase in fluorescence.
Wild type MmRce1 and mutants were purified in Buffer B, as described above, at a stock concentration of 1 μM. The peptide stock was diluted in Buffer B to a final range of concentrations of 10 - 90 μM. Assays were performed with a 96-well opaque microplate (Nunc) using an Omega-POLARstar plate reader (BMG Labtech). The reactions were performed at 25 °C and 200 measurements were obtained for each run with 5 sec delay between each measurement. The rate of substrate hydrolysis was determined by monitoring the fluorescence as a function of time (excitation λ, 330 nm; emission λ, 490 nm). Neither peptide nor MmRce1 showed any significant changes in fluorescence over the time period of the assays when incubated alone.
The Relative Fluorescence Units (RFU) obtained from these assays were converted to Concentration Units (μM) by measuring the total fluorescence change of substrate during long reaction times with enzyme, which allowed for nearly complete conversion of the known concentrations of substrate to product. Data were analyzed using PRISM software (Graphpad). Graphs were plotted after subtraction of the uncatalyzed peptide control data and were fitted to a non-linear regression one-phase decay equation, since hydrolysis of the peptide by MmRce1 obeyed Michaelis-Menten kinetics. The apparent binding constant (Km) was 19.7 ± 1.0 μM and the apparent turnover constant (kcat) was 0.175 ± 0.0027 sec−1.
FRET assays (Fig. 1a, 3d, Extended Data Fig. 4 and Extended Data Fig. 8) were performed at the following concentrations: MmRce1, MmRce1-Fab and all mutants: 1 μM; farnesylated and non-farnesylated peptides: 50 μM; AFC, ZnSO4, EDTA, 1,7 Phenanthroline and 1,10 Phenanthroline: 300 μM and 5 mM.
Liquid chromatography-mass spectrometry analysis
The RhoA peptide (50 μM) and MmRce1 (1 μM) were mixed in Buffer B at 25 °C for 2 hours prior to mass spectrometry analysis.
Reversed phase chromatography was performed using an HP1200 platform (Agilent, Wokingham, UK). Peptide reaction solutions, incubated with and without enzyme, were diluted to 1 in 50 and 5 μL were injected for analysis (estimated 5 pmol initial unreacted peptide loaded on column). Peptides were resolved on a 75 μm I.D. 15 cm C18 packed emitter column (3 μm particle size; Nikkyo Technos Co., Ltd., Tokyo, Japan) over 30 min using a linear gradient of 96:4 to 30:70 buffer LC-A:LC-B (buffer LC-A: 2% acetonitrile/0.1% formic acid; buffer LC-B: 80% acetonitrile/0.1% formic acid) at 250 nL/min. Peptides were ionised by electrospray ionisation using 2.3 kV applied immediately pre-column via a microtee built into the nanospray source. Sample was infused into an LTQ Velos Orbitrap mass spectrometer (Thermo Fisher Scientific, Hemel Hempstead, UK) directly from the end of the tapered tip silica column (6-8 μm exit bore). The ion transfer tube was heated to 200 °C and the S-lens set to 60%. MS2 scans were acquired using data dependent acquisition based on a full 30,000 resolution FT-MS scan (280-1800 m/z range) to sequence the top 10 most intense ions using HCD fragmentation and 7,500 resolution FT-MS2 Orbitrap scans, with a single repeat count (5 s repeat duration) followed by a 10 s dynamic exclusion with a 10 ppm mass window based on a maximal exclusion list of 500 entries. Automatic gain control was set to 1,000,000 for FT-MS and 50,000 for FT-MS2, full FT-MS maximum inject time was 500 ms and normalized collision energy was set to 35% with an activation time of 10 ms. MS2 was targeted towards specific MS1 precursor ions 361.87921 and 542.31518 m/z, corresponding to the triply and doubly charged ions of the putative truncated peptide AKSGAKASGC(farnesyl), using an MS2 acquisition inclusion list.
Liquid chromatography-mass spectrometry analysis of geranylgeranylated peptides was performed as above, except that a sharper linear gradient of 96:4 to 30:80 buffer LC-A:LC-B was used.
Data were analysed in Xcalibur 2.1 Qual browser (Thermo Fisher Scientific, Hemel Hempstead, UK) and fragmentation peaks were assigned manually by comparison against theoretical fragmentation values. This was performed since the fragmentation profiles of the farnesylated peptides can score poorly using traditional proteomic database searching algorithms (data not shown). Semi-quantitative analysis of selected peptides was performed by assembling extracted ion chromatograms (XICs) of the first and second isotopes of doubly and triply charged ions corresponding to each target peptide using Xcalibur v2.1 (Thermo Scientific, Hemel Hempstead, UK). Chromatograms were smoothed using the Gaussian algorithm with 5 iterations, and the selected ions extracted with a mass error tolerance of 5 ppm. The XICs were integrated using the ICIS peak picking and integration algorithm in the Xcalibur software.
Ligand modelling and docking
A farnesylated peptide [GAKASGC(farnesyl)LVS)] was built with COOT 39. To remove model bias due to the starting conformation of the ligand, different conformers of the ligand with varying φ, ψ angles of the peptide and χ angles of farnesyl carbon chain, in 20° steps, were generated with InsightII® (http://accelrys.com/). The ligand was docked into MmRce1 using the ROSETTALIGAND module of ROSETTA 41. ROSETTALIGAND was allowed to search different available conformations of the ligand. Conformers with inter-atomic clashes were excluded from the docking. We selected 7,898 conformers for the docking study. Based on the total ROSETTA score (which is a function of the overall ROSETTA energy for the receptor-ligand complex) 10 docked structures were selected and subjected to further minimization. These docked conformer-protein complexes were minimized and subjected to 1ns MD simulations in a lipidic environment with GROMACS 4.6 (ref. 42), by employing the GROMOS96 43a1 force field. Before the MD simulations, the ligand-protein complexes were soaked in POPC lipid bilayer using InflateGor (http://moose.bio.ucalgary.ca/index.php?page=Translate_lipdis) and GROMACS 4.6. During minimization and MD, positional restraints were applied to the protein atoms. Selection of final model was based on the interaction protein-ligand interaction energy and conformational stability of the ligand during the MD simulations.
Modelling human Rce1
A model of human Rce1 was determined using MODELLER 43,44 based on the MmRce1 coordinates. The model was further refined using Gromacs 4.6 (ref. 42). A model for a geranylgeranylated peptide [GAKASGC(gg)LVS)] was based on the farnesylated peptide described above and docked into MmRce1 and human Rce1 model at the site of the farnesylated peptide [GAKASGC(farnesyl)LVS)] in MmRce1. The complex was subject to energy minimization as described above for the MmRce1-[GAKASGC(farnesyl)LVS)] peptide.
Circular Dichroism (CD)
CD experiments on wild type MmRce1 and six mutants (E140A, H173A, H227A, N231A, L45W and L132W) were performed using a Jasco J-715 spectropolarimeter at 25 °C in 20 mM MES [pH 6.5], 100 mM KF, 0.029% (w/v) UDM and 0.86% (w/v) OG. The CD spectra for secondary structure determination were recorded between 190 nm and 300 nm, using a 0.01 mm path length cell at protein concentration of 1 mg/mL. Three spectra were recorded for each protein in 0.5 nm increments and averaged.
The analysis of CD spectra was performed by the programs CONTIN/LL 45 and SELCON3 (refs 46,47) incorporated in the software DICHROWEB 48 using SP175 as the reference protein set 49. The CD spectra were zeroed between 260 nm and 300 nm and are presented as Δε (liter mol−1 cm−1) against wavelength (nm).
CPM-based thermostability assays
CPM-based thermostability assays on wild type MmRce1 and all mutants were carried out as previously described 50. Ten microliters of purified MmRce1 at 1 mg/mL were added to 140 microliters of buffer containing 20 mM MES [pH 6.5], 200 mM NaCl, 0.029% (w/v) UDM and 0.86% (w/v) OG in a 96-well black Nunc plate. N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) dye at 4 mg/mL in DMSO was diluted 100-fold in the same buffer and warmed to room temperature. Three microliters of dye at 40 μg/mL were added to the protein and almost immediately fluorescence emission was measured at 463 nm (excitation 387 nm) on the SpectraMax2e plate reader (Molecular Devices) at 40 °C. Recordings were measured every 5 min for 2 hours with a 15 sec shaking interval between each measurement. A single exponential decay curve was plotted for each protein run and fitted to Boltzmann sigmoidal equation using Prism software (GraphPad). For wild type, N231A and N231D mutants, the screen was also performed at pH 8 in 20 mM HEPES [pH 8.0], 200 mM NaCl, 0.029% (w/v) UDM and 0.86% (w/v) OG. The mean and standard deviation of 3 experiments were considered for data analysis.
Thermal Shift Screen
Thermal shift assays for wild type MmRce1 and all mutants were carried out using an Applied Biosystems 7500 Fast RTPCR instrument. All available excitation and emission wavelengths of the instrument were used during each run. Ten microliters of purified MmRce1 or mutants at 1 mg/mL were added to 50 microliters of buffer containing 20 mM MES [pH 6.5], 200 mM NaCl, 0.029% (w/v) UDM and 0.86% (w/v) OG in an ABgene® SuperPlate™ Skirted 96-well PCR plate (Thermo-Scientific). CPM dye at 4 mg/mL in DMSO was diluted 10-fold in the same buffer and 7 microliters of dye at 100 μg/mL were added to the protein. The plate was sealed with a micro-seal ‘B’ clear adhesive seal (Biorad) and almost immediately fluorescence emission was measured. The samples were heated from 10 °C to 95 °C at a rate of 1 °C/min. Ten microliters of purified MmRce1 at 30 μM concentration were added to 50 microliters of buffer containing 20 mM MES [pH 6.5], 200 mM NaCl, 0.029% (w/v) UDM, 0.86% (w/v) OG and either 5 mM 1,7-Phenanthroline, or 5 mM 1,10-Phenanthroline, or increasing amounts of urea (1 M to 5 M at 1 M-increments). The CPM dye was added and the experiments were performed as described above. The data were analyzed by the Protein Thermal Shift™ software using the dFluorescence derivative method. The Tm values were taken as the minima in the derivative plots (derivative melt profiles). For wild type, N231A and N231D mutants, the screen was also performed at pH 8 in 20 mM HEPES [pH 8.0], 200 mM NaCl, 0.029% (w/v) UDM and 0.86% (w/v) OG. The mean and standard deviation of 3 experiments were considered for data analysis.
Geranylgeranyl peptide synthesis and assays
Solid phase peptide synthesis of three geranylgeranylated peptides was performed as described 51. The amino acid sequence of two peptides was based on the C-terminus of RhoA (ARSGAKASGC(geranylgeranyl)LVS) and the sequence of the third peptide was based on the C-terminal sequence of the yeast a-factor (YIIKGVFWDPAC(geranylgeranyl)VIA). One RhoA peptide was further labelled with DABCYL and EDANS to resemble the farnesylated FRET RhoA peptide described above. All three lyophilized peptides were dissolved in DMSO and stored at a final concentration of 10 mM at −80 °C.
The two non-fluorescent peptides (RhoA and yeast a-factor) were used for mass spectrometry analysis (as described above), whereas the FRET RhoA peptide was diluted to a range of concentrations (10 μM to 60 μM) in Buffer B and its proteolytic cleavage by MmRce1 (1 μM to 5 μM) was assayed by FRET (assay and conditions described above).
Acknowledgments
This work was funded by a Cancer Research UK grant to D.B. Part of this work was supported by the research acceleration program of the Japan Science and Technology agency and by the BBSRC BB/G023425/1 (S.I.). We thank staff at I04-1 Diamond Light Source for help with data collection, Jing Yang (ICR) for advice and discussions, Isabel De Moraes (Membrane Protein Laboratory at Diamond Light Source) for support and Tina Daviter (ISMB Biophysics Centre at Birkbeck, University of London) for help with the CD experiments.
Footnotes
Author information. The coordinates have been deposited with the RCSB under accession number: 4cad. The authors declare no competing financial interests.
References
- 1.Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 2.Prior IA, Hancock JF. Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci U S A. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaelis S, Barrowman J. Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol Mol Biol Rev. 2012;76:626–651. doi: 10.1128/MMBR.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 8.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 9.Li X, et al. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 10.Pryor EE, Jr., et al. Structure of the integral membrane protein CAAX protease Ste24p. Science. 2013;339:1600–1604. doi: 10.1126/science.1232048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quigley A, et al. The structural basis of ZMPSTE24-dependent laminopathies. Science. 2013;339:1604–1607. doi: 10.1126/science.1231513. [DOI] [PubMed] [Google Scholar]
- 12.Hu J, Xue Y, Lee S, Ha Y. The crystal structure of GXGD membrane protease FlaK. Nature. 2011;475:528–531. doi: 10.1038/nature10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei J, Grishin NV. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci. 2001;26:275–277. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- 14.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. The abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol. 2010;192:2068–2076. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry. 2000;39:4096–4104. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- 16.Plummer LJ, et al. Mutational analysis of the ras converting enzyme reveals a requirement for glutamate and histidine residues. J Biol Chem. 2006;281:4596–4605. doi: 10.1074/jbc.M506284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergo MO, et al. Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol Cell Biol. 2002;22:171–181. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlstrom AM, et al. Rce1 deficiency accelerates the development of K-RAS-induced myeloproliferative disease. Blood. 2007;109:763–768. doi: 10.1182/blood-2006-05-024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergo MO, et al. On the physiological importance of endoproteolysis of CAAX proteins: heart-specific RCE1 knockout mice develop a lethal cardiomyopathy. J Biol Chem. 2004;279:4729–4736. doi: 10.1074/jbc.M310081200. [DOI] [PubMed] [Google Scholar]
- 20.Christiansen JR, Kolandaivelu S, Bergo MO, Ramamurthy V. RAS-converting enzyme 1-mediated endoproteolysis is required for trafficking of rod phosphodiesterase 6 to photoreceptor outer segments. Proc Natl Acad Sci U S A. 2011;108:8862–8866. doi: 10.1073/pnas.1103627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto JC, Kim E, Young SG, Casey PJ. Cloning and characterization of a mammalian prenyl protein-specific protease. J Biol Chem. 1999;274:8379–8382. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 22.Hollander I, Frommer E, Mallon R. Human ras-converting enzyme (hRCE1) endoproteolytic activity on K-ras-derived peptides. Analytical biochemistry. 2000;286:129–137. doi: 10.1006/abio.2000.4795. [DOI] [PubMed] [Google Scholar]
- 23.Hollander IJ, Frommer E, Aulabaugh A, Mallon R. Human Ras converting enzyme endoproteolytic specificity at the P2′ and P3′ positions of K-Ras-derived peptides. Biochim Biophys Acta. 2003;1649:24–29. doi: 10.1016/s1570-9639(03)00150-x. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 25.Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci U S A. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Ha Y. Open-cap conformation of intramembrane protease GlpG. Proc Natl Acad Sci U S A. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujinaga M, Cherney MM, Oyama H, Oda K, James MN. The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum. Proc Natl Acad Sci U S A. 2004;101:3364–3369. doi: 10.1073/pnas.0400246101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vinothkumar KR, et al. The structural basis for catalysis and substrate specificity of a rhomboid protease. The EMBO journal. 2010;29:3797–3809. doi: 10.1038/emboj.2010.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trueblood CE, et al. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol Cell Biol. 2000;20:4381–4392. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day PW, et al. A monoclonal antibody for G protein-coupled receptor crystallography. Nat Methods. 2007;4:927–929. doi: 10.1038/nmeth1112. [DOI] [PubMed] [Google Scholar]
- 32.Hino T, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans P. Scaling and assessment of data quality. Acta crystallographica. Section D, Biological crystallography. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 36.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowtan K. Recent developments in classical density modification. Acta crystallographica. Section D, Biological crystallography. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic acids research. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raveh B, London N, Zimmerman L, Schueler-Furman O. Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One. 2011;6:e18934. doi: 10.1371/journal.pone.0018934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Der Spoel D, et al. GROMACS: fast, flexible, and free. Journal of computational chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 43.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 44.Eswar N, et al. Comparative protein structure modeling using Modeller. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] 2006 Oct; doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 46.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Analytical biochemistry. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 47.Sreerama N, Venyaminov SY, Woody RW. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein science: a publication of the Protein Society. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic acids research. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lees JG, Miles AJ, Wien F, Wallace BA. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 50.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Riou P, et al. 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell. 2013;153:640–653. doi: 10.1016/j.cell.2013.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]