Summary
Ubiquitin receptors connect substrate ubiquitylation to proteasomal degradation. HHR23a binds proteasome subunit 5a (S5a) through a surface that also binds ubiquitin. We report that S5a's UIM2 binds preferentially to hHR23a over polyubiquitin and provide a model for the ternary complex, which we expect represents one of the mechanisms used by the proteasome to capture ubiquitylated substrates. Furthermore, we demonstrate that hHR23a is surprisingly adept at sequestering the ubiquitin moieties of a polyubiquitin chain, and provide evidence that it and the ubiquitylated substrate are committed to each other after binding.
Keywords: ubiquitin receptors, proteasome subunit S5a, hHR23a, Rad23, proteasomal degradation
Introduction
Ubiquitin signaling regulates an astounding array of cellular events and remains essential throughout the life cycle of a cell. In its most established role ubiquitylation targets proteins for degradation by the 26S proteasome,1 a process important for controlling the lifespan of regulatory proteins, removing misfolded proteins,2 producing immunocompetent peptides,3 activating and repressing transcription,4; 5 and regulating cell cycle progression.6 Ubiquitin-mediated protein degradation begins with an enzymatic cascade that culminates in the attachment of polyubiquitin to a protein substrate. Substrates conjugated with chains linked by ubiquitin's K487 or K638 are degraded by the proteasome. Little is known of the pathway(s) that connects ubiquitylation to proteasomal degradation; however, ubiquitin receptors undoubtedly play key roles in this process. Two proteasomal ubiquitin receptors have been identified including S5a9 and S6′.10 S5a contains two functional elements, an N-terminal 188-amino acid von Willebrand A (VWA) domain and two ubiquitin interacting motifs (UIMs) that bind ubiquitin (Figure 1).11 The VWA domain is conserved in S5a's yeast homologue Rpn10, which is truncated after UIM1. Therefore, this N-terminal domain is expected to mediate S5a/Rpn10 proteasome association.
Figure 1.
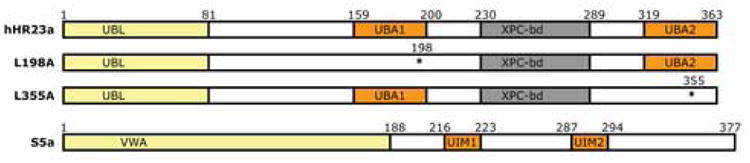
Domain architecture of ubiquitin receptors hHR23a and S5a. Regions that bind proteasome components or ubiquitin are shaded in yellow or orange respectively. The effect of the point mutations used in this manuscript, namely L198A and L355A, are illustrated by the loss of the corresponding UBA domain. Each of these mutations destroys UBA domain structural integrity as well as its capacity to bind (poly)ubiquitin.
In addition to binding ubiquitin, S5a's UIMs bind members of the UBL/UBA ubiquitin receptor family, which exist largely free of the proteasome.12; 13 The UBL/UBA family of proteins are defined by ubiquitin-associated (UBA) domains that bind ubiquitin14; 15; 16 and ubiquitin-like (UBL) domains that bind the proteasome.12; 13; 17; 18 They have attracted much attention for their ability to regulate the lifespans of other proteins. The three UBL/UBA family members in Saccharomyces cerevisiae, Rad23 (hHR23a/b in humans), Dsk2 (hPLIC-1/2 in humans) and Ddi1, recruit ubiquitylated substrates to the proteasome for degradation.19; 20; 21; 22; 23; 24 Importantly, mice lacking the two Rad23 homologues (mHR23a/b) die as embryos,25 suggesting that it plays an essential role in delivering one or more ubiquitylated substrates to the proteasome in mammals. Experiments in yeast demonstrated that Rad23 provides an Rpn10-independent pathway to the proteasome.20; 23
In previous studies, we determined the structure of hHR23a to reveal that its UBL domain interacts dynamically with each of its UBA domains (Figure 1).26 These interactions are abrogated in the presence of ubiquitin,16 the preferred UBA domain binding partner, or S5a,26 the preferred UBL domain binding partner. Indeed, the binding constant for hHR23b binding to S5a's UIM2 is 3.4 μM27 and structures of the hHR23a/UIM228 or hHR23b/UIM227 complexes demonstrate strong hydrophobic and polar contacts between these two proteins. S5a and hHR23a/b each have two ubiquitin binding regions that are connected by flexible linkers, and this shared attribute may contribute to their preference for longer polyubiquitin chains.16
Very little is known of how polyubiquitin behaves in an environment of multiple receptors, yet such knowledge is essential to determine how substrates are delivered to the proteasome. The structure of K48-linked tetraubiquitin revealed its ubiquitin subunits to pack against each other;29 however, K48-linked tetraubiquitin binds multiple Rad23 molecules,30 indicating that it adopts opened conformations when bound to this ubiquitin receptor.
In this manuscript, we demonstrate that hHR23a is surprisingly efficient at sequestering the ubiquitin moieties of a chain such that the binding of additional receptors is strongly disfavored, even for octaubiquitin. HHR23a changes its own conformation26 to bind S5a,12 but through a surface on S5a that also binds ubiquitin. S5a contains two UIMs (Figure 1), the second of which (UIM2) exhibits a 5-fold stronger binding affinity for monoubiquitin31 and binds hHR23a.12 Intriguingly, we reveal that S5a binds hHR23a in the presence of tetraubiquitin and use NMR spectroscopy to define the resulting ternary complex. Our findings yield a mechanistic model for how the proteasome captures its substrates.
Results
HHR23a UBA domains sequester polyubiquitin
By using a previously established assay,30 we determined that the UBA domains of hHR23a are highly efficient at sequestering the ubiquitin moieties of K48- and K63-linked tetraubiquitin. Purified GST-tagged hHR23a bound to glutathione S-sepharose resin was exposed to K48- (Figure 2(a)) or K63- (Figure 2(b)) linked chains followed by untagged hHR23a protein (Materials and Methods). Surprisingly, K48- and K63-linked tetraubiquitin supported the binding of only one hHR23a molecule. This result was observed even with hHR23a variants containing only one intact UBA domain (Figure 2(a) and (b)), namely those with the L355A or L198A mutation incorporated (Figure 1).15; 16
Figure 2.
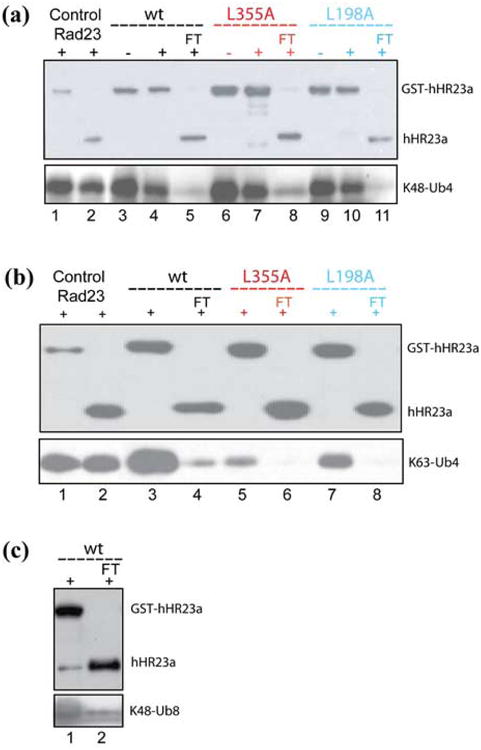
HHR23a's UBA domains sequester the ubiquitin subunits of polyubiquitin. Western blot analysis demonstrates that two hHR23a molecules cannot bind a common K48- (a) or K63-linked (b) tetraubiquitin. Glutathione S-sepharose resin (20 μl) pre-incubated with 0.1 nmoles GST-hHR23a wild-type, L355A, or L198A was mixed with 0.03 nmoles K48- (a) or K63-linked (b) tetraubiquitin without (labeled “-“) or with (labeled “+”) 0.1 nmoles of untagged hHR23a wild-type (black), L355A (red), and L198A (blue), washed extensively and then probed with anti-hHR23a (top panel) or anti-ubiquitin (bottom panel). No ternary complex involving either K48- (a) or K63-linked (b) tetraubiquitin was observed, and greater than 90% of the untagged protein was recovered in the flow through (FT). As positive controls, Ni-NTA resin (20 μl) pre-incubated with 0.1 nmoles 6*His-tagged Rad23 was mixed with 0.03 nmoles of K48- (a) or K63-linked (b) tetraubiquitin and 0.1 nmoles of GST tagged or untagged hHR23a. (c) Octaubiquitin exhibits a strong preference for binding only one hHR23a molecule. 20 μl of glutathione S-sepharose resin pre-incubated with 0.1 nmoles of GST-hHR23a was mixed with 0.03 nmoles K48-linked octaubiquitin and 0.1 nmoles of untagged hHR23a. The flow through (FT) was loaded into lane 2 and the resin washed extensively and loaded into lane 1. Western blot analysis was performed with anti-hHR23a (top panel) or anti-ubiquitin (bottom panel) to reveal untagged hHR23a largely in the flow through rather than retained in a ternary complex with GST-hHR23a and octaubiquitin.
Furthermore, hHR23a binding to tetraubiquitin remained stoichiometric even when either protein was at large molar excess, as measured by analytical ultracentrifugation. In particular, the maximum value in the plot of sw versus tetraubiquitin:hHR23a molar ratio is close to 1:1 stoichiometry (Figure 3(a)). Furthermore, our data match closely with a theoretical plot for 1:1 binding stoichiometry (Figure 3(a), red). The experimental data is slightly above the theoretical line at higher tetraubiquitin:hHR23a molar ratios due to the presence of small amounts of aggregates under those conditions. For this analysis, we used a previously determined sedimentation coefficient for hHR23a of 2.22S (data not shown). The sedimentation coefficient for tetraubiquitin was obtained from the DcDt+ analysis of samples containing tetraubiquitin:hHR23a at molar ratios where tetraubiquitin was in excess, with a resulting value of 2.5S. This value was supported by c(s) analysis (data not shown). Finally, c(s) analysis of each mixture revealed the presence of two peaks, a slower one in the region expected for the component in excess, and a faster one in the region expected for the 1:1 complex (∼3.8 S). A representative c(s) profile is provided in Figure 3(b) for the sample with a tetraubiquitin:hHR23a molar ratio of 0.64:1. The relative peak areas for this sample were 18% for the peak at 2.26 S and 82% for the peak at 3.7 S. These values are quite close to the expected values of 23% and 77% for tetraubiquitin and hHR23a forming a 1:1 complex. The SEDPHAT analysis of the data for the two samples containing tetraubiquitin:hHR23a at molar ratios nearest to 1:1 yielded a molecular weight of 76.2 ± 1.6 kDa and a corrected sedimentation coefficient, s20,w, of 3.8 ± 0.07 S. This result matches closely with the sequence molecular weight for the 1:1 molar complex of hHR23a and tetraubiquitin, which is 74.3 kDa.
Figure 3.
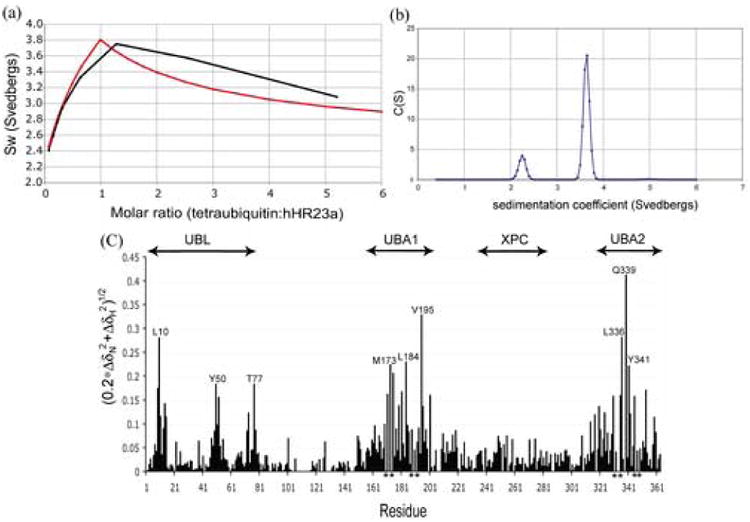
HHR23a's UBA domains are highly efficient at binding tetraubiquitin. (a) The weight average sedimentation coefficient as a function of tetraubiquitin to hHR23a molar ratio reveals 1:1 binding stoichiometry. Experimentally determined values obtained by using sedimentation velocity analysis (black) and a theoretical plot (Materials and Methods) for 1:1 binding stoichiometry (red) are included. (b) Sedimentation velocity results for the sample containing a 0.64:1 molar ratio of tetraubiquitin:hHR23a. The c(s) distribution was obtained using SEDFIT, with the slower peak corresponding to excess hHR23a and the faster peak to the tetraubiquitin:hHR23a 1:1 complex. (c) Chemical shift perturbation data analyzed according to Equation 2 reveals that each of hHR23a's UBA domains bind K48-linked tetraubiquitin. For this analysis, the amide chemical shift values of hHR23a alone were compared to its values in the presence of 2-fold molar excess K48-linked tetraubiquitin.
We tested whether two hHR23a proteins bind octaubiquitin and found that its binding to a second hHR23a molecule is unexpectedly weak (Figure 2(c)). In particular, only small quantities of untagged hHR23a are retained in a ternary complex with GST-hHR23a and octaubiquitin (Figure 2(c), lane 1 of upper panel). Altogether, these data suggest that the hHR23a UBA domains are highly efficient at sequestering the ubiquitin moieties of tetraubiquitin and that two tetraubiquitins do not bind hHR23a simultaneously, suggesting that hHR23a commits to a single ubiquitylated substrate.
The data of Figure 2(a) and (b) suggest that either UBA domain of hHR23a can support binding to polyubiquitin, as the L198A and L355A variants bind tetraubiquitin. It is worth noting that tetraubiquitin is not retained on GST-bound resin30 and that the observed interaction is therefore not a result of non-specific binding to the resin. To further validate this result, we acquired [ 1H,15N] HSQC spectra on 15N-labeled hHR23a alone and in the presence of 2-fold molar excess K48-linked tetraubiquitin. The effect of adding tetraubiquitin to hHR23a is summarized in Figure 3(c), which was derived as described in Materials and Methods. As expected, each of hHR23a's UBA domains exhibited significant chemical shift perturbations in the presence of tetraubiquitin (Fig. 3(c)). The UBL domain is also affected, as UBL/UBA domain interactions are disturbed. This result was also observed when hHR23a's UBA domains bound monoubiquitin.16 Altogether, these data provide strong evidence that each of hHR23a's UBA domains bind K48-linked tetraubiquitin.
In a previous manuscript, we demonstrated that tetraubiquitin can bind simultaneously to Rad23 and hHR23a.30 This experiment is used as a positive control in Figures 2(a) and (b). In these experiments, tetraubiquitin is incubated with his-tagged Rad23 bound to Ni-NTA agarose resin. After washing the resin extensively, GST-tagged or untagged hHR23a is added, and the resin again washed extensively. In this experiment, each of the hHR23a species are retained on the resin, only when His-tagged Rad23 is added.30 This data suggests that Rad23 uses a different mechanism to bind tetraubiquitin or changes the tetraubiquitin binding mode of hHR23a. Indeed, UBA2 of Rad23 differs significantly from hHR23a's UBA domains, as it mediates Rad23 dimerization.30; 32 Further experiments are required to determine whether hHR23a or Rad23 exhibit preference for different ubiquitin subunits in a polyubiquitin chain.
S5a binds preferentially to hHR23a over tetraubiquitin
We used NMR spectroscopy to test whether it is mechanistically plausible for hHR23a to deliver polyubiquitin to S5a. The hHR23a/S5a interaction occurs via S5a's UIM2,12 which also binds ubiquitin.11 Surprisingly, the hHR23a/S5a interaction was found to be unaffected by tetraubiquitin. In particular, the hHR23a UBL domain (Figure 4(a)) and S5a's UIM2 (Figure 4(b)) bind each other in an identical manner whether tetraubiquitin is present or not. This phenomenon is demonstrated in Figure 4(a), which contains an expanded region of [ 1H,15N] HSQC spectra of 15N-labeled hHR23a by itself (black), mixed with equimolar quantities of S5a (196-306) (blue), or with equimolar quantities of S5a (196-306) and tetraubiquitin (red). HHR23a UBL domain resonances exhibit identical chemical shift perturbations due to S5a addition in the presence of tetraubiquitin.
Figure 4.
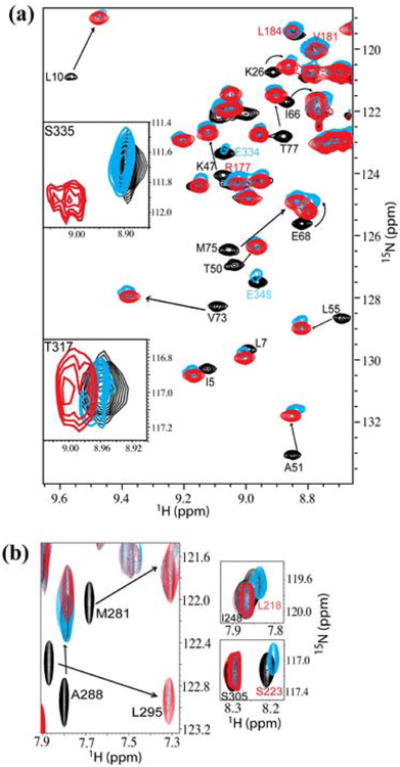
Characterizing the hHR23a/S5a/tetraubiquitin complex by NMR. (a) HHR23a binds S5a and tetraubiquitin simultaneously. An expanded region of [ 1H,15N] -HSQC spectra of 15N-labeled hHR23a alone (black), with equimolar quantities of S5a (196-306) (blue), or with equimolar amounts of S5a (196-306) and K48-linked tetraubiquitin (red) illustrates that hHR23a binding to S5a is unaffected by tetraubiquitin. The resonances of E334 and E348 (labeled in blue) are severely broadened and those of S335 and T317 shifted by tetraubiquitin, whereas those of R177, V181, and L184 (labeled in red) are unaffected. The UBL, UBA1 and UBA2 domains of hHR23a span residues V3-K78, S161-T200, T317-N359, respectively. For spectra acquired on hHR23a, S5a and tetraubiquitin (red), equimolar quantities of K48-linked tetraubiquitin was added to 15N-labeled hHR23a. Unlabeled S5a was added to this mixture and spectra recorded at 1:1:0 (not shown), 1:1:0.5 (not shown), 1:1:1 (red), and 1:1:2 (not shown) molar ratio of hHR23a: tetraubiquitin: S5a (196-306). The experiments were recorded on 0.32 mM 15N-labeled hHR23a in 20 mM sodium phosphate (pH 6.5), 30 mM NaCl, 0.1% NaN3.
(b) S5a's UIM2 binds preferentially to the UBL domain of hHR23a. Expanded regions of [ 1H,15N] -HSQC spectra of 15N-labeled S5a (196-306) alone (black), with equimolar quantities of hHR23a (blue), or with equimolar amounts of hHR23a and K48-linked tetraubiquitin (red) indicates that S5a's UIM2 binds hHR23a (left panel) and that its UIM1 is attenuated due to tetraubiquitin binding (right panels). The experiments were performed with 0.25 mM 15N-labeled S5a (196-306) in 20 mM sodium phosphate (pH 6.5), 30 mM NaCl, 0.1% NaN3.
Conversely, Figure 4(b) shows an expanded region of [ 1H,15N] HSQC spectra acquired on 15N-labeled S5a by itself (black), mixed with equimolar quantities of hHR23a (blue), or with equimolar quantities of hHR23a and tetraubiquitin (red). The comparisons reveal that S5a's UIM2 resonances exhibit identical chemical shift changes from hHR23a addition when tetraubiquitin is present. Furthermore, when the mixture containing equimolar quantities of the three proteins is subjected to size exclusion chromatography, the three proteins co-elute with no higher or lower molecular weight species (Figure 5). Altogether, these results provide strong evidence that S5a's UIM2 binds preferentially to hHR23a's UBL domain over tetraubiquitin.
Figure 5.
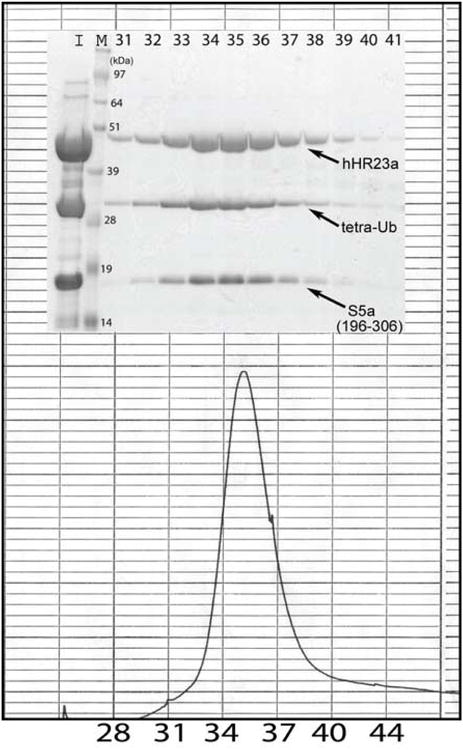
An expanded region of a chromatogram and accompanying gel demonstrate that hHR23a, S5a and tetraubiquitin co-elute after size exclusion chromatography. Equimolar quantities of hHR23a, S5a (196-306) and K48-linked tetraubiquitin were incubated for 1 hour at 4 °C, and passed through an FPLC system equipped with a Hiload 16/60 Superdex 200 prep grade column (Amersham Pharmacia Biotech). Only one peak was observed which was confirmed to contain all three components by gel electrophoresis and Coomassie staining. The retention volume of the fractions containing hHR23a, S5a and tetraubiquitin spanned 59.5 to 65.5 ml, which is reasonable for the 87 kDa complex as albumin (Mr 67 KDa) elutes at ∼75 ml. Lanes labeled I and M contain an aliquot of the mixture prior to FPLC injection and a molecular weight marker, respectively.
HHR23a's UBA2 and S5a's UIM1 bind tetraubiquitin
Surprisingly, our data indicate that in the presence of S5a, hHR23a UBA2 residues bind tetraubiquitin, whereas those of UBA1 do not (Figure 4(a)). In particular, resonances derived from UBA2 residues exhibit severe broadening (such as E334 or E348) or chemical shift perturbations (such as S335 or T317) in the samples containing tetraubiquitin and S5a. In contrast, UBA1 resonances exhibit only minor perturbations, as demonstrated in Figure 4(a) for R177, V181, and L184.
S5a's UIM1 binds ubiquitin11 but not hHR23a.12 Our NMR experiments reveal that as UIM2 binds hHR23a's UBL domain, UIM1 residues bind tetraubiquitin (Figure 4(b), right panels). In particular, resonances originating from UIM1 residues exhibit severe resonance broadening in the samples containing hHR23a and tetraubiquitin. No effect is observed when tetraubiquitin is absent.
Discussion
Altogether, our results indicate that hHR23a's UBA2 domain binds tetraubiquitin as its UBL domain binds S5a's UIM2, which facilitates UIM1 binding to the polyubiquitin. These findings lead to a model of the ternary complex that provides insight into how ubiquitylated substrates are captured by the proteasome (Figure 6). In particular, UIM1 is proximal to S5a's 188-residue VWA domain, which is expected to mediate its proteasome association. Therefore, UIM1 is expected to be deeper within the proteasome and its interaction with polyubiquitin, facilitated by the UIM2/UBL domain interaction, offers a mechanism for moving substrates into the proteasome.
Figure 6.
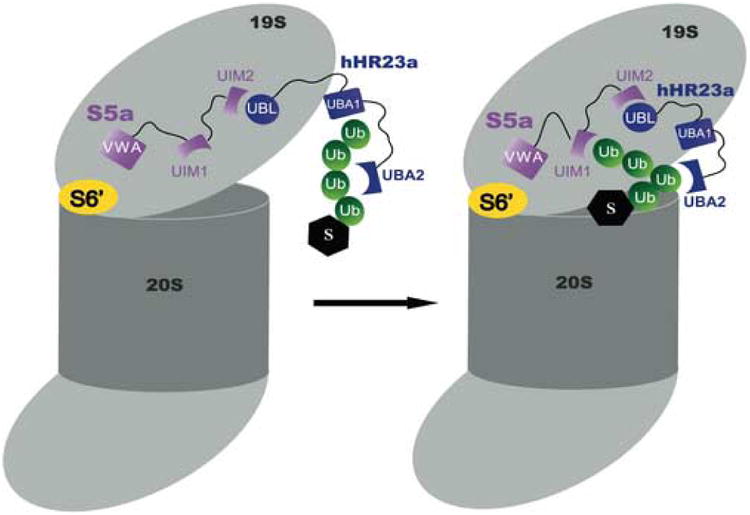
Proposed model for how hHR23a and S5a bind to tetraubiquitin. HHR23a recruits the chain via its UBA2 domain as its UBL domain binds proteasome component S5a. UIM1 of S5a is able to receive the chain.
We demonstrate that hHR23a binding to tetraubiquitin is stoichiometric over a wide range of molar ratios, which include either protein being at greater than 5-fold molar excess over the other. There are two mechanisms by which hHR23a could block or dramatically reduce the binding of another molecule to tetraubiquitin or octaubiquitin, respectively. In particular, a preferred binding orientation could exist whereby binding to certain subunits, such as the terminal ones, is not desirable, or multiple ubiquitin subunits could bind the UBA domains simultaneously. These two possibilities are supported by the significantly weaker binding observed to monoubiquitin33 and by the structure of hHR23a's UBA2 domain complexed with diubiquitin.34 In this structure, the UBA domain is sandwiched between the two ubiquitin subunits. HHR23a's inclination to bind only one polyubiquitin chain and its exclusion of additional proteins suggests that it becomes committed to a single ubiquitylated substrate. Furthermore, this property seems to have evolved in eukaryotic family members, as Rad23 (yeast ortholog of hHR23a/b) binds to Rad23-30 or hHR23a-bound (Figure 2(a) and (b)) tetraubiquitin.
S5a was identified for its role as a proteasome bound ubiquitin receptor, and later found to bind hHR23a12 and other ubiquitin receptors.13 The amino acid residues used to bind ubiquitin, however, overlap with those that bind hHR23a11; 12 and therefore it was not previously known whether this interaction occurs in the presence of a ubiquitylated substrate. Here, we demonstrate that hHR23a's UBA2 domain binds tetraubiquitin as its UBL domain binds S5a's UIM2. Within this complex, S5a's UIM1 binds polyubiquitin, and when S5a is assembled into the proteasome, this interaction could play an important role in capturing substrates (Figure 6). In summary, we expect that the model presented in Figure 6 represents one mechanism by which ubiquitylated substrates are captured by the proteasome.
Materials and Methods
Sample preparation
Tagged and untagged versions of hHR23a wild-type protein as well as variants with the L198A or L355A mutations incorporated were expressed and purified from E. coli as described previously.16; 26 K48- and K63-linked tetraubiquitin was purchased (Boston Biochem Inc.) or synthesized using a previously published protocol,35 which was also used to produce K48-linked octaubiquitin. S5a (196-306) was expressed and purified as described previously.31
Western blot analysis
The experiments to determine whether two hHR23a proteins bind a common polyubiquitin chain were performed by using a previously described method.30 0.1 nmoles of purified GST-tagged hHR23a or its L198A, L355A mutants were bound to 20 μl of pre-washed glutathione S-sepharose resin respectively. Each resin was allowed to mix at 4°C overnight with K48- or K63-linked tetraubiquitin (Boston Biochem Inc.), or octaubiquitin, and then washed extensively with buffer A (20 mM sodium phosphate (pH 6.5), 50 mM NaCl, 0.5% (v/v) Triton X-100). The resin was next incubated with untagged hHR23a wild-type or mutated protein for one hour at 4°C. Each resin was spun down and then washed extensively with buffer A. As a control, 0.1 nmoles of purified His-tagged Rad23 was bound to 20 μl pre-washed Ni-NTA resin (Qiagen), and mixed with K48-linked (Figure 2(a)) or K63-linked (Figure 2(b)) tetraubiquitin and GST-tagged hHR23a or untagged hHR23a under the same conditions, and washed extensively with buffer B (50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 20 mM imidazole, 10% glycerol). In all cases, proteins that were retained on the resin were fractioned by electrophoresis, whereas proteins in supernatant were precipitated by 10% TCA, resolved by SDS-PAGE, transferred to a PVDF membrane, and probed with anti-hHR23a antibody (Abcam) and anti-ubiquitin antibody (Invitrogen). Visualization was performed using anti-rabbit or anti-mouse horseradish peroxidase and ECL.
Sedimentation velocity analysis
Stock samples of hHR23a and K48-linked tetraubiquitin were prepared at concentrations of 21.8 and 20.7 mg/ml respectively in 20 mM sodium phosphate buffer containing 30 mM NaCl at pH 6.5. Seven concentration ratios of the proteins were prepared, ranging from ∼12:1 to ∼1:6 at a constant total protein concentration of ∼30 μM. The dilutions were then subjected to sedimentation velocity experiments at 20 °C and 50,000 rpm using a Beckman-Coulter XLI analytical ultracentrifuge. Interference scans were acquired at 1 minute intervals for 4½ hours. A g(s) analysis of the data for each mixing ratio was performed using the program DcDt+36; 37. The weight average sedimentation coefficient, sw, for each mixture was calculated by integration of the g(s) profile over the range of sedimentation coefficients covered by the analysis. The plot of sw versus the molar ratio of the two components will have a maximum value at the correct stoichiometric ratio of the complex. Data were also analyzed by the c(s) method using SEDFIT38.
Theoretical weight average sedimentation coefficient (Sw) values were calculated by using Equation 1, in which the c(i) and s(i) are the weight concentrations and sedimentation coefficients of each species, respectively.
| (1) |
The data for the two samples with molar ratios closest to 1:1 were also analyzed using the program SEDPHAT39 with a model that allows characterization of the predominant species present in solution.
NMR spectroscopy
All NMR samples were dissolved in 20 mM NaPO4 (pH 6.5), 30 mM NaCl, 0.1% NaN3, and 10% D2O. Spectra were acquired at 25°C on Varian NMR spectrometers operating at 800 MHz with a cryogenically cooled probe. Processing was performed in NMRPipe40 and the resulting spectra visualized in XEASY.41 Protein concentrations were calculated by using extinction coefficients based on amino acid composition and absorbance at 280 nm for protein dissolved in 6M guanidine-HCl.
Chemical shift perturbation (CSP) data for hHR23a binding to tetraubiquitin were obtained for each amino acid residue by comparing the amide chemical shift values of hHR23a alone with those of hHR23a in the presence of 2-fold molar excess tetraubiquitin. Values were calculated according to Equation 2.
| (2) |
In this equation ΔδN and ΔδH represent the changes in the amide nitrogen and proton chemical shifts (in parts per million), respectively.
Acknowledgments
Ultracentrifugation was performed at the University of Connecticut's National Analytical Ultracentrifugation Facility in Storrs, CT (James L. Cole, Director). We are grateful to Cecile Pickart for the E2-25k construct as well as to Hiroshi Matsuo and Deanna Koepp for helpful discussions. NMR data were acquired in the UMN NMR facility (NSF BIR-961477), spectra processed and interpreted in the MSI BSCL, and the work supported by the National Institutes of Health CA097004-01A1 (KJW).
Abbreviations used
- hHR23
human homologue of Rad23
- HSQC
heteronuclear single quantum coherence
- NMR
nuclear magnetic resonance
- UBA
ubiquitin-associated
- UBL
ubiquitin-like
- UIM
ubiquitin interacting motif
- GST
glutathione-S-transferase
- VWA
von Willebrand A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 2.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 3.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 4.Conaway RC, Brower CS, Conaway JW. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 5.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi R, Dutta A. Proteasome inhibitors alter the orderly progression of DNA synthesis during S-phase in HeLa cells and lead to replication of DNA. Exp Cell Res. 2000;261:271–283. doi: 10.1006/excr.2000.5053. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 9.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 10.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 11.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 12.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 13.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 15.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Goh AM, Howley PM, Walters KJ. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry. 2003;42:13529–13535. doi: 10.1021/bi035391j. [DOI] [PubMed] [Google Scholar]
- 17.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 18.Saeki Y, Sone T, Toh-e A, Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem Biophys Res Commun. 2002;296:813–819. doi: 10.1016/s0006-291x(02)02002-8. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 21.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 22.Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin- dependent proteolysis. Biochem Biophys Res Commun. 2002;293:986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- 23.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Kaplun L, Tzirkin R, Bakhrat A, Shabek N, Ivantsiv Y, Raveh D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol Cell Biol. 2005;25:5355–5362. doi: 10.1128/MCB.25.13.5355-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng JM, Vrieling H, Sugasawa K, Ooms MP, Grootegoed JA, Vreeburg JT, Visser P, Beems RB, Gorgels TG, Hanaoka F, Hoeijmakers JH, van der Horst GT. Developmental defects and male sterility in mice lacking the ubiquitin- like DNA repair gene mHR23B. Mol Cell Biol. 2002;22:1233–1245. doi: 10.1128/MCB.22.4.1233-1245.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci U S A. 2003;100:12694–12699. doi: 10.1073/pnas.1634989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J Biol Chem. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 28.Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. Embo J. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook WJ, Jeffrey LC, Kasperek E, Pickart CM. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol. 1994;236:601–609. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- 30.Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol. 2006;356:1027–1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains mediate protein-protein interactions between two DNA damage- inducible proteins. J Mol Biol. 2001;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- 33.Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 34.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Raasi S, Pickart CM. Ubiquitin chain synthesis. Methods Mol Biol. 2005;301:47–55. doi: 10.1385/1-59259-895-1:047. [DOI] [PubMed] [Google Scholar]
- 36.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 37.Philo JS. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 38.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]


