Abstract
Living systems are replete with complex, stimuli-responsive nanoscale materials and molecular self-assemblies. There is an ever increasing and intense interest within the chemical sciences to understand, mimic and interface with these biological systems utilizing synthetic and/or semi-synthetic tools. Our aim in this review is to give perspective on this emerging field of research by highlighting examples of polymeric nanoparticles and micelles that are prepared utilizing biopolymers together with synthetic polymers for the purpose of developing nanomaterials capable of interacting and responding to biologically relevant stimuli. It is expected that with the merging of evolved biological molecules with synthetic materials, will come the ability to prepare complex, functional devices. A variety of applications will become accessible including self-healing materials, self-replicating systems, biodiagnostic tools, drug targeting materials and autonomous, adaptive sensors. Most importantly, the success of this type of strategy will impact how biomolecules are stabilized and incorporated into synthetic devices and at the same time, will influence how synthetic materials are utilized within biomedical applications.
Introduction
Biomolecules are attractive as synthons in the preparation of complex synthetic materials. The advantage of such an approach is that by incorporating biomolecules, one can impose evolutionarily derived properties on artificial structures. Therefore, biohybrid materials have the potential to respond to natural biochemical signals including those associated with certain disease states via dramatic switches in their physical morphology and/or chemical structure. In a biological context, morphology transitions are common responses to patterns of specific stimuli making possible many of the processes necessary for life. Certainly, a variety of materials capable of programmed shape and size change are found in biological systems, from allosteric enzymes to pseudopodium formation during chemotaxis, and endosomes during cell-uptake processes. By contrast, programmable synthetic supramolecular assemblies of this type are in their infancy.1–9 Mimicking and understanding nanomaterials that can undergo changes in morphology in response to stimuli are expected to have broad utility in a range of applications. Furthermore, synthetic elements bring chemical diversity and in turn, guide the behavior of the biomolecules themselves and could lead to greater stability of the biomolecules in extreme, abiotic conditions. Hence, the preparation of nanostructured, semi-synthetic and biohybrid materials may have future utility in applications not traditionally accessible to biological systems. This is an attractive strategy because, inherent to the biological molecules are desirable properties including order at the nanometer length scale and well-defined patterns of selective recognition elements. Uniting these two types of chemical systems is the goal of much of the work we describe in this perspective, with a focus on stimuli-responsive materials with potential biological relevance.
The most important advances in nanoscale materials have involved their characterization and the development of strategies for controlling structure at this challenging length scale. It is certainly no secret that the bottom-up synthesis of soft nanomaterials in particular, is difficult. This problem lends itself inherently to approaches utilizing supramolecular assembly of complex materials from simpler building blocks. These include copolymer amphiphiles, which are well suited for the development of functional supramolecular systems, as changes in the chemical or physical nature of the hydrophilic portion of an amphiphile can lead to formation, destruction, or morphology transition of the assemblies.10–16 This type of self-assembly will be a main focus of this review because a significant body of research exists describing efforts to trigger and manipulate the morphology of discrete assemblies of amphiphiles10 utilizing stimuli such as pH,17,18 temperature,19 small molecules or ions,20,21 enzymes,7,22,23 and light.24,25 In turn, there is an increasing focus on systems capable of responding to stimuli inherent to biological systems such as enzymatic reactions, protein expression patterns, DNA sequences, and cell-surface receptor recognition. Moreover, biochemical stimuli constitute programmed, specific interactions that are incredibly efficient, and underutilized in general within chemical and/or biochemical applications. Future designs taking these properties into account will no doubt make synthetic, stimuli-responsive nanomaterials more useful in a range of contexts, including in vivo applications.
This review will focus on biomolecules as programming tools in the assembly and manipulation of nanoscale polymeric nanoparticles and micelles (summary in Table 1). The goal is to introduce the reader to the field through several highlighted examples for a variety of different types of materials and stimuli. In addition, we will describe systems intended for biological applications that utilize polymeric structures that are not themselves biological, but are responsive to biologically relevant stimuli. Therefore, the review is split into two main sections; 1) polymeric nanoscale particles made from a combination of synthetic and biomolecular moieties capable of responding to stimuli and, 2) polymeric nanoscale particles that are entirely synthetic but respond to biologically relevant stimuli. The use of biomolecular interactions to increase complexity will be the main focus, with systems in which degradation is mediated by selective processes being described for perspective. Additionally, inorganic particles programmed with biomolecules26 will be excluded as comprehensive reviews already exist for such materials including bioorganic/inorganic hybrids.27–29
Table 1.
Summary of biohybrid or abiotic polymeric particles and their response to biologically relevant stimuli.
| Polymeric nanoparticles and micelles designed to respond to biologically relevant stimuli | Stimuli |
|||||
|---|---|---|---|---|---|---|
| DNA | Enzymes | Protein binding | Temperature | pH | Redox | |
| DNA micelles/nanoparticles | Morphology switch | Morphology switch, particle growth and degradation | — | Morphology switch | — | — |
| Peptide micelles/nanoparticles | — a | Morphology switch | — | Micelle assembly and reversed micelle switch | Morphology switch, micelle assembly, and reversed micelle switch | — |
| Protein micelles/nanoparticles | — | — | — | Morphology switch | Morphology switch | — |
| Sugar-based micelles | — | Morphology switch and disassembly | — | — | — | — |
| Abiotic polymers and particles | — | Structure destabilization, degradation and assembly | Structure destabilization, and surface modificatio | Morphology switch | Morphology switch and particle expansion | Morphology switch and vesicular burst |
To our knowledge, no examples have been demonstrated in the context of the scope of this review.
Biological and synthetic biohybrid polymeric nanoparticles and micelles
The biohybrid nanomaterials that will be the focus of this section consist of synthetic and biological building blocks. The use of biological molecules together with synthetic polymers brings together the programmability of biomolecules with the chemical diversity inherent to and enabled by synthetic organic chemistry. Certainly, biological molecules have been programmed by evolution to respond to and interact with very specific stimuli and hence their incorporation into nanostructures means they can respond to specific biologically relevant stimuli including pH differentials,18,30,31 ionic strength changes,32–34 the presence of given metabolites or other small molecules,21,35,36 protein-recognition events,37–39 DNA-sequence recognition,40–42 and enzymatic activity.7,43–45 The biohybrid materials of interest here generally contain a polymeric backbone with covalently linked nucleic acids, sugars, peptides, or proteins. In these systems both synthetic and biological polymers are utilized to impart structural features and functional properties on the materials. That is, the biomolecules are intrinsic parts of the polymers that form nanostructures. It should be noted that while non-covalent biomolecule–polymer systems such as polyplexes of high molecular weight DNA and polycationic polymers are of great interest, they are beyond the scope of this review.
DNA-containing polymeric nanoparticles and micelles
Nucleic acids are unique informational molecules capable of highly predictable sequence-specific recognition by other synthetic or natural nucleic acids, enzymes and proteins. Additionally, nucleic acids are capable of multiple modes of selective binding with small molecules as observed for aptameric systems46–49 and via intercalation or DNA sequence-specific binding.50,51 In addition, nucleic acids are known to occur naturally as catalysts52 and can be evolved in vitro53–56 to behave as such. This functional diversity arises because of natural evolution of function in combination with non-natural function facilitated by modern synthetic molecular evolution and selection processes.57 At the heart of these capabilities is the information content of nucleic acids. This is the cornerstone of the genetic code and is increasingly of value in the construction of synthetic nanoscale materials. Indeed, the field of DNA nanotechnology has blossomed in recent years with DNA increasingly employed in the construction of complex nanoscale architectures made entirely from nucleic acids3,58–63 or for the decoration of both organic64–66 and inorganic nanoparticles.67,68 By contrast, its use as a building block in the synthesis of hybrid polymeric materials surprisingly remains rare69,70 with several examples highlighted here that take advantage of synthetic amphiphilic block copolymers capable of self-assembly into nanoscale biomolecule-responsive micellar materials.4,41,71–73 Amphiphilic DNA-based biohybrid polymers have the ability to be manipulated via two key approaches: 1) isothermal and thermal DNA hybridization, and 2) response to enzymatic activity. Given the broad range of interactions and chemistry available to nucleic acids, such materials are expected to provide a rich source of interesting phenomena moving forward.
Manipulation of morphology via isothermal and thermally controlled DNA hybridization
While it remains a challenge to control the morphologies and dimensions of soft materials in general, it has been found that by using the unique properties of DNA hybridization one can encode structural information within nanoscale polymer-based particles.4 This process can be performed reversibly via isothermal or thermal approaches, as highlighted here.
In 2007, Caruso and coworkers74 described an interesting example of isothermal DNA hybridization controlled morphology. In this work, a shrinking capsule was created from a layer-by-layer (LbL) assembled particle. Initially, DNA is electrostatically adsorbed on an amine-functionalized silica particle. Various DNA sequences of either homopolymeric repeating units of adenine and guanine (AG) and/or thymine and cytosine (TC), or longer complementary sequences were hybridized onto silica creating 2 to 9 layers. Upon removal of the silica core, the particle reduced in size by up to 50% depending on the sequence and order of hybridization on the silica particle. This type of programmable control of size on the micrometer scale shows promise for increasing the concentration of a payload entrapped inside capsules. Additionally, this concept has been extended to control the enzymatic degradation of microcapsules utilizing restriction enzymes (vide infra).75
In addition to control over size, control of the morphology of materials is also of great interest.41,76,77 Recently, our group has shown that using the property of isothermal DNA hybridization, one can manipulate the morphology of DNA-brush copolymers.41 In this system a spherical micelle can be converted to a cylindrical, or fibromicelle structure, and back to a spherical morphology reversibly (Fig. 1). The fibromicelle was formed via reaction with a DNAzyme that recognizes a target DNA sequence in the shell of the spherical micelle, hybridizes, and then cleaves at a specific RNA base. After the fibromicelle was formed, addition of a longer complementary strand allowed reversible conversion back to a spherical structure by formation of a more thermodynamically stable DNA duplex. In addition to relying on sequence selective reactions to isothermally modulate morphology, these systems are capable of morphology changes via thermally controlled DNA hybridization. This provides an alternative route by melting duplexes and reannealing them in order to reversibly manipulate structural morphology.
Fig. 1.
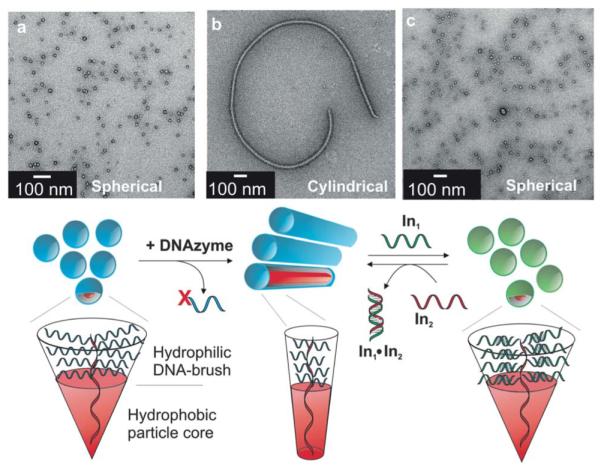
Assembly of DNA-brush copolymers into micelles with spherical or cylindrical morphologies. Amphiphile structures are represented as cones for each respective morphology. The hydrophobic domain is highlighted in red. Transmission electron microscopy (TEM) images of (a) 25 nm spherical micelles assembled from initial DNA-brush copolymers; (b) cylindrical morphology formed following DNAzyme addition to spheres; (c) spherical micelles (green) formed after addition of In1 to cylinders. Adapted from ref. 41. Copyright 2010 Wiley-VCH Verlag GmbH & Co.
Herrmann and coworkers42 have demonstrated control over morphology simply by varying the length of a sequence of complementary DNA added to a micelle formed from a DNA block copolymer with a poly(propylene oxide) (PPO) backbone. The PPO-DNA block copolymer initially formed a spherical micelle. Upon addition of a short complementary strand of DNA, the micelle structure remained intact. However, after adding a longer complementary strand of DNA, short rod-like structures were observed (Fig. 2). Furthermore, in a powerful demonstration of this approach to controlling nanoscale architecture, control over the length of the rod-like micelle was accomplished by varying the number of nucleotides in the template DNA sequence.
Fig. 2.
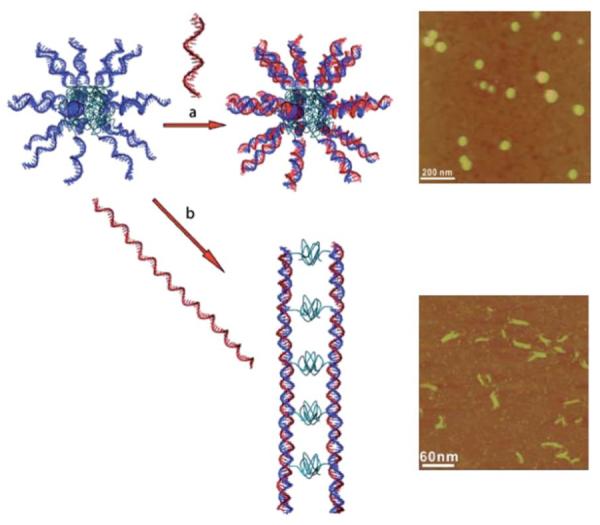
Schematic representation of hybridization of ssDNA-b-PPO micelles with different DNA molecules. (a) Base pairing with a short complementary sequence yields micelles with a double stranded hydrophilic shell while maintaining almost identical structural properties of the ssDNA aggregates. (b) Hybridization with long DNA templates results in rod-like micelles consisting of two parallel-aligned double helices. Reprinted from ref. 73. Copyright 2008 Wiley-VCH Verlag GmbH & Co.
Enzymatic control over DNA-based nanomaterials
Given the number of enzymes capable of sequence-selective DNA recognition, approaches that include reactions such as strand cleavage, ligations, error correction, and abasic site removal are potentially viable for manipulation of DNA-containing materials. Various enzymes have been reported in the literature for controlling the size and shape of micellar nanostructures including DNA polymerases,78,79 DNAzymes,41 and restriction enzymes.75 Several examples using the unique properties of enzymes in controlling the morphology of DNA biohybrid materials will be discussed in this section.
Terminal deoxynucleotidyl transferase (TdT) is an enzyme that belongs to the family of nucleic acid polymerases and is able to catalyze the addition of deoxyribonucleotides to the 3′ hydroxyl end of DNA in a template-independent fashion.80 Recently, Herrmann and coworkers81 showed the in situ growth of a DNA-b-PPO diblock copolymer micelle using TdT as a “particle growth” enzyme. The micelle was immobilized on a mica surface and growth of the micelle was analyzed by scanning force microscopy (SFM). The initial height of the DNA-b-PPO aggregates was approximately 5 nm and was controllably increased by addition of 2′-deoxythymidine-5′-triphosphate (dTTP) mononucleotide with the enzyme (TdT). This example demonstrates the controlled growth of a micelle using an enzyme on a mica surface.
In addition to size change, recent literature has shown the morphology of micellar structures can be altered via enzymatic activity as described above for the addition of a DNAzyme to DNA-brush copolymer micelles.41,77,82 These DNAzymes are not traditional enzymes, but rather evolved catalytic DNA sequences making them easily tunable with regards to their sequence selectivity. On the other hand, the inherent specificity and efficiency of naturally occurring endonucleases makes them very attractive as tools for manipulating nanostructures formed from nucleic acids. Caruso and coworkers75 utilized the specificity of restriction enzymes to selectively degrade a layered (LbL) DNA capsule. The assembly process was driven by the hybridization of single stranded oligonucleotides. A portion of each oligonucleotide was designed to contain the GAATTC EcoRI cut site. Upon treatment with the restriction enzyme, selective degradation of the LbL was observed. These enzymatic processes were monitored by quartz crystal microbalance (QCM), fluorescence microscopy and flow cytometry. The data showed EcoRI degraded the particles in a selective fashion with only minor non-selectively cleaved products formed. The majority of the DNA capsules that contain the EcoRI cut site were degraded and decreased in size from 2.7 μm to 0.8 μm. These DNA capsules were also studied for their propensity to encapsulate and release guest molecules in a sequence-selective fashion. In these studies, a fluorescently labeled BSA protein was encapsulated and up to 90% was subsequently released in the presence of EcoRI. These studies demonstrate an example of the programmed degradation of a micro to nanoscale material made by combining synthetic DNA-nanostructures with naturally evolved sequence-selective enzymes.
Peptide containing polymeric nanoparticles and micelles
Natural and non-natural amino acids offer greater chemical diversity than nucleic acids, but have less predictable binding and recognition. At the same time they are susceptible to optimization via molecular evolution strategies,83,84 and are capable of selective targeting,85,86 signaling,87–89 receptor binding90,91 and behaving as substrates for specific enzymes.92–95 Therefore, the incorporation of amino acids and peptides into polymeric materials, as functional and structural building blocks, is of great interest and has tremendous potential utility by virtue of making synthetic particles susceptible to the inherent biological characteristics of the peptide sequence.96–98 Herein, we focus on peptide-polymeric materials that take advantage of the solution properties of biohybrid architectures. Of particular interest is how these peptides can be used for coupling the nanostructures to biologically relevant signals. We will describe several stimuli in this respect: 1) pH, 2) temperature, 3) dual-responsive pH and temperature systems, and 4) enzymes.
pH responsive systems
Smart peptide-polymeric materials that are susceptible to variations in pH have been the focus of much attention.30,99–105 This stimulus is of particular interest within biomedical applications due to the pH variation between diseased and healthy tissues,106–108 and within particular biological compartments such as endosomes.109 Herein, we describe several recent examples of peptide copolymer based materials that are sensitive to acidic and basic conditions that undergo pH driven assembly or disassembly.
Poly(glutamic acid) (PLG) is one of the polyamino acid examples that has attracted particular interest. It is capable of undergoing reversible transformation from a rod-like α-helical conformation to a coil with varying temperature.100,110 PLG is also an interesting candidate for pH sensitive systems due to the ability for the surrounding solution to determine the hydrophobicity or hydrophilicity of the material. An interesting example using PLG shows the successful generation of a reversible process for turning a micelle inside-out.111 The diblock copolymer consisted of 15 repeating units of poly(l-glutamic acid)-b-poly(l-lysine) (PLG15-b-PLL15). At neutral pH the zwitterionic polymer consisted of unique unimers. Under acidic conditions a micellar structure with PLL as the hydrophilic shell and PLG as the hydrophobic core was formed. At basic pH the reverse vesicle was formed with PLG as the hydrophilic shell and PLL as the hydrophobic core, as determined by 1H NMR.
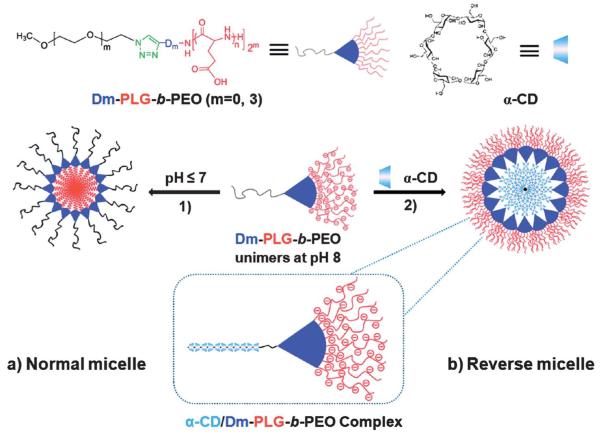
Taking advantage of the unique hydrophobic–hydrophilic transition of PLG in response to pH coupled with the docking of cyclodextran, Chen et al. demonstrated an example of a normal and reversed micelle (Fig. 3).112 In this study a linear and Dendron-like/linear poly(l-glutamic acid)-b-poly(ethylene oxide) (Dm-PLG-b-PEO, m = 0 and 3) block copolymer was synthesized. Under acidic conditions the PLG-b-PEO self-assembles to form a micelle with PLG as the hydrophobic core. Upon addition of α-cyclodextrin (α-CD) at high pH a new micelle is formed with the anionic PLG as the hydrophilic corona and α-CD/PEO as the hydrophobic core. Additionally, doxorubicin (DOX) was loaded into the micelles and sustained approximately a 70 day drug release period in vitro.
Fig. 3.
Aqueous self-assembly of (a) polypeptide-dendritic micelles and (b) reversed micelles generated from Dm-PLG-b-PEO copolymers. Reaction (1) indicates micelle formation of the copolymer in neutral or acidic solution and (2), the reversed micelle formation of copolymer after adding α-CD in alkaline solution. Reprinted with permission from Y. Chen and C.-M. Dong, J. Phys. Chem. B, 2010, 114, 7461–7468. Copyright 2010 American Chemical Society.
Contrary to the previous example describing a change in structure without a change in particle shape, pH can also be utilized to induce a change in morphology.18 In this work, an amphiphilic β-cyclodextrin vesicle (CDV) was decorated with an adamantane modified octapeptide (Fig. 4). The octapeptide binds to the surface of CDV with a binding constant of 3.5 × 104 M−1 and a stoichiometry of approximately 1 : 1. At pH 7.4, the peptide does not adopt a β-sheet conformation and the particle remains spherical. However, at pH 5.0 the peptide undergoes a random coil-to-β-sheet transition as determined by circular dichroism, resulting in an overall cylindrical structure of the CDV peptide complex. To test the hypothesis that these materials may perform as small molecule delivery systems, a pyrene dye was encapsulated into the CPV. The dye did not leach out of the vesicle structure as long as the pH was maintained. However, a decrease in pH showed a drop in pyrene fluorescence intensity, indicating that dye is released from the vesicle.
Fig. 4.
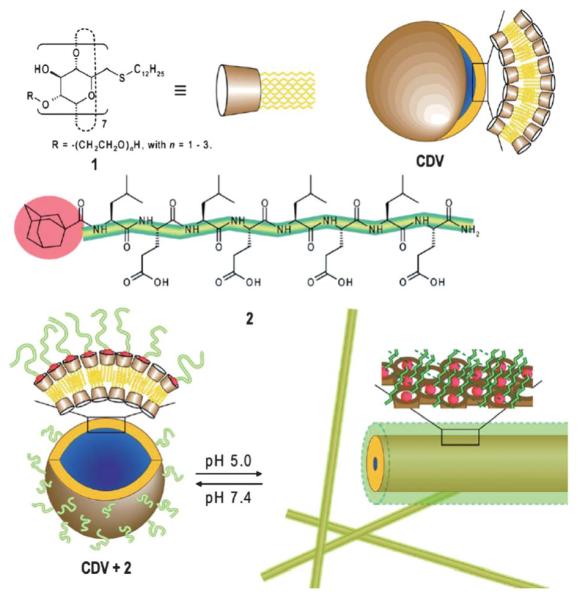
Chemical structures of β-CD derivative (1), which self-assembles into cyclodextrin vesicles (CDVs), and the adamantane modified octapeptide (2) that binds to the CDV. Octapeptide 2 adopts a random coil conformation at pH 7.4 while bound to CDV. Upon acidification, 2 rearranges into a β-sheet, inducing a morphology switch from vesicle to a fibril structure. Reprinted with permission from F. Versluis, I. Tomatsu, S. Kehr, C. Fregonese, A. W. J. W. Tepper, M. C. A. Stuart, B. J. Ravoo, R. I. Koning and A. Kros, J. Am. Chem. Soc., 2009, 131, 13186–13187. Copyright 2009 American Chemical Society.
Temperature responsive systems
Similar to the pH responsive systems described above, nanoscale micelles can also be designed to respond to temperature variation through changes in hydration state, switches in morphology, and aggregation state through polymer assembly or disassembly. These processes are mediated by adjusting the solution temperature appropriately above and below the lower critical solution temperature (LCST) of the polymer blocks.113,114 These materials have potential in many biological applications, including the encapsulation of drugs and diagnostic agents. While natural variations in body temperature specific to diseased versus normal tissue are small,115–117 it is plausible that temperature responsive particles may be useful coupled with localized heating from co-targeted inorganic particles.118–122 Organic peptide copolymer materials capable of temperature transformations will be discussed here.
As mentioned before, PLG is capable of undergoing reversible transformation from rod-like α-helical conformations to a coil with varying stimuli such as pH. Similarly, these conformation changes are induced with temperature variation.100,110 Commercial “Jeffamine” consists of an amino terminated random copolymer of ethylene oxide and propylene oxide and is known to be thermoresponsive with a LCST of 30 °C in water.113 A recent example by Lecammandoux and coworkers113 coupled the known thermoresponsive Jeffamine material to create a stimuli-responsive double hydrophilic Jeffamine-b-PLG copolymer. The hydrophilic block copolymer became amphiphilic and self-assembled at 51 °C. At a temperature of 66 °C the core became dehydrated as determined by small-angle neutron scattering (SANS). Additionally, the size of the core decreased with increasing temperature as a result of the dehydration of the Jeffamine block. This has interesting potential in the future manipulation of payloads in the core of micelles.
Elastin-like polypeptides (ELPs) are an interesting class of biopolymers since they are both biocompatible and thermoresponsive.123,124 In recent work by Chilkoti and coworkers,125 an ELP was used as a building block to create a linear AB-block copolymer capable of self-assembly at approximately 40 ° C. The ELP block copolymer consisted of an N-terminal ELP[V1A8G7-n] which is hydrophilic at higher temperatures and a C-terminal ELP[V5-n] that is hydrophobic at lower temperatures. When the hydrophilic-to-hydrophobic ratio is between 1 : 2 and 2 : 1, the material goes through two phase transitions: the first transition is from unimer to spherical micelle at an intermediate temperature of 36–46 °C; the second, at temperatures ranging between 49–51 °C whereby the micelles form micrometer aggregates as determined by optical fluorescence spectroscopy, dynamic light scattering (DLS) and cryogenic transmission electron microscopy (TEM). Lastly the corona of the micelles were modified with RGD and NGR peptide motifs and were capable of cellular uptake at physiological relevant temperatures.
Chaikof and coworkers126 demonstrated that amphiphilic diblock polypeptides could be designed and expressed to undergo temperature-dependent transitions. These polypeptides contain a hydrophilic block and a temperature responsive block that aggregates in aqueous solution at elevated temperatures (Fig. 5). At temperatures above 10 °C, the material begins an endothermic transition as determined by differential scanning calorimetry (DSC), and circular dichroism (CD). At temperatures above 25 °C it is believed the material has completed the transition to the micellar phase as shown by 1H NMR and DLS. Importantly, to help stabilize the structure, disulfide bonds add extra cross-linking stability allowing for lower critical micelle concentrations (CMCs) than reported for other ELP diblock copolymers.
Fig. 5.
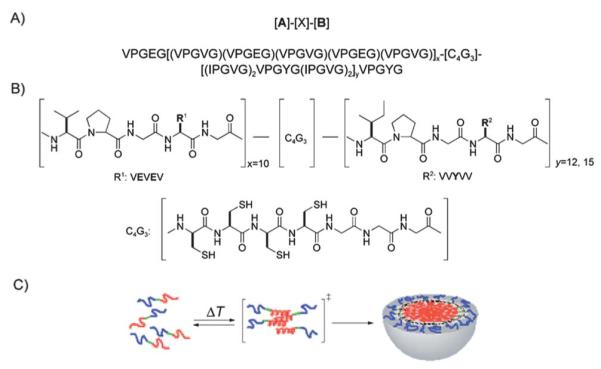
(A) Amino acid sequence of elastin diblock polypeptides (ADP1 (x10y12) and ADP2 (x10y15)) and (B) structures of ADP1 and ADP2. (C) Thermally responsive micelle formation utilizing elastin diblock polypeptide. Reprinted from ref. 126. Copyright 2010 Wiley-VCH GmbH & Co.
pH and temperature responsive systems
The ability for a polymeric material to be responsive to both pH and temperature increases its complexity and potential versatility.11,31,127–133 As already discussed, PLG is known to respond to pH and temperature. This and/or PLL as a copolymer with known thermoresponsive polymers such as poly(N-isopropylacrylamide) (PNIPAM)134 or poly[2-(dimethylamino) ethyl methacrylate] (PDMAEMA)135 is capable of forming double hydrophilic block copolymers (DBHCs). DBHCs are a class of polymers that are initially soluble in water and in response to specific stimuli, become amphiphilic and often self-assemble into micellar nanomaterials.
Zhang et al.136 reported the synthesis of a PLG-b-PNIPAM DBHC that is responsive to both pH and temperature. The polymer was synthesized by a combination of ring-opening polymerization (ROP) and reversible addition-fragmentation chain transfer (RAFT). The aggregation behavior was analyzed via 1H NMR, DLS, and TEM. It was found that the PLG block had no interference on the collapse of the PNIPAM block, however it did shift the aggregation process to a higher temperature. Fig. 6 shows TEM images of pH and temperature dependence on the size and shape of aggregates. Additionally, the authors note that at a temperature above 50 °C, the aggregates shrink due to further dehydration. As the pH increased, larger aggregates were formed but had no affect on the collapse of the PNIPAM block. This is one of the first examples of control over the morphology of a material using two stimuli.
Fig. 6.
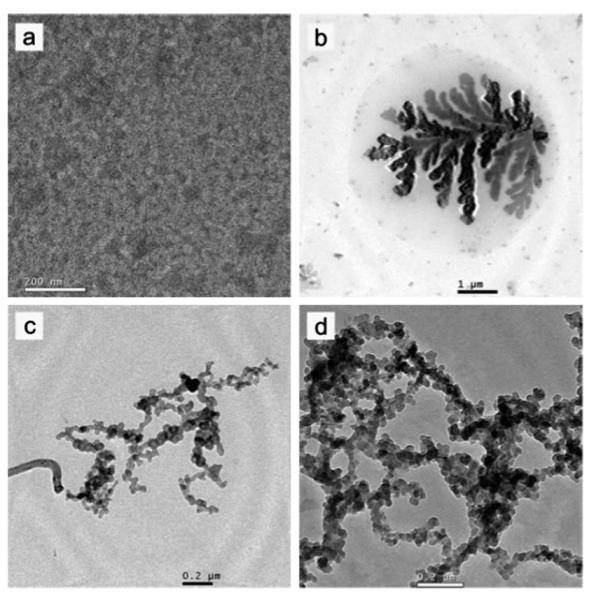
TEM images for PLG57PNIPAM228: (a) unimer at pH 8.0, room temperature; (b) aggregate formation at pH 8.0, 50 °C; (c) cylinders and network structure formation at pH 9.0, 50 °C; (d) network formation at pH 10.0, 50 °C. Reprinted with permission from X. Zhang, J. Li, W. Li and A. Zhang, Biomacromolecules, 2007, 8, 3557–3567. Copyright 2007 American Chemical Society.
Another interesting report by Jing and coworkers129 found that by varying pH and temperature, they could alter whether the PNIPAM block was in the hydrophobic core or the hydrophilic shell when polymerized with poly(l-lysine). Using cloud point measurements, it was found that the LCST of the copolymer decreased at higher pH and increased at lower pH. Further DLS measurements indicated that at pH 5.0 (25 °C), no aggregate formation was observed. However, at pH 12.5 (25 °C), aggregates of 65 nm were formed. At pH 5.0 (45 °C), aggregates of 100 nm had formed. The authors concluded that at low pH and high temperature, PLL is in the hydrophilic corona, whereas at high pH and low temperature, PNIPAM is in the hydrophilic corona.
Additionally, Lecommandoux and coworkers130 published a pH and temperature responsive DBHC capable of forming micelles and undergoing subsequent switching of corona and core to form reversed structures. The material consisting of PDMAEMA-b-PLG was studied by varying the relative lengths of the PLG block while keeping the PDMAEMA block constant at a block size of 85. At low pH and all temperatures, the material remained as unimers, regardless of the length of PGA. As the pH increased from 4 to 7 at low temperatures, electrostatic vesicles formed with PDMAEMA85-b-PLG77, where PDMAEMA was the hydrophilic corona. At high temperatures in the same pH range PDMAEMA85-b-PLG186 formed vesicles with PLG now forming the hydrophilic shell of the vesicle. At pH 7 to 9 the material only formed soluble complexes when the PLG block size is 186 and at high temperatures. However, at pH greater than 9, with all block sizes of PLG, the material only formed aggregates at higher temperatures. For PDMAEMA85-b-PLG37 and PDMAEMA85-b-PLG77, the vesicular structures were formed with the PLG block in the corona. However for PDMAEMA85-b-PLG186, a thermoresponsive micelle was formed. Control over the morphology via correct placement of particular blocks shows great promise for designing various stimuli-responsive elements into a given material.
Enzyme-responsive systems
Enzymes play a critical role in biology through a myriad of natural processes involved in the manipulation of nanoscale self-assemblies including the replication of nucleic acids,137 decomposition of biomaterials including extracellular matrices,138,139 and in the assembly of viruses.140,141 This has inspired efforts to create materials that respond to very specific enzymatic triggers by building them out of or incorporating peptide substrates. To date, the majority of research into organic enzyme-responsive systems has focused on phosphorylation, dephosphorylation, and other enzymatic reactions of peptide-only nanostructures to control their formation.7,142–144 We highlight here research into the utility of enzymatic reactions to control the structural characteristics of peptide-polymer conjugates. The use of enzymatic reactions to build and manipulate organic nanostructures is the subject of a recent review.7 Here we aim to highlight several examples in which morphology changes, or unusual degradation processes are demonstrated for both biohybrid systems (this section) and later, in abiotic polymeric particles (vide infra).
α-Chymotrypsin is a serine protease responsible for cleavage on the C-terminal side of tyrosine, phenylalanine and tryptophan.145 With this in mind, Cenker and coworkers146 designed a peptide–polymer conjugate capable of hydrolysis upon treatment with α-chymotrypsin. Conjugation of PEG3000 to peptide βAβAKLVFF led to the formation of spherical micelles approximately 10 nm in diameter where the peptide is contained in the hydrophobic core and PEG is located on the hydrophilic shell. After treatment with α-chymotrypsin, the enzyme cleaves between the F–F residue on the peptide leaving a F–PEG3000 and βAβAKLVF behind as determined by mass spectrometry. The released peptide fragment and F–PEG showed no noticeable secondary nanostructures. This type of example highlights the potential for the delivery of therapeutic peptide fragments in response to specific enzymes.
Phosphatases are a class of enzymes that remove phosphoryl groups attached to serine, threonine or tyrosine residues.145 These enzymes play a vital role in turning off signaling pathways that are activated by kinases. Borner and coworkers147 recently utilized an acid phosphatase to manipulate a peptide–polymer conjugate (Fig. 7). The polymer consists of a PEO block linked to a repeating peptide segment of threonine and valine diads (TV)5. This repeating segment of TV is known to form β-sheet aggregates in water.148 The introduction of three phosphothreonine units into the (TV)5 peptide aggregator results in a double hydrophilic block copolymer. Only upon dephosphorylation with an acid phosphatase was it possible to form the nanofibrils.
Fig. 7.
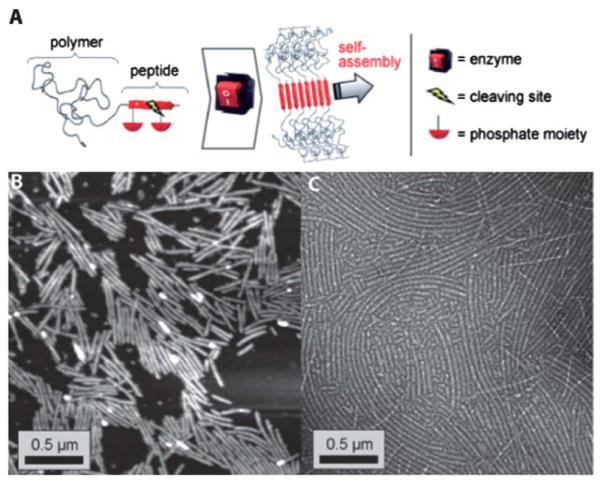
(A) Schematic representation of enzyme-switchable PEO–peptide conjugates. Microstructures formed by enzyme triggered self-assembly of PEO–peptide conjugates: (B) visualized by AFM 7 days after enzyme treatment and (C) TEM micrograph of structures stained with uranyl acetate, after 10 days. Adapted from ref. 147. Copyright 2009 Wiley-VCH Verlag GmbH & Co.
Inspired by the utility of enzymes as catalytic amplification tools and selective protagonists in natural systems, our group recently developed peptide–polymer amphiphiles (PPAs) capable of forming well-defined enzyme-responsive spherical micelles.22 These micelles undergo responses to several enzymes demonstrating in situ, selective, reversible and user-defined shifts in micellar nanoparticle morphology. Utilizing the recognition properties of a substrate for selective enzymatic cleavage, and or phosphorylation/dephosphorylation, information stored in the micelle shell can be read and manipulated in several modes causing dramatic, and sometimes reversible changes in morphology and particle size. Fig. 8 illustrates the reversible morphological change observed for the peptide material in response to sequential phosphorylation/dephosphorylation cycles that occur in remarkably high yield.
Fig. 8.

Response of peptide polymer particles to sequential additions of protein kinase A (PKA) and protein phosphatase 1 (PP1). Reprinted with permission from T. H. Ku, M. P. Chien, M. P. Thompson, R. S. Sinkovits, N. H. Olson, T. S. Baker and N. C. Gianneschi, J. Am. Chem. Soc., 2011, 133, 8392–8395. Copyright 2011 American Chemical Society.
Protein-containing polymeric nanoparticles and micelles
Proteins provide the ultimate complexity in evolved three-dimensional structures at the nanoscale. They can have well-defined recognition properties for a range of molecules including other proteins,149 peptides,150 DNA,151 and small molecules152 and operate as enzymes on natural and synthetic substrates. Perhaps the most intriguing examples of relevance to organic nanomaterials come from selective protein recognition events responsible for virus capsid packaging and assembly.153 These features make proteins very attractive for incorporation into semi-synthetic nanoscale architectures,154,155 metal-directed assembly of proteins,156–158 protein tiling assemblies159 and protein amphiphile aggregates.160–162 Again, by incorporating proteins into such structures, one imposes evolutionarily derived properties on the synthetic material. Moreover, the synthetic structures bring chemical diversity and guide the functionality of the protein for use in the desired context. Herein we describe examples of stimuli-responsive synthetic protein–polymer nanoparticles.
pH responsive protein–polymer nanomaterials
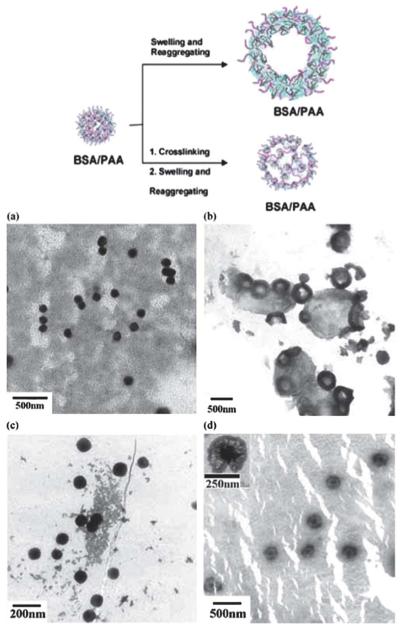
One of the only examples to our knowledge of a pH responsive protein–polymer based nanomaterial comes from Wang et al.163 In this example, bovine serum albumin–poly(acrylic acid) (BSA/PAA) initially self assembles into spherical nanoparticles. If placed in the dark for 7 days the material forms a capsule as evidenced by TEM (Fig. 9) and DLS. The material was also cross-linked with glutaraldehyde, which initially forms nanospheres, and again capsules if incubated in the dark for one week. These “nanocapsules” were then loaded with either Rhodamine B (RB) or doxorubicin hydrochloride (Dox·HCl) and the release profile was measured over a pH gradient. For both crosslinked and non-crosslinked nanocapsules at 20 °C, the percent release of RB is greater at pH 7.4 than pH 3.6. At 37 °C this trend was reversed with a higher percent release at low pH. Additionally, the overall rate of release was faster for the non-cross-linked nanocapsule and was therefore used to measure the release of Dox. At 20 °C, the percent release of Dox was greater at pH 3.6 than pH 7.4. However, the material itself was not characterized further in response to various pH changes.
Fig. 9.
Schematic representation of the formation of BSA/PAA nano-spheres and nanocapsules. TEM images of (a) BSA/PAA, (b) BSA/PAA nanocapsule, (c) crosslinked BSA/PAA nanospheres, (d) crosslinked BSA/PAA nanocapsules. Adapted from ref. 163. Copyright 2010 John Wiley & Sons, Ltd.
Temperature responsive protein biohybrid materials
In the context of complex polymeric conjugation reactions, one of the major concerns is purity of the material post-modification. For this reason, Boyer et al.164 set out to synthesize a BSA–PNIPAM polymer in situ via RAFT polymerization. A water-soluble N-isopropylacrylamide monomer was conjugated to BSA via the free thiol group on the Cys-34 residue. This material was then grown via RAFT and characterized by 1H NMR, SEC, MALDITOF, and PAGE. Considering the thermoresponsive nature of PNIPAM, temperature studies were performed to analyze the LCST of the material. DLS showed that at temperatures below 35 °C, both fragments are hydrophilic and therefore, no aggregate formation is observed. Above the LCST, aggregates of 250–300 nm were formed.
Glutathione peroxidase (GPx) is an antioxidative enzyme that protects cellular membranes and various cellular components from oxidative damage.165 Considering its biological importance, several research groups have set out to create GPx mimics.166,167 Addition of these mimics to a polymeric structure can allow for the design of an amphiphilic material that is capable of self-assembling into micellar structures. The conjugation of proteins to known temperature responsive polymeric material such as PNIPAM is particularly useful since it undergoes a volume phase transition at physiological temperature.168–173 Huang et al.165 were capable of mimicking GPx activity by synthesizing a polyacrylamide-b-PNIPAM-Te (PAA-b-PNIPAM-Te) and PAA-Te-b-PNIPAM polymer. DLS determined that as the temperature was increased from 15 to 45 °C the aggregate size also increased from 12 to 200 nm. Aggregate formation started for PAA-b-PNIPAM-Te at 33 °C with an average size of approximately 25 nm. However as the temperature increased, so did the aggregate size, reaching a thermodynamic minimum of approximately 200 nm. The aggregates were determined to be spherical in shape as determined by SEM. For PAA-Te-b-PNIPAM, spherical aggregates began to form at 35 °C with no smaller aggregates formed at lower temperatures. Additionally, the authors determined the optimal catalytic activity was for a PAA-b-PNIPAMTe polymer over PAAm-Te-b-PNIPAM block copolymer.
Sugar-based polymeric micellar and vesicular particles and their response to enzymes
The examples shown in previous sections have described nucleic acids, peptides and proteins as structural, and programming elements within responsive polymeric materials. Of course, there are other important biological polymers including sugars of various types. In an early example of enzyme responsive systems, Akiyoshi and coworkers174 utilized enzyme-responsive carbohydrate polymerizations to induce morphology changes in vesicles formed from standard surfactants. In this study, amylose-primer surfactants with various alkyl groups (C8, C12, C16) linked to the reduced terminus of maltopentaose (MP) can be elongated by reaction with glucose phosphate via phosphorylase catalysis. This carbohydrate polymerization increased the hydrophilicity and critical micelle concentration of the surfactants leading to the disassembly of the micelles (Fig. 10A). In addition, they also investigated a micelle-to-vesicle phase transition controlled by polymerization of the amylose-primer surfactants. In this case, a mixture of l-α-dipalmytoyl phosphatidylcholine (DPPC) with the C12MP surfactant forms a small aggregate. Upon polymerization, the increase in hydrophilicity of C12MP causes it to depart, resulting in an expansion of aggregate size and formation of vesicles (Fig. 10B). Furthermore, they demonstrated this micelle-to-vesicle transition by an enzyme stimulus is effective for reconstitution and refolding of membrane proteins. In these studies, Bacteriorhodopsin (BR), a model membrane protein, was mixed with the C12MP/DPPC-micelle solution and upon micelle-to-vesicle transition, BR was reconstituted in DPPC liposomes.
Fig. 10.
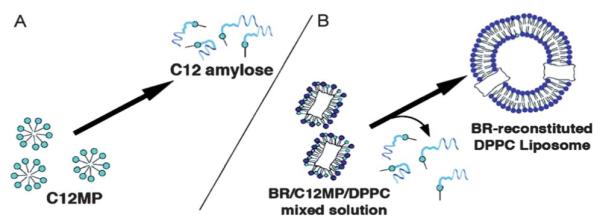
(A) C12MP dissociation under polymerization of a sugar moiety. (B) Micelle-to-vesicle morphology transition of DPPC/C12MP micelle mixture under enzymatic elongation. Reprinted with permission from N. Morimoto, N. Ogino, T. Narita, S. Kitamura and K. Akiyoshi, J. Am. Chem. Soc. 2006, 129, 458–459. Copyright 2006 American Chemical Society.
Abiotic polymeric materials responsive to biological stimuli
Stimuli-responsive polymeric nanomaterials have attracted increasing attention for biomedical applications including drug delivery because of their potential to switch function and release their contents upon interaction with disease-associated signals. Herein, we will discuss fully abiotic structures formed from synthetic polymers only, that are not themselves biohybrid polymeric materials but are nevertheless designed to recognize biologically relevant stimuli. We discuss specific nanoscale materials and their response to a range of endogenous stimuli associated with cells, endosomes and tissue, enzymes and pH, in addition to the possibility of exogenously applied signals such as small molecules and foreign proteins for manipulating materials selectively. We will not describe efforts to utilize light as a stimulus for manipulating nanomaterials in vivo as it is non-biological when applied externally. However, a number of examples in the literature suggest that light, when used as a non-invasive, exogenously applied signal, may be effective in manipulating materials for biomedical applications.25,175–180 Chemical or physical stimuli endogenous to biological systems have been explored for interacting with nanoscale materials intended for biomedical applications. Such stimuli include low pH environments (as found within late endosomes), overexpressed cell-surface receptors, excreted disease-associated enzymes, and oxidative micro-environments. As described here, materials capable of responding to such stimuli are not limited to biological, or biohybrid-based systems as discussed in the earlier sections of this perspective. Here, particles are categorized into six sections according to type of stimulus: 1) pH, 2) enzymes, 3) oxidation/reduction processes, 4) protein binding/recognition, 5) temperature change, and 6) small molecules. Again, we will predominantly focus on systems where the stimuli induces a morphology change and/or cause an increase in complexity rather than degradation processes. There are several potential benefits for systems such as these as opposed to degradable particles, and some of these will be described in the sections below.
pH-responsive micelles and nanoparticles
pH-responsive systems have attracted attention as some of the most studied due to their promise in biomedical applications.181–193 Such systems often aim to take advantage of the natural decrease of pH within endosomes to facilitate release of drugs following cell targeting and successful uptake. Materials capable of disintegration or degradation such as polymers with pH-sensitive linkages (esters,194–197 carbamates,198,199 hydrazones,200–202 and acetals194,203) in the backbone have been developed by many research groups and will be beyond the scope of this review. Herein, pH responsive materials without biomolecular moieties will be addressed with a particular interest in the capability of undergoing morphology changes in response to pH.
pH as a stimulus resulting in morphology transitions
The preparation of particles responsive to environmental pH changes is a very promising and active area of research.111,181,183–188,190–193,204–206 Multiple examples in the literature describe morphology switches of micellar particles in response to changes in solution pH.17,111,133,204,205,207,208 However, little attention has been devoted to stimuli-responsive shape switchable, non-micellar polymeric nanoparticles. Recently, Mitragotri and coworkers described shape switchable PLGA (poly(lactide-co-glycolide)) particles capable of changing from an elliptical disk (ED) morphology to a spherical morphology in response to pH, temperature and chemical signals (Fig. 11).209 To assess the ability of pH to induce shape switches, EDs were prepared using PLGA-acid (PLGA with terminal carboxylic acids). Specifically, the surface charge of PLGA-acid nanoparticles can be changed through ionization of the terminal carboxylic acid groups, and thus the interfacial tension of PLGA EDs can be controlled. At physiological pH, the carboxyl groups of PLGA (pKa = 3.85) are largely deprotonated, causing low hydrophobicity and thus low interfacial tension. Lowering the pH (<3.85) protonates the acid groups and results in a morphology switch concomitant with the increase in interfacial tension.
Fig. 11.
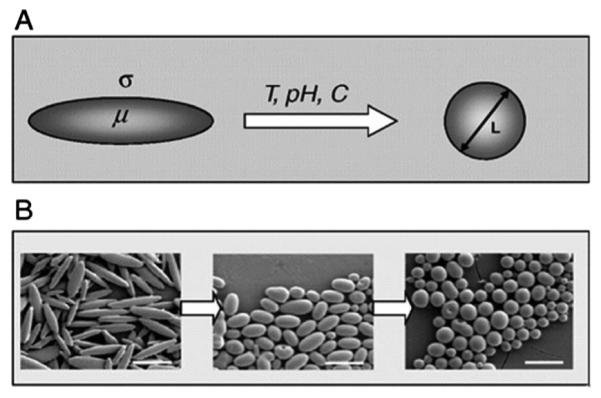
Design of stimuli-responsive morphology-switchable PLGA particles. (A) Shape change in response to temperature (T), pH, or a chemical stimuli (C). (B) Shape switching of PLGA particles. Scale bar = 5 μm. Reprinted from ref. 209 with permission from National Academy of Sciences.
pH as a stimulus resulting in particle expansion
In another example of switchable nanoparticles, Grinstaff and coworkers have shown that nanoparticles can expand several hundred-fold in volume (Fig. 12) from nanometers to micrometers in diameter, in response to a pH change.210 Importantly, these so-called “expansile nanoparticles” release their contents upon undergoing this expansion in size. The approach utilizes an initially hydrophobic nanoparticle that, upon cellular internalization and acidification within endosomes, undergoes a transition to a hydrophilic polymeric system. The hydrophilic structure formed in response to the decreased pH (~5) within the endosome undergoes expansion and pre-encapsulated drugs leak from the particle. Chemically, this was achieved by preparing the particles utilizing hydrophobic monomers in which the hydroxyl groups were masked by an acid-labile 2,4,6-trimethoxybenzaldehyde protecting group. Therefore, at neutral pH the nanoparticles are stable and do not release the encapsulant, but the drop in pH cleaves the protecting group, revealing the hydroxyl groups, and inducing a transformation from hydrophobic to hydrophilic structures.
Fig. 12.
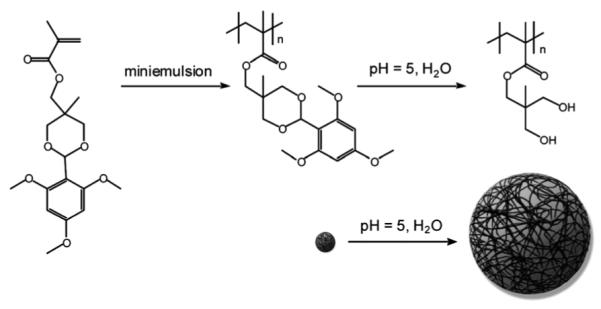
Synthesis of pH-responsive expansile nanoparticles. The nano-particles can expand in volume from nanometers to micrometers in diameter at pH 5. Reprinted with permission from A. P. Griset, J. Walpole, R. Liu, A. Gaffey, Y. L. Colson and M. W. Grinstaff, J. Am. Chem. Soc., 2009, 131, 2469–2471. Copyright 2009 American Chemical Society.
To assess the ability of expansile nanoparticles to unload a functional encapsulant, nanoparticles were loaded with paclitaxel and were evaluated for the efficiency of drug release and efficacy within an in vivo model for lung cancer. A notable degree of tumor inhibition was observed in mice injected with paclitaxel-loaded expansile nanoparticles in comparison to paclitaxel alone. pH driven degradation processes could also result in the release of encapsulated drugs. However, systems such as this offer several potential nuances within drug delivery and could impact other applications. For example, if reversible, such a system could be utilized in the uptake and release of small molecules. In the context of endosome escape, expansion could aid in release by disrupting membrane integrity. Certainly, polymers that do not degrade during the release process may aid in reducing any toxicity inherent to the carrier itself, broadening the chemical systems that may be employed for these purposes.
Enzyme responsive micelles and nanoparticles
The response of a material to a given stimulus constitutes a detection event. In the case of enzymatically responsive materials these detection events are catalytic, selective, and in some cases specific to a particular disease state of a given tissue or cell.138,211–219 Therefore, materials capable of responding in a dramatic fashion to enzymes may be applicable in catalytically-amplified in vitro schemes for enzyme detection, or in selective therapeutic and/or diagnostic delivery in vivo. Enzymes, such as proteases, kinases, phosphatases, and oxidases, recognize proteins or peptides as substrates to catalyze various reactions. Here, enzymatic reactions on abiotic substrates resulting in morphology changes and catalyzed degradation of micelles and/or nanoparticles, will be highlighted.
Enzymes as stimuli resulting in structural destabilization
Encapsulation of enzymes for the purpose of in vivo delivery within vesicles provides protection from proteases present in biological fluids and prolongs the lifetime of the enzyme by slowing the denaturation process. Furthermore, incorporation of an enzyme such as glucose oxidase (GOx) may provide controlled access to the substrate and release of the product via disruption of the barrier function of the membrane. Hubbell et al. described glucose oxidase (GOx) encapsulated within PEG–PPS–PEG ((poly(ethylene glycol))–(poly(propylene sulfide))–PEG) polymersome.220 This enzyme-loaded polymersome is permeable to glucose resulting in intravesicular formation of H2O2 upon generation of gluconic acid. Peroxide generation causes polymersome destabilization (Fig. 13) and particle destruction. This type of enzyme-amplified approach to particle degradation may have utility in drug delivery and the detection of biological analytes.
Fig. 13.
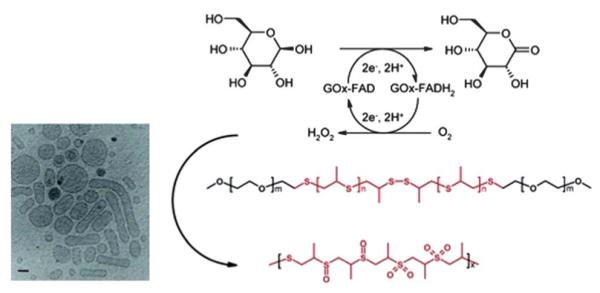
GOx-catalyzed oxidation of d-glucose to gluconolactone (gluconic acid), producing H2O2 in the presence of oxygen, resulting in membrane disruption from the inside out. A cryo-TEM micrograph shows the unilamellar structure of the enzyme-loaded polymersomes. Scale bar = 100 nm. Reprinted with permission from Langmuir, 2004, 20, 3487–3491. Copyright 2004 American Chemical Society.
Enzyme-driven assembly of nanoparticles and micelles
The majority of examples of enzymatically driven morphology switches utilizing polymer-based particles have been described in the previous section. In addition, a range of materials including hydrogels have been demonstrated to undergo enzyme-driven assembly processes.7 However, to our knowledge there is only one example of the direct assembly of a polymeric nanoparticle or micelle in response to an enzymatic stimulus. In their recent study, Hawker and coworkers developed an enzyme-responsive system where self-assembly of purely abiotic block copolymers can be triggered in the presence of a phosphatase under physiological conditions.23 This approach utilizes a water-soluble block polymer consisting of monomers containing phosphate moieties and a PEG polymer. The phosphate moieties are cleaved upon addition of acid phosphatase (APase) generating an amphiphilic block copolymer in situ (Fig. 14). Subsequently, the polymers aggregate to generate well-defined polymeric micelles.
Fig. 14.
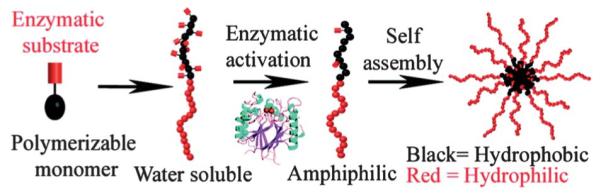
Schematic representation of the enzymatic activation of a water-soluble copolymer to give an amphiphilic copolymer and subsequent self-assembly into colloidal nanostructures. Reprinted with permission from J. Am. Chem. Soc., 2009, 131, 13949–13951. Copyright 2009 American Chemical Society.
Redox-responsive micelles and nanoparticles
Oxidation and reduction (redox) reactions can be utilized as a stimulus to trigger the release of encapsulated materials from nanocarriers, or to induce the phase transition of nano-particles.220–223 Oxidative environments exist physiologically in extracellular fluids and pathophysiologically in inflamed or tumor tissues where high concentrations of oxidants including superoxide and hydrogen peroxide can cause damage to DNA and proteins contributing to the carcinogenesis process.224–226 In addition, reductive environments within the cell, specifically the endosome, can be exploited as a trigger to destabilize nanocarriers and release drugs. The destabilization of these vehicles in the early endosome prior to lysosomal fusion can prevent drugs traveling to harsh lysosomal environments instead of escaping the endosome. The following section will present an overview of redox-responsive materials as diagnostic and/or drug delivery tools.
Oxidation as a stimulus resulting in a morphology change
Recently, Hubbell and coworkers developed a system for tuning the amphiphilicity of a polymer by oxidation.222 The system utilizes a tunable amphiphilic ABA triblock copolymer where the B block contains a hydrophobic poly(propylene sulphide) (PPS) block flanked by two hydrophilic PEG blocks (A block). The polymeric material formed an initial vesicular structure (Fig. 15). After addition of a chemical stimulus, such as H2O2, the hydrophobic PPS block is oxidized to a sulfone, resulting in a morphology switch from vesicular to cylindrical.
Fig. 15.
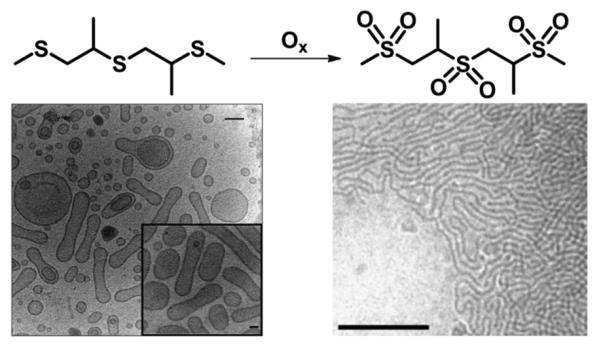
Oxidation-responsive vesicles. Through oxidation by hydrogen peroxide, the vesicles formed by an ABA triblock copolymer PEG–PPS–PEG can be transformed into cylindrical micelles. Left image, scale bar = 100 nm; inset scale bar = 50 nm; right image, scale bar = 250 nm. Modified figure from ref. 222. Copyright 2004 Nature Publishing Group.
Reduction as a stimulus resulting in structural destabilization
Reductive environments within the endosome can be exploited as a trigger to destabilize nano- and microcarriers, ultimately resulting in the release of guest molecules into the early endosome, prior to lysosomal fusion. Hubbell and coworkers developed a vesicle-forming polymeric amphiphile that can undergo a vesicular burst within a few minutes of entering the early endosome during endocytosis.220,221,223 The material is triggered by a reductive stimulus, providing an alternative route to controlling drug release under physiologically relevant conditions. Similar to the oxidation-induced morphology switch previously mentioned, the design of the block copolymer uses a PEG and PPS block copolymer linked by a reduction-sensitive disulfide bond (PEG17–SS–PPS30).223 Vesicular disruption is triggered within the endosome whereby an influx of cysteine leads to a reducing environment and subsequent cleavage of the disulfide bond. The structure and function of the encapsulated polymersome is outlined in Fig. 16. Also shown is the reduction-sensitive intracellular release of a guest fluorescent molecule, Calcein, from the polymersome in vitro (Fig. 16A) as compared to a stimuli-resistant polymersome (Fig. 16B). This rapid vesicular burst occurs in the presence of the reducing environment in the endosome prior to lysosomal fusion.
Fig. 16.
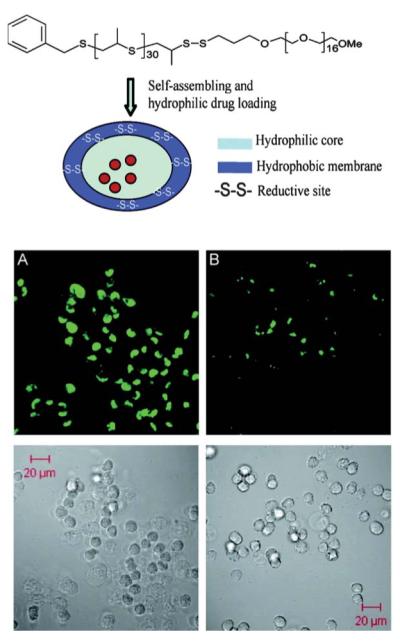
Structure and formation of reduction-sensitive polymers (top). (A) In vitro experiment of intracellular release of the quenched fluorophore, Calcein, from the reduction-sensitive polymersomes, after 2 h. (B) Initial release from reduction-insensitive polymersomes, after 2 h. Reprinted with permission from S. Cerritelli, D. Velluto and J. A. Hubbell, Biomacromolecules, 2007, 8, 1966–1972. Copyright 2007 American Chemical Society.
Protein-responsive micelles and nanoparticles
Protein recognition is a process by which protein macromolecules interact with each other or with smaller ligands to form a specific complex. Protein recognition differentiates itself from other types of interactions because it is able to distinguish highly specific from less specific binding. For example, signaling pathways, enzyme regulation and the immune response all involve processes that are based on selective interactions between proteins involved in cellular responses. In this section, protein recognition as a trigger for selective micelle disassembly and liposomal clearance will be discussed.
Protein recognition as an exogenously added stimulus to an in vivo system
The ability to predict and/or control the pharmacokinetics of nanomaterials is critical when moving towards in vivo applications. With this in mind, Mulder and coworkers reported an avidin protein as a trigger, administered as a chase following prior injection of biotinylated liposomes, to change their pharmacokinetic profile and to clear them from circulation.38 The biotinylated liposome is coated with PEG to increase blood circulation in addition to carrying paramagnetic and fluorescent agents to create a bimodal liposome. This work demonstrated that the clearance properties of avidin can be used to rapidly clear long circulating PEG coated, biotinylated liposomes from blood circulation. Fig. 17 shows the biotinylated paramagnetic liposomes treated with avidin as prepared for electron microscopy, in vitro. This ability to rapidly clear circulating contrast agents opens up exciting possibilities to study targeting kinetics, and for the potential to increase the specificity of molecular MRI while enabling in vivo control over pharmacokinetics.
Fig. 17.
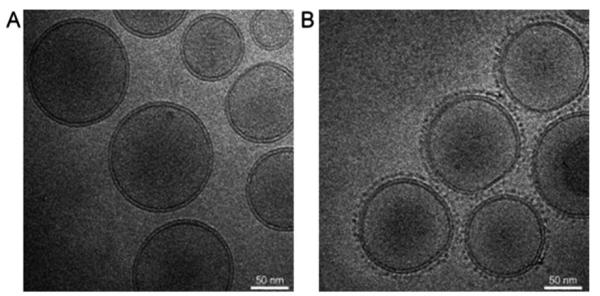
Cryo-TEM images of biotinylated paramagnetic liposomes (A) without avidin and (B) after overnight incubation with 2 μM avidin. The surface structure of liposomes was changed (B) after addition of avidin compared to the original liposomes (A). Reprinted from ref. 38. Copyright 2008 Wiley-Liss, Inc.
Protein recognition as a stimulus resulting in supramolecular assembly and disassembly
Protein–ligand recognition can be used as a trigger to release encapsulants from micelles and vesicles. These supramolecular assemblies of surfactant molecules are of interest because of their ability to selectively contain and in some cases release guest molecules. Thayumanavan and coworkers reported a supramolecular assembly in which an amphiphilic structure was obtained through noncovalent interactions.39 In this work, they developed a method for detecting analytes using protein recognition to induce the disassembly of the supramolecular structure. The design of the material allows polyelectrolytes to interact with complementary small molecule surfactants. The combination can provide supramolecular assemblies with apolar interiors that can sequester hydrophobic guest molecules, i.e. fluorescent molecules, in water. Globular proteins, such as β-glucosidase (β-Glu), are known to effectively bind polyelectrolytes and have the potential to release guest molecules due to protein recognition and binding (Fig. 18). In this study, the authors utilized the polymer poly(potassium acrylate) (PPA) and the surfactant cetyltrimethylammonium bromide (CTAB) to form a noncovalent amphiphilic micelle. The micelle was loaded with pyrene and released through disassembly of the micelle upon addition of β-Glu. Also, by changing the structures of the polymer and the surfactant, different responses to various proteins could be achieved. The simplicity of the design utilizing noncovalent interactions of the receptor assembly makes this approach highly versatile. It would be intriguing to see if this type of responsive system could be translated into a material for in vivo use taking advantage of specific, overexpressed cell-surface receptors.
Fig. 18.
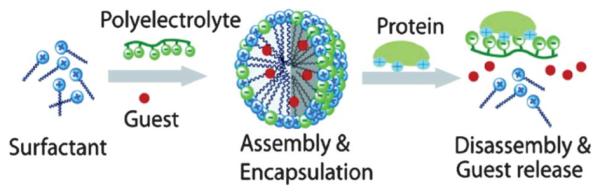
Schematic of the assembly and disassembly of particles upon protein binding. Reprinted with permission from E. N. Savariar, S. Ghosh, D. C. Gonzalez and S. Thayumanavan, J. Am. Chem. Soc., 2008, 130, 5416–5417. Copyright 2008 American Chemical Society.
Temperature-responsive micelles and nanoparticles
As is clear from the myriad of examples described so far, there are a variety of stimuli that can act as a trigger for stimuli-responsive nanomaterials. The development of temperature-responsive materials in the context of biohybrid systems was described in the previous section. Here, we focus on several examples that utilize entirely synthetic polymers. It has been reported that tumor tissues tend to be hyperthermic relative to normal tissues to various degrees in the narrow range of 0.5 to 2 °C.117 Developing materials that undergo well-defined changes in morphology over such a narrow temperature range is not a trivial problem. However, in order to successfully use temperature-responsive materials in a controlled manner, mild hyperthermia may be induced locally utilizing particles that respond to an external stimulus such as a magnetic field to selectively elevate the temperature. This can lead to a phase transition of various materials, which has been demonstrated in several cases including polymer-coated inorganic particles and inorganic micro-hyperthermic systems.120–122 Here, temperature-responsive polymeric materials with thermal phase transitions at various temperatures will be discussed.
Temperature as a stimulus resulting in a morphology change
There are several abiotic polymeric particle based temperature switches that result in morphology transition in the literature.19,227–231 Some of these are amenable to responses in biologically relevant applications and temperature ranges, possibly coupled with some mechanism for inducing local hyperthermic conditions. For example, O'Reilly and coworkers have demonstrated a system capable of a reversible, thermally-induced morphology transition between micelles and vesicles.19 This system utilized a thermoresponsive polymer block, poly(N-isopropyl acrylamide) or PNIPAM, located between a hydrophobic block, poly(tert-butylacrylate) or PtBuA and a hydrophilic, charged quaternary amino group. This copolymer was dissolved in tetrahydrofuran (THF) followed by dialysis against water to form micellar particles. These micelles were then heated to 65 °C (above LCST) in order to switch the morphology to form vesicles. Spherical micelles were able to reform upon cooling back to 25 °C (Fig. 19).
Fig. 19.
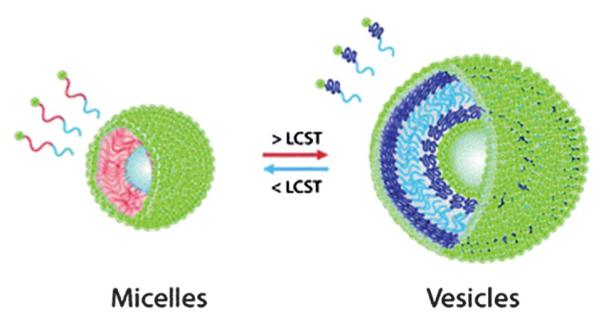
Schematic representation of a thermally-induced micelle-tovesicle morphology transition. Reproduced from ref. 19.
In another example, the Sheiko group has demonstrated a reversible, thermoresponsive polymeric material capable of morphology transitions between cylinders, vesicles and spheres (Fig. 20).227 In this example, at 25 °C, a sample of polystyrene-b-polyisoprene diblock micelles with a polystyrene block of 20.6 kDa and a polyisoprene block of 6 kDa, was observed to form cylindrical micelles that undergo a temperature-sensitive morphology transition to spherical micelles upon heating to 35 °C. Furthermore, cylindrical micelles were formed when the spherical structures were cooled back to 25 °C. In addition, a reversible transition from vesicles to cylindrical micelles, upon heating from 25 to 40 °C, was observed for a second diblock sample with the same polystyrene block (20.6 kDa) and a shorter polyisoprene block of 4.3 kDa. This process is possible due to the increase of temperature, which causes an increase in the solubility of the polyisoprene block concomitant with a decrease in the surface energy of the polystyrene core. Although these materials were designed for use in a hydrophobic solvent, this concept should be amenable to adaption for aqueous environments over this same temperature range. In contrast to the use of tunable amphiphilicity to control particle morphology, another example of a temperature-responsive material has been described in the context of multi-stimuli responsive polymeric nanoparticles described by Mitragotri and coworkers (Fig. 11). It is intriguing to consider the possible application of materials capable of sensing biologically relevant temperature changes and responding by a very well-defined shape transition or switch in surface chemistry.
Fig. 20.
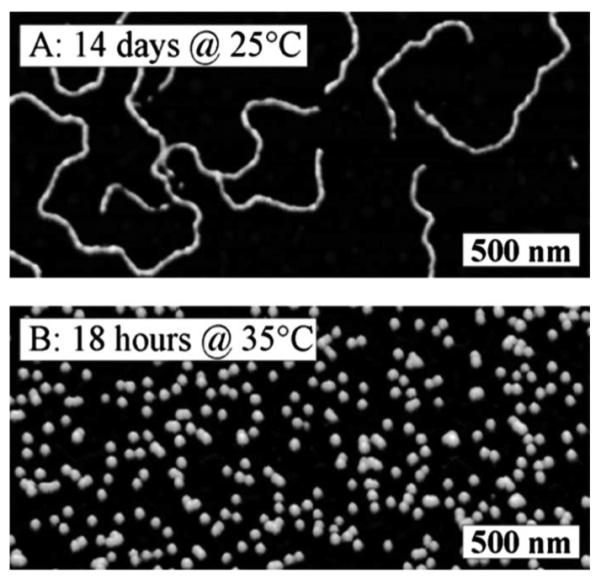
AFM height micrographs of PS-b-PI micelles. (A) After a solution of sample had been equilibrated at 25 °C for 2 weeks. (B) after 18 h at 35 °C. Reprinted with permission from I. LaRue, M. Adam, M. Pitsikalis, N. Hadjichristidis, M. Rubinstein and S. S. Sheiko, Macromolecules, 2006, 39, 309–314. Copyright 2006 American Chemical Society.
Potential for the use of small molecules as selective triggers for tunable morphology
Examples described in the first section of this review focused on the utility of nucleic acids and their propensity for selective hydrogen-bonding driven recognition as programming elements. In an analogous fashion, several interesting systems have been described that make use of selective hydrogen bonding elements incorporated into synthetic polymers, to control particle morphology.232 In particular, examples include approaches whereby diaminopyridine (DAP) and thymine (THY) have been used as hydrogen bonding motifs responsive to small molecules and polymers.21,233–235 Rotello and coworkers report a DAP-functionalized polystyrene able to self-assemble into a micrometer-scale spherical aggregate.235 Upon addition of a THY-functionalized polymer, the spherical structure undergoes a phase transition into a vesicular structure. These newly formed vesicles can be reversibly transitioned back to a spherical structure after addition of a DAP-functionalized polymer.
In addition, the disassembly of a structure upon small molecule recognition via selective H-bonding has been studied. For example, Sleiman and coworkers have shown that synthesis of an ABC triblock ROMP copolymer containing both a DAP and a THY block is able to undergo complete destruction upon the addition of a THY-containing small guest molecule.21 However, addition of a DAP-containing guest molecule does not destroy the micelle but instead results in a new smaller micellar aggregate with a different selectivity profile.
A further expansion of this method is the inclusion of a polymerizable unit on a small molecule stimulus. The new hydrogen bonding interactions can be used as a template for polymerization with high molecular weight and low polydispersity. As shown by Sleiman and coworkers (Fig. 21) a DAP containing ROMP copolymer will form spherical aggregates, however addition of a THY monomeric unit will disrupt the colloidal structure.234 The DAP polymer is now capable of acting as a template for the polymerizaton of the THY monomer due to the newly formed DAP–THY hydrogen bonded systems. In contrast, addition of an adenine monomer will not break up the DAP polymeric aggregate and will be unable to undergo a nucleobase-templated polymerization.
Fig. 21.
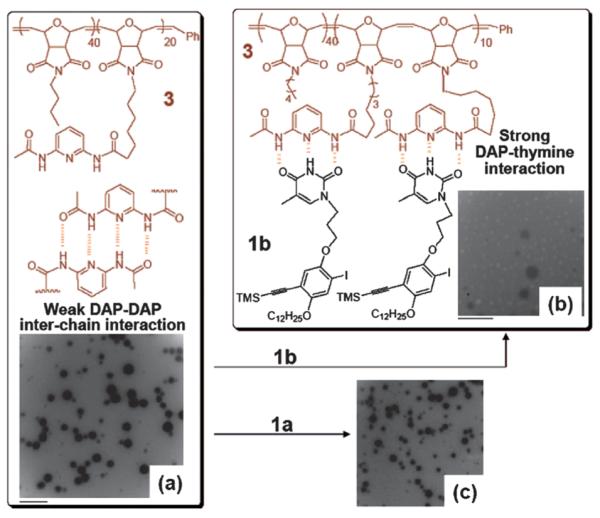
TEM images of (a) template 3, (b) 3 with 1b, and (c) 3 with noncomplementary 1a (adenine unit) in CHCl3. Scale bar is 500 nm with an average colloidal aggregate of 240 nm. Reprinted with permission from P. K. Lo and H. F. Sleiman, J. Am. Chem. Soc., 2009, 131, 4182–4183. Copyright 2009 American Chemical Society.
These abiotic polymeric systems utilize biomolecule-like recognition elements akin to those used in DNA- and peptide-based materials described in the first section of this review. Certainly, an intriguing but as yet unexplored mechanism for modulating polymeric nanoparticles and micelles with small molecules is the use of small molecule nucleic acid interactions via aptameric-type recognition. Aptamers are DNA or RNA molecules that are selected through in vitro evolution for their ability to selectively bind other molecules. To date, DNA-only nanostructures have been described that incorporate aptamer recognition, however this has not been translated into DNA-polymeric systems.236–239 Moreover, simpler approaches utilizing interchelators and/or metal complexes for binding DNA have yet to be explored and should be adaptable for selective manipulation of such materials. Given the multiple binding modes, and selective reactions possible with nucleic acids and peptides, small molecules should find increasing utility in the manipulation of nanoparticles and micelles. Certainly, such materials may enable the development of nanoparticle probes capable of recognizing specific metabolites and/or exogenously added small molecules including drugs.
Perspectives and conclusion
There is a growing body of work describing materials capable of interfacing with biological systems by responding to relevant biochemical signals in a stimuli-responsive manner. Certainly, the development of nanomaterials capable of responding in a predictable fashion to biological stimuli is of increasing interest for a range of in vivo, biomedical applications. This type of approach is complementary to efforts to develop probes that can be induced to switch ON, or OFF in response to externally added stimuli including light and ultrasound. From a purely basic science perspective, the development of preprogrammed, autonomous materials is a significant challenge, and is one that must be addressed if we aim to utilize truly smart, targeted systems in complex environments.
Tremendous challenges face researchers aiming to utilize nanoparticles for in vivo applications, the most important of which is the development of materials capable of selective targeting and possessing the required stability and degradability in complex biological milieu. These are obviously desirable capabilities, but it is less obvious how this can be achieved in a rational fashion, with all the required properties built into a single delivery vehicle. However, stimuli-responsive materials such as expanding nanoparticles,210 offer new routes to materials that function in clever, and novel ways to achieve multiple goals from efficient tissue targeting to effective endosome release. There is little doubt that the need to interface man-made synthetic nanomaterials with biological stimuli for biomedical applications, and the great potential for autonomously responsive and adaptive materials to impact a range of applications, will drive this field towards ever more complex and interesting chemical systems. More importantly, many of these approaches are beginning to yield true value as they become functional.
Acknowledgements
The authors acknowledge support from NIH through 1R01EB011633-01, and via a Director's New Innovator Award, 1DP2OD008724-01. Furthermore, we acknowledge the generous support from AFOSR through a PECASE (FA9550-11-1-0105) and the support of the ARO (W911NF-11-1-0264). N. C. G. acknowledges the Henry & Camille Dreyfus Foundation for a New Faculty Award.
References
- 1.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 2.Gothelf KV, LaBean TH. Org. Biomol. Chem. 2005;3:4023–4037. doi: 10.1039/b510551j. [DOI] [PubMed] [Google Scholar]
- 3.Seeman NC. Mol. Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alemdaroglu FE, Herrmann A. Org. Biomol. Chem. 2007;5:1311–1320. doi: 10.1039/b617941j. [DOI] [PubMed] [Google Scholar]
- 5.Aldaye Faisal A, Palmer Alison L, Sleiman Hanadi F. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 6.Turberfield AJ, Mitchell JC, Yurke B, Mills AP, Jr., Blakey MI, Simmel FC. Phys. Rev. Lett. 2003;90:118102. doi: 10.1103/PhysRevLett.90.118102. [DOI] [PubMed] [Google Scholar]
- 7.Hahn ME, Gianneschi NC. Chem. Commun. 2011:11814–11821. doi: 10.1039/c1cc15220c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frezza BM, Cockroft SL, Ghadiri MR. J. Am. Chem. Soc. 2007;129:14875–14879. doi: 10.1021/ja0710149. [DOI] [PubMed] [Google Scholar]
- 9.Ashkenasy G, Ghadiri MR. J. Am. Chem. Soc. 2004;126:11140–11141. doi: 10.1021/ja046745c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Xu H, Zhang X. Adv. Mater. 2009;21:1–16. doi: 10.1002/adma.200901237. [DOI] [PubMed] [Google Scholar]
- 11.Buetuen V, Liu S, Weaver JVM, Bories-Azeau X, Cai Y, Armes SP. React. Funct. Polym. 2006;66:157–165. [Google Scholar]
- 12.Discher DE, Eisenberg A. Science. 2002;297:967–973. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- 13.Hawker CJ, Wooley KL. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]
- 14.Smart T, Lomas H, Massignani M, Flores-Merino MV, Perez LR, Battaglia G. Nano Today. 2008;3:38–46. [Google Scholar]
- 15.Jain S, Bates FS. Science. 2003;300:460–464. doi: 10.1126/science.1082193. [DOI] [PubMed] [Google Scholar]
- 16.Fuks G, Mayap Talom R, Gauffre F. Chem. Soc. Rev. 2011;40:2475–2493. doi: 10.1039/c0cs00085j. [DOI] [PubMed] [Google Scholar]
- 17.Lee NS, Lin LY, Neumann WL, Freskos JN, Karwa A, Shieh JJ, Dorshow RB, Wooley KL. Small. 2011;7:1998–2003. doi: 10.1002/smll.201100567. [DOI] [PubMed] [Google Scholar]
- 18.Versluis F, Tomatsu I, Kehr S, Fregonese C, Tepper AWJW, Stuart MCA, Ravoo BJ, Koning RI, Kros A. J. Am. Chem. Soc. 2009;131:13186–13187. doi: 10.1021/ja9026264. [DOI] [PubMed] [Google Scholar]
- 19.Moughton AO, O'Reilly RK. Chem. Commun. 2010;46:1091–1093. doi: 10.1039/b922289h. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Yu K, Eisenberg A. Science. 1996;272:1777–1779. doi: 10.1126/science.272.5269.1777. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara Y, Bazzi HS, Toader V, Godin F, Sleiman HF. Chem.–Eur. J. 2007;13:4560–4570. doi: 10.1002/chem.200601423. [DOI] [PubMed] [Google Scholar]
- 22.Ku TH, Chien MP, Thompson MP, Sinkovits RS, Olson NH, Baker TS, Gianneschi NC. J. Am. Chem. Soc. 2011;133:8392–8395. doi: 10.1021/ja2004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amir RJ, Zhong S, Pochan DJ, Hawker CJ. J. Am. Chem. Soc. 2009;131:13949–13951. doi: 10.1021/ja9060917. [DOI] [PubMed] [Google Scholar]
- 24.Zou J, Tao F, Jiang M. Langmuir. 2007;23:12791–12794. doi: 10.1021/la702815h. [DOI] [PubMed] [Google Scholar]
- 25.Fomina N, McFearin C, Sermsakdi M, Edigin O, Almutairi A. J. Am. Chem. Soc. 2010;132:9540–9542. doi: 10.1021/ja102595j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C-L, Zhang P, Rosi NL. J. Am. Chem. Soc. 2008;130:13555–13557. doi: 10.1021/ja805683r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-Romero P. Adv. Mater. 2001;13:163–174. [Google Scholar]
- 28.Storhoff JJ, Mirkin CA. Chem. Rev. 1999;99:1849–1862. doi: 10.1021/cr970071p. [DOI] [PubMed] [Google Scholar]
- 29.Ofir Y, Samanta B, Rotello VM. Chem. Soc. Rev. 2008;37:1814–1825. doi: 10.1039/b712689c. [DOI] [PubMed] [Google Scholar]
- 30.Chécot F, Rodríguez-Hernáandez J, Gnanou Y, Lecommandoux S. Biomol. Eng. 2007;24:81–85. doi: 10.1016/j.bioeng.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Tu Y-L, Wang C-C, Chen C-Y. J. Polym. Sci., Part A: Polym. Chem. 2011;49:2866–2877. [Google Scholar]
- 32.Carrick LM, Aggeli A, Boden N, Fisher J, Ingham E, Waigh TA. Tetrahedron. 2007;63:7457–7467. [Google Scholar]
- 33.Reguera J, Urry DW, Parker TM, McPherson DT, Rodriguez-Cabello JC. Biomacromolecules. 2007;8:354–358. doi: 10.1021/bm060936l. [DOI] [PubMed] [Google Scholar]
- 34.Cho Y, Zhang Y, Christensen T, Sagle LB, Chilkoti A, Cremer PS. J. Phys. Chem. B. 2008;112:13765–13771. doi: 10.1021/jp8062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haghpanah JS, Yuvienco C, Roth EW, Liang A, Tu RS, Montclare JK. Mol. BioSyst. 2010;6:1662–1667. doi: 10.1039/c002353a. [DOI] [PubMed] [Google Scholar]
- 36.Peppas NA, Huang Y. Pharm. Res. 2002;19:578–587. doi: 10.1023/a:1015389609344. [DOI] [PubMed] [Google Scholar]
- 37.Shi H, Tsai W-B, Garrison MD, Ferrari S, Ratner BD. Nature. 1999;398:593–597. doi: 10.1038/19267. [DOI] [PubMed] [Google Scholar]
- 38.van Tilborg GA, Strijkers GJ, Pouget EM, Reutelingsperger CP, Sommerdijk NA, Nicolay K, Mulder WJ. Magn. Reson. Med. 2008;60:1444–1456. doi: 10.1002/mrm.21780. [DOI] [PubMed] [Google Scholar]
- 39.Savariar EN, Ghosh S, González DC, Thayumanavan S. J. Am. Chem. Soc. 2008;130:5416–5417. doi: 10.1021/ja800164z. [DOI] [PubMed] [Google Scholar]
- 40.Mao Y, Luo C, Deng W, Jin G, Yu X, Zhang Z, Ouyang Q, Chen R, Yu D. Nucleic Acids Res. 2004;32:e144. doi: 10.1093/nar/gnh145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chien MP, Rush A, Thompson M, Gianneschi N. Angew. Chem., Int. Ed. 2010;49:5076–5080. doi: 10.1002/anie.201000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding K, Alemdaroglu F, Börsch M, Berger R, Herrmann A. Angew. Chem., Int. Ed. 2007;46:1172–1175. doi: 10.1002/anie.200603064. [DOI] [PubMed] [Google Scholar]
- 43.Kim J-H, Lee S, Park K, Nam HY, Jang SY, Youn I, Kim K, Jeon H, Park R-W, Kim I-S, Choi K, Kwon IC. Angew. Chem., Int. Ed. 2007;46:5779–5782. doi: 10.1002/anie.200700767. [DOI] [PubMed] [Google Scholar]
- 44.Azagarsamy MA, Sokkalingam P, Thayumanavan S. J. Am. Chem. Soc. 2009;131:14184–14185. doi: 10.1021/ja906162u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dongen SFM, de Hoog H-PM, Peters RJRW, Nallani M, Nolte RJM, van Hest JCM. Chem. Rev. 2009;109:6212–6274. doi: 10.1021/cr900072y. [DOI] [PubMed] [Google Scholar]
- 46.Farokhzad OC, Jon S, Khademhosseini A, Tran T-NT, LaVan DA, Langer R. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 47.Thomson K, Amin I, Morales E, Winters-Hilt S. BMC Bioinformatics. 2007;8:S11. doi: 10.1186/1471-2105-8-S7-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi T, Matile S. J. Am. Chem. Soc. 2009;131:18048–18049. doi: 10.1021/ja908909m. [DOI] [PubMed] [Google Scholar]
- 49.Brody EN, Gold L. J. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 50.Persil O, Hud NV. Trends Biotechnol. 2007;25:433–436. doi: 10.1016/j.tibtech.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Dervan PB. Science. 1986;232:464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- 52.Baum DA, Silverman SK. Cell. Mol. Life Sci. 2008;65:2156–2174. doi: 10.1007/s00018-008-8029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaschke A, Seelig B. Curr. Opin. Chem. Biol. 2000;4:257–262. doi: 10.1016/s1367-5931(00)00086-7. [DOI] [PubMed] [Google Scholar]
- 54.Jaschke A. Curr. Opin. Struct. Biol. 2001;11:321–326. doi: 10.1016/s0959-440x(00)00208-6. [DOI] [PubMed] [Google Scholar]
- 55.Santoro SW, Joyce GF. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breaker RR, Joyce GF. Chem. Biol. 1995;2:655–660. doi: 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 57.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 58.Aldaye FA, Palmer AL, Sleiman HF. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 59.Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. Nano Lett. 2009;9:2747–2750. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]
- 60.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 61.Zhang C, Su M, He Y, Zhao X, Fang P.-a., Ribbe AE, Jiang W, Mao C. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10665–10669. doi: 10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dirks RM, Lin M, Winfree E, Pierce NA. Nucleic Acids Res. 2004;32:1392–1403. doi: 10.1093/nar/gkh291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 64.Bertin PA, Gibbs JM, Shen CK-F, Thaxton CS, Russin WA, Mirkin CA, Nguyen ST. J. Am. Chem. Soc. 2006;128:4168–4169. doi: 10.1021/ja056378k. [DOI] [PubMed] [Google Scholar]
- 65.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feldkamp U, Sacca B, Niemeyer CM. Angew. Chem., Int. Ed. 2009;48:5996–6000. doi: 10.1002/anie.200902285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 68.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr., Schultz PG. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 69.Aldaye FA, Sleiman HF. J. Am. Chem. Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 70.Prigodich AE, Alhasan AH, Mirkin CA. J. Am. Chem. Soc. 2011;133:2120–2123. doi: 10.1021/ja110833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z, Zhang Y, Fullhart P, Mirkin CA. Nano Lett. 2004;4:1055–1058. [Google Scholar]
- 72.Chen X-J, Sanchez-Gaytan BL, Hayik SEN, Fryd M, Wayland BB, Park S-J. Small. 2010;6:2256–2260. doi: 10.1002/smll.201001185. [DOI] [PubMed] [Google Scholar]
- 73.Alemdaroglu FE, Alemdaroglu NC, Langguth P, Herrmann A. Macromol. Rapid Commun. 2008;29:326–329. [Google Scholar]
- 74.Johnston A, Caruso F. Angew. Chem., Int. Ed. 2007;46:2677–2680. doi: 10.1002/anie.200605135. [DOI] [PubMed] [Google Scholar]
- 75.Johnston APR, Lee L, Wang Y, Caruso F. Small. 2009;5:1418–1421. doi: 10.1002/smll.200900075. [DOI] [PubMed] [Google Scholar]
- 76.Thompson MP, Chien MP, Ku TH, Rush AM, Gianneschi NC. Nano Lett. 2010;10:2690–2693. doi: 10.1021/nl101640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chien MP, Gianneschi NC. Small. 2011;7:2041–2046. doi: 10.1002/smll.201101014. [DOI] [PubMed] [Google Scholar]
- 78.Alemdaroglu FE, Wang J, Boersch M, Berger R, Herrmann A. Angew. Chem., Int. Ed. 2008;47:974–976. doi: 10.1002/anie.200703466. [DOI] [PubMed] [Google Scholar]
- 79.He F, Liu L, Li L. Adv. Funct. Mater. 2011;21:3143–3149. [Google Scholar]
- 80.Fowler JD, Suo Z. Chem. Rev. 2006;106:2092–2110. doi: 10.1021/cr040445w. [DOI] [PubMed] [Google Scholar]
- 81.Wang J, Alemdaroglu FE, Prusty DK, Herrmann A, Berger R. Macromolecules. 2008;41:2914–2919. [Google Scholar]
- 82.Chien M-P, Thompson MP, Gianneschi NC. Chem. Commun. 47:167–169. doi: 10.1039/c0cc02291h. [DOI] [PubMed] [Google Scholar]
- 83.Azzazy HME, Highsmith WE. Clin. Biochem. 2002;35:425–445. doi: 10.1016/s0009-9120(02)00343-0. [DOI] [PubMed] [Google Scholar]
- 84.Bratkovic T. Cell. Mol. Life Sci. 2010;67:749–767. doi: 10.1007/s00018-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duvshani-Eshet M, Keren H, Oz S, Radzishevsky IS, Mor A, Machluf M. J. Gene Med. 2008;10:1150–1159. doi: 10.1002/jgm.1235. [DOI] [PubMed] [Google Scholar]
- 86.Ruoslahti E. BioMEMS Biomed. Nanotechnol. 2006;3:127–136. [Google Scholar]
- 87.Boonen K, Creemers JW, Schoofs L. BioEssays. 2009;31:300–314. doi: 10.1002/bies.200800055. [DOI] [PubMed] [Google Scholar]
- 88.Molkentin JD. J. Clin. Invest. 2003;111:1275–1277. doi: 10.1172/JCI18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawamura K, Oishi J, Kang J-H, Kodama K, Sonoda T, Murata M, Niidome T, Katayama Y. Biomacromolecules. 2004;6:908–913. doi: 10.1021/bm0493887. [DOI] [PubMed] [Google Scholar]
- 90.Huang J, Ding J. Soft Matter. 2010;6:3395–3401. [Google Scholar]
- 91.Ruoslahti E. Annu. Rev. Cell Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 92.Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galande AK, Hilderbrand SA, Weissleder R, Tung C-H. J. Med. Chem. 2006;49:4715–4720. doi: 10.1021/jm051001a. [DOI] [PubMed] [Google Scholar]
- 94.Harris TJ, von Maltzahn G, Derfus AM, Ruoslahti E, Bhatia SN. Angew. Chem., Int. Ed. 2006;45:3161–3165. doi: 10.1002/anie.200600259. [DOI] [PubMed] [Google Scholar]
- 95.Von Maltzahn G, Harris TJ, Park J-H, Min D-H, Schmidt AJ, Sailor MJ, Bhatia SN. J. Am. Chem. Soc. 2007;129:6064–6065. doi: 10.1021/ja070461l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bull SR, Guler MO, Bras RE, Meade TJ, Stupp SI. Nano Lett. 2005;5:1–4. doi: 10.1021/nl0484898. [DOI] [PubMed] [Google Scholar]
- 97.Cui H, Webber MJ, Stupp SI. Biopolymers: Peptide Science. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palmer LC, Stupp SI. Acc. Chem. Res. 2008;41:1674–1684. doi: 10.1021/ar8000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chécot F, Lecommandoux S, Gnanou Y, Klok HA. Angew. Chem., Int. Ed. 2002;41:1339–1343. doi: 10.1002/1521-3773(20020415)41:8<1339::aid-anie1339>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 100.Checot F, Brulet A, Oberdisse J, Gnanou Y, Mondain-Monval O, Lecommandoux S. Langmuir. 2005;21:4308–4315. doi: 10.1021/la0468500. [DOI] [PubMed] [Google Scholar]
- 101.Lavignac N, Lazenby M, Franchini J, Ferruti P, Duncan R. Int. J. Pharm. 2005;300:102–112. doi: 10.1016/j.ijpharm.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 102.Rodríguez-Hernández J, Chécot F, Gnanou Y, Lecommandoux S. Prog. Polym. Sci. 2005;30:691–724. [Google Scholar]
- 103.Sutthasupa S, Shiotsuki M, Matsuoka H, Masuda T, Sanda F. Macromolecules. 2010;43:1815–1822. [Google Scholar]
- 104.Wu XL, Kim JH, Koo H, Bae SM, Shin H, Kim MS, Lee B-H, Park R-W, Kim I-S, Choi K, Kwon IC, Kim K, Lee DS. Bioconjugate Chem. 2010;21:208–213. doi: 10.1021/bc9005283. [DOI] [PubMed] [Google Scholar]
- 105.Wang C, Wang G, Wang Z, Zhang X. Chem.–Eur. J. 2011;17:3322–3325. doi: 10.1002/chem.201003502. [DOI] [PubMed] [Google Scholar]
- 106.Engin K, Leeper DB, Cater JR, Thistlethwaite AJ, Tupchong L, McFarlane JD. Int. J. Hyperthermia. 1995;11:211–216. doi: 10.3109/02656739509022457. [DOI] [PubMed] [Google Scholar]
- 107.Gerweck LE, Seetharaman K. Cancer Res. 1996;56:1194–1198. [PubMed] [Google Scholar]
- 108.Ojugo ASE, McSheehy PMJ, McIntyre DJO, McCoy C, Stubbs M, Leach MO, Judson IR, Griffiths JR. NMR Biomed. 1999;12:495–504. doi: 10.1002/(sici)1099-1492(199912)12:8<495::aid-nbm594>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 109.Bareford LM, Swaan PW. Adv. Drug Delivery Rev. 2007;59:748–758. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kricheldorf HR. Angew. Chem., Int. Ed. 2006;45:5752–5784. doi: 10.1002/anie.200600693. [DOI] [PubMed] [Google Scholar]
- 111.Rodriguez-Hernandez J, Lecommandoux S. J. Am. Chem. Soc. 2005;127:2026–2027. doi: 10.1021/ja043920g. [DOI] [PubMed] [Google Scholar]
- 112.Chen Y, Dong C-M. J. Phys. Chem. B. 2010;114:7461–7468. doi: 10.1021/jp100399d. [DOI] [PubMed] [Google Scholar]
- 113.Agut W, Brulet A, Taton D, Lecommandoux S. Langmuir. 2007;23:11526–11533. doi: 10.1021/la701482w. [DOI] [PubMed] [Google Scholar]
- 114.Chen C, Wang Z, Li Z. Biomacromolecules. 2011;12:2859–2863. doi: 10.1021/bm200849m. [DOI] [PubMed] [Google Scholar]
- 115.Whittle IR, Stavrinos N, Akil H, Yau Y, Lewis SC. Br. J. Neurosurg. 2010;24:447–453. doi: 10.3109/02688691003746290. [DOI] [PubMed] [Google Scholar]
- 116.He Y, Shirazaki M, Liu H, Himeno R, Sun Z. Comput. Biol. Med. 2006;36:1336–1350. doi: 10.1016/j.compbiomed.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 117.Thrall DE, Page RL, Dewhirst MW, Meyer RE, Hoopes PJ, Kornegay JN. Cancer Res. 1986;46:6229–6235. [PubMed] [Google Scholar]
- 118.Park J-H, von Maltzahn G, Ong LL, Centrone A, Hatton TA, Ruoslahti E, Bhatia SN, Sailor MJ. Adv. Mater. 2010;22:880–885. doi: 10.1002/adma.200902895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Park J-H, Derfus AM, Segal E, Vecchio KS, Bhatia SN, Sailor MJ. J. Am. Chem. Soc. 2006;128:7938–7946. doi: 10.1021/ja0612854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hasan W, Stender CL, Lee MH, Nehl CL, Lee J, Odom TW. Nano Lett. 2009;9:1555–1558. doi: 10.1021/nl803647n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Singh N, Karambelkar A, Gu L, Lin K, Miller JS, Chen CS, Sailor MJ, Bhatia SN. J. Am. Chem. Soc. 2011;133:19582–19585. doi: 10.1021/ja206998x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen S, Li Y, Guo C, Wang J, Ma J, Liang X, Yang L-R, Liu H-Z. Langmuir. 2007;23:12669–12676. doi: 10.1021/la702049d. [DOI] [PubMed] [Google Scholar]
- 123.Gao W, Xu D, Lim DW, Craig SL, Chilkoti A. Polym. Chem. 2011;2:1561–1566. [Google Scholar]
- 124.MacEwan SR, Chilkoti A. Biopolymers. 2010;94:60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 125.Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A. J. Am. Chem. Soc. 2007;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim W, Thévenot J, Ibarboure E, Lecommandoux S, Chaikof E. Angew. Chem., Int. Ed. 2010;49:4257–4260. doi: 10.1002/anie.201001356. [DOI] [PubMed] [Google Scholar]
- 127.Rao J, Luo Z, Ge Z, Liu H, Liu S. Biomacromolecules. 2007;8:3871–3878. doi: 10.1021/bm700830b. [DOI] [PubMed] [Google Scholar]
- 128.He C, Zhao C, Guo X, Guo Z, Chen X, Zhuang X, Liu S, Jing X. J. Polym. Sci., Part A: Polym. Chem. 2008;46:4140–4150. [Google Scholar]
- 129.Zhao C, Zhuang X, He C, Chen X, Jing X. Macromol. Rapid Commun. 2008;29:1810–1816. [Google Scholar]
- 130.Agut W, Brulet A, Schatz C, Taton D, Lecommandoux S. Langmuir. 2010;26:10546–10554. doi: 10.1021/la1005693. [DOI] [PubMed] [Google Scholar]
- 131.Naik SS, Ray JG, Savin DA. Langmuir. 2011;27:7231–7240. doi: 10.1021/la200882f. [DOI] [PubMed] [Google Scholar]
- 132.Lee R-S, Chen W-H, Huang Y-T. Polymer. 2010;51:5942–5951. [Google Scholar]
- 133.Roy D, Cambre JN, Sumerlin BS. Chem. Commun. 2009:2106–2108. doi: 10.1039/b900374f. [DOI] [PubMed] [Google Scholar]
- 134.Schild HG. Prog. Polym. Sci. 1992;17:163–249. [Google Scholar]
- 135.Fournier D, Hoogenboom R, Thijs HML, Paulus RM, Schubert US. Macromolecules. 2007;40:915–920. [Google Scholar]
- 136.Zhang X, Li J, Li W, Zhang A. Biomacromolecules. 2007;8:3557–3567. doi: 10.1021/bm700729t. [DOI] [PubMed] [Google Scholar]
- 137.Lehman IR, Bessman MJ, Simms ES, Kornberg A. J. Biol. Chem. 1958;233:163–170. [PubMed] [Google Scholar]
- 138.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weaver Alissa M. Clin. Exp. Metastasis. 2006;23:97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- 140.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kang H, Yu J, Jung G. Biochem. J. 2008;416:47–54. doi: 10.1042/BJ20080724. [DOI] [PubMed] [Google Scholar]
- 142.Signarvic RS, DeGrado WF. J. Mol. Biol. 2003;334:1–12. doi: 10.1016/j.jmb.2003.09.041. [DOI] [PubMed] [Google Scholar]
- 143.Yang Z, Liang G, Wang L, Xu B. J. Am. Chem. Soc. 2006;128:3038–3043. doi: 10.1021/ja057412y. [DOI] [PubMed] [Google Scholar]
- 144.Lowik DWPM, Leunissen EHP, van den Heuvel M, Hansen MB, van Hest JCM. Chem. Soc. Rev. 2010;39:3394–3412. doi: 10.1039/b914342b. [DOI] [PubMed] [Google Scholar]
- 145.Berg JM, Tymoczko JL, Stryer L. Biochemistry. W.H. Freeman and Company; New York: 2007. [Google Scholar]
- 146.Castelletto V, McKendrick JE, Hamley IW, Olsson U, Cenker C. Langmuir. 2010;26:11624–11627. doi: 10.1021/la101806z. [DOI] [PubMed] [Google Scholar]
- 147.Kuhnle H, Borner HG. Angew. Chem., Int. Ed. 2009;48:6431–6434. doi: 10.1002/anie.200805768. [DOI] [PubMed] [Google Scholar]
- 148.Janek K, Behlke J, Zipper J, Fabian H, Georgalis Y, Beyermann M, Bienert M, Krause E. Biochemistry. 1999;38:8246–8252. doi: 10.1021/bi990510+. [DOI] [PubMed] [Google Scholar]
- 149.Conte LL, Chothia C, Janin J. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 150.Lee H-J, Zheng J. Cell Commun. Signaling. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stormo GD, Zhao Y. Nat. Rev. Genet. 2010;11:751–760. doi: 10.1038/nrg2845. [DOI] [PubMed] [Google Scholar]
- 152.Wilson AJ. Chem. Soc. Rev. 2009;38:3289–3300. doi: 10.1039/b807197g. [DOI] [PubMed] [Google Scholar]
- 153.Twarock R. J. Theor. Biol. 2004;226:477–482. doi: 10.1016/j.jtbi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 154.Huber V, Sengupta S, Wuerthner F. Chem.–Eur. J. 2008;14:7791–7807. doi: 10.1002/chem.200800764. [DOI] [PubMed] [Google Scholar]
- 155.Hoffman AS, Stayton PS. Prog. Polym. Sci. 2007;32:922–932. [Google Scholar]
- 156.Salgado EN, Faraone-Mennella J, Tezcan FA. J. Am. Chem. Soc. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Salgado EN, Radford RJ, Tezcan FA. Acc. Chem. Res. 2010;43:661–672. doi: 10.1021/ar900273t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Salgado EN, Lewis RA, Faraone-Mennella J, Tezcan FA. J. Am. Chem. Soc. 2008;130:6082–6084. doi: 10.1021/ja8012177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Padilla JE, Colovos C, Yeates TO. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dirks AJ, Nolte RJM, Cornelissen JJLM. Adv. Mater. 2008;20:3953–3957. [Google Scholar]
- 161.Boerakker MJ, Hannink JM, Bomans PHH, Frederik PM, Nolte RJM, Meijer EM, Sommerdijk NAJM. Angew. Chem., Int. Ed. 2002;41:4239–4241. doi: 10.1002/1521-3773(20021115)41:22<4239::AID-ANIE4239>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 162.Velonia K, Rowan AE, Nolte RJM. J. Am. Chem. Soc. 2002;124:4224–4225. doi: 10.1021/ja017809b. [DOI] [PubMed] [Google Scholar]
- 163.Wang R-M, Li G, Zhang H-F, He Y-F, He N-P, Lei Z. Polym. Adv. Technol. 2010;21:685–690. [Google Scholar]
- 164.Boyer C, Bulmus V, Liu J, Davis TP, Stenzel MH, Barner-Kowollik C. J. Am. Chem. Soc. 2007;129:7145–7154. doi: 10.1021/ja070956a. [DOI] [PubMed] [Google Scholar]
- 165.Huang X, Yin Y, Jiang X, Tang Y, Xu J, Liu J, Shen J. Macromol. Biosci. 2009;9:1202–1210. doi: 10.1002/mabi.200900208. [DOI] [PubMed] [Google Scholar]
- 166.Mugesh G, du Mont W-W, Sies H. Chem. Rev. 2001;101:2125–2180. doi: 10.1021/cr000426w. [DOI] [PubMed] [Google Scholar]
- 167.Dong Z, Liu J, Mao S, Huang X, Yang B, Ren X, Luo G, Shen J. J. Am. Chem. Soc. 2004;126:16395–16404. doi: 10.1021/ja045964v. [DOI] [PubMed] [Google Scholar]
- 168.Heredia KL, Bontempo D, Ly T, Byers JT, Halstenberg S, Maynard HD. J. Am. Chem. Soc. 2005;127:16955–16960. doi: 10.1021/ja054482w. [DOI] [PubMed] [Google Scholar]
- 169.Hong C-Y, Pan C-Y. Macromolecules. 2006;39:3517–3524. [Google Scholar]
- 170.Kulkarni S, Schilli C, Grin B, Muller AHE, Hoffman AS, Stayton PS. Biomacromolecules. 2006;7:2736–2741. doi: 10.1021/bm060186f. [DOI] [PubMed] [Google Scholar]
- 171.Kushner AM, Vossler JD, Williams GA, Guan Z. J. Am. Chem. Soc. 2009;131:8766–8768. doi: 10.1021/ja9009666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pelah A, Bharde A, Jovin TM. Soft Matter. 2009;5:1006–1010. [Google Scholar]
- 173.Li M, Li H, De P, Sumerlin BS. Macromol. Rapid Commun. 2011;32:354–359. doi: 10.1002/marc.201000619. [DOI] [PubMed] [Google Scholar]
- 174.Morimoto N, Ogino N, Narita T, Kitamura S, Akiyoshi K. J. Am. Chem. Soc. 2006;129:458–459. doi: 10.1021/ja065966a. [DOI] [PubMed] [Google Scholar]
- 175.Lee H. i., Wu W, Oh J, Mueller L, Sherwood G, Peteanu L, Kowalewski T, Matyjaszewski K. Angew. Chem., Int. Ed. 2007;46:2453–2457. doi: 10.1002/anie.200604278. [DOI] [PubMed] [Google Scholar]
- 176.Vlassiouk I, Park C-D, Vail SA, Gust D, Smirnov S. Nano Lett. 2006;6:1013–1017. doi: 10.1021/nl060313d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu X, Jiang M. Angew. Chem., Int. Ed. 2006;45:3846–3850. doi: 10.1002/anie.200504364. [DOI] [PubMed] [Google Scholar]
- 178.Tong X, Wang G, Soldera A, Zhao Y. J. Phys. Chem. B. 2005;109:20281–20287. doi: 10.1021/jp0524274. [DOI] [PubMed] [Google Scholar]
- 179.Wang G, Tong X, Zhao Y. Macromolecules. 2004;37:8911–8917. [Google Scholar]
- 180.Lee CT, Smith KA, Hatton TA. Macromolecules. 2004;37:5397–5405. [Google Scholar]
- 181.Mano JF. Adv. Eng. Mater. 2008;10:515–527. [Google Scholar]
- 182.Du J, O'Reilly RK. Soft Matter. 2009;5:3544–3561. [Google Scholar]
- 183.Devalapally H, Shenoy D, Little S, Langer R, Amiji M. Cancer Chemother. Pharmacol. 2007;59:477–484. doi: 10.1007/s00280-006-0287-5. [DOI] [PubMed] [Google Scholar]
- 184.Gillies ER, Fréchet JMJ. Bioconjugate Chem. 2005;16:361–368. doi: 10.1021/bc049851c. [DOI] [PubMed] [Google Scholar]
- 185.Kiser PF, Wilson G, Needham D. Nature. 1998;394:459–462. doi: 10.1038/28822. [DOI] [PubMed] [Google Scholar]
- 186.Lee ES, Oh KT, Kim D, Youn YS, Bae YH. J. Control. Release. 2007;123:19–26. doi: 10.1016/j.jconrel.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lynn DM, Amiji MM, Langer R. Angew. Chem., Int. Ed. 2001;40:1707–1710. [PubMed] [Google Scholar]
- 188.Paramonov SE, Bachelder EM, Beaudette TT, Standley SM, Lee CC, Dashe J, Fréchet JMJ. Bioconjugate Chem. 2008;19:911–919. doi: 10.1021/bc7004472. [DOI] [PubMed] [Google Scholar]
- 189.Sawant RM, Hurley JP, Salmaso S, Kale A, Tolcheva E, Levchenko TS, Torchilin VP. Bioconjugate Chem. 2006;17:943–949. doi: 10.1021/bc060080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schmaljohann D. Adv. Drug Delivery Rev. 2006;58:1655–1670. doi: 10.1016/j.addr.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 191.Shin J, Shum P, Thompson DH. J. Control. Release. 2003;91:187–200. doi: 10.1016/s0168-3659(03)00232-3. [DOI] [PubMed] [Google Scholar]
- 192.Wang C-H, Hsiue G-H. J. Control. Release. 2005;108:140–149. doi: 10.1016/j.jconrel.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 193.Yang SC, Bhide M, Crispe IN, Pierce RH, Murthy N. Bioconjugate Chem. 2008;19:1164–1169. doi: 10.1021/bc700442g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cordes EH, Bull HG. Chem. Rev. 1974;74:581–603. [Google Scholar]
- 195.Zhu J, Munn RJ, Nantz MH. J. Am. Chem. Soc. 2000;122:2645–2646. [Google Scholar]
- 196.Guo X, Andrew MacKay J, Szoka FC., Jr Biophys. J. 2003;84:1784–1795. doi: 10.1016/s0006-3495(03)74986-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Guo X, Szoka FC. Bioconjugate Chem. 2001;12:291–300. doi: 10.1021/bc000110v. [DOI] [PubMed] [Google Scholar]
- 198.Liu D, Hu i., Qiao W, Li Z, Zhan S, Cheng L. Lipids. 2005;40:839–848. doi: 10.1007/s11745-005-1446-5. [DOI] [PubMed] [Google Scholar]
- 199.Bertin PA, Smith D, Nguyen ST. Chem. Commun. 2005:3793–3795. doi: 10.1039/b504643b. [DOI] [PubMed] [Google Scholar]
- 200.Rihová B, Etrych T, Pechar M, JelInková M, Stastný M, Hovorka O, Kovár M, Ulbrich K. J. Control. Release. 2001;74:225–232. doi: 10.1016/s0168-3659(01)00320-0. [DOI] [PubMed] [Google Scholar]
- 201.Etrych T, Jelinkova M, Rihova B, Ulbrich K. J. Control. Release. 2001;73:89–102. doi: 10.1016/s0168-3659(01)00281-4. [DOI] [PubMed] [Google Scholar]
- 202.Kaneko T, Willner D, Monkovic I, Knipe JO, Braslawsky GR, Greenfield RS, Vyas DM. Bioconjugate Chem. 1991;2:133–141. doi: 10.1021/bc00009a001. [DOI] [PubMed] [Google Scholar]
- 203.Tang X, Pan CY. J. Biomed. Mater. Res., Part A. 2008;86A:428–438. doi: 10.1002/jbm.a.31515. [DOI] [PubMed] [Google Scholar]
- 204.Liu F, Eisenberg A. J. Am. Chem. Soc. 2003;125:15059–15064. doi: 10.1021/ja038142r. [DOI] [PubMed] [Google Scholar]
- 205.Hentschel J, Krause E, Börner HG. J. Am. Chem. Soc. 2006;128:7722–7723. doi: 10.1021/ja060759w. [DOI] [PubMed] [Google Scholar]
- 206.Schilli CM, Zhang M, Rizzardo E, Thang SH, Chong YK, Edwards K, Karlsson G, Müller AHE. Macromolecules. 2004;37:7861–7866. [Google Scholar]
- 207.Du J, Tang Y, Lewis AL, Armes SP. J. Am. Chem. Soc. 2005;127:17982–17983. doi: 10.1021/ja056514l. [DOI] [PubMed] [Google Scholar]
- 208.Cai Y, Armes SP. Macromolecules. 2004;37:7116–7122. [Google Scholar]
- 209.Yoo J-W, Mitragotri S. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11205–11210. doi: 10.1073/pnas.1000346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Griset AP, Walpole J, Liu R, Gaffey A, Colson YL, Grinstaff MW. J. Am. Chem. Soc. 2009;131:2469–2471. doi: 10.1021/ja807416t. [DOI] [PubMed] [Google Scholar]
- 211.Sawyers CL. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 212.Vartak DG, Gemeinhart RA. J. Drug Targeting. 2007;15:1–20. doi: 10.1080/10611860600968967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Raffetto JD, Khalil RA. Biochem. Pharmacol. 2008;75:346–359. doi: 10.1016/j.bcp.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liotta LA, Tryggvason K, Garbisa S, Hart I, Foltz CM, Shafie S. Nature. 1980;284:67–68. doi: 10.1038/284067a0. [DOI] [PubMed] [Google Scholar]
- 215.Davies B, Waxman J, Wasan H, Abel P, Williams G, Krausz T, Neal D, Thomas D, Hanby A, Balkwill F. Cancer Res. 1993;53:5365–5369. [PubMed] [Google Scholar]
- 216.MacDougall JR, Bani MR, Lin Y, Muschel RJ, Kerbel RS. Br. J. Cancer. 1999;80:504–512. doi: 10.1038/sj.bjc.6690385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.MacDougall JR, Bani MR, Lin Y, Rak J, Kerbel RS. Cancer Res. 1995;55:4174–4181. [PubMed] [Google Scholar]
- 218.Jinga DC, Blidaru A, Condrea I, Ardeleanu C, Dragomir C, Szegli G, Stefanescu M, Matache C. J. Cell. Mol. Med. 2006;10:499–510. doi: 10.1111/j.1582-4934.2006.tb00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Maatta M, Santala M, Soini Y, Turpeenniemi-Hujanen T, Talvensaari-Mattila A. Acta Obstet. Gynecol. Scand. 2010;89:380–384. doi: 10.3109/00016340903555990. [DOI] [PubMed] [Google Scholar]
- 220.Napoli A, Boerakker MJ, Tirelli N, Nolte RJM, Sommerdijk NAJM, Hubbell JA. Langmuir. 2004;20:3487–3491. doi: 10.1021/la0357054. [DOI] [PubMed] [Google Scholar]
- 221.Napoli A, Tirelli N, Wehrli E, Hubbell JA. Langmuir. 2002;18:8324–8329. [Google Scholar]
- 222.Napoli A, Valentini M, Tirelli N, Muller M, Hubbell JA. Nat. Mater. 2004;3:183–189. doi: 10.1038/nmat1081. [DOI] [PubMed] [Google Scholar]
- 223.Cerritelli S, Velluto D, Hubbell JA. Biomacromolecules. 2007;8:1966–1972. doi: 10.1021/bm070085x. [DOI] [PubMed] [Google Scholar]
- 224.Halliwell B, Clement MV, Long LH. FEBS Lett. 2000;486:10–13. doi: 10.1016/s0014-5793(00)02197-9. [DOI] [PubMed] [Google Scholar]
- 225.Gius D, Spitz DR. Antioxid. Redox Signaling. 2006;8:1249–1252. doi: 10.1089/ars.2006.8.1249. [DOI] [PubMed] [Google Scholar]
- 226.Ohshima H, Tatemichi M, Sawa T. Arch. Biochem. Biophys. 2003;417:3–11. doi: 10.1016/s0003-9861(03)00283-2. [DOI] [PubMed] [Google Scholar]
- 227.LaRue I, Adam M, Pitsikalis M, Hadjichristidis N, Rubinstein M, Sheiko SS. Macromolecules. 2006;39:309–314. [Google Scholar]
- 228.Ramzi A, Rijcken, Veldhuis, Schwahn D, Hennink WE, van Nostrum CF. J. Phys. Chem. B. 2008;112:784–792. doi: 10.1021/jp073673d. [DOI] [PubMed] [Google Scholar]
- 229.Sundararaman A, Stephan T, Grubbs RB. J. Am. Chem. Soc. 2008;130:12264–12265. doi: 10.1021/ja8052688. [DOI] [PubMed] [Google Scholar]
- 230.De P, Gondi SR, Sumerlin BS. Biomacromolecules. 2008;9:1064–1070. doi: 10.1021/bm701255v. [DOI] [PubMed] [Google Scholar]
- 231.Lee H.-i., Ah Lee J, Poon Z, Hammond PT. Chem. Commun. 2008:3726–3728. doi: 10.1039/b807561a. [DOI] [PubMed] [Google Scholar]
- 232.Gonzalez-Rodriaguez D, Schenning APHJ. Chem. Mater. 2011;23:310–325. [Google Scholar]
- 233.Nakade H, Jordan BJ, Uzun O, LaPointe NL, Coughlin EB, Rotello VM. Chem. Commun. 2005:3271–3273. doi: 10.1039/b502929e. [DOI] [PubMed] [Google Scholar]
- 234.Lo PK, Sleiman HF. J. Am. Chem. Soc. 2009;131:4182–4183. doi: 10.1021/ja809613n. [DOI] [PubMed] [Google Scholar]
- 235.Uzun O, Sanyal A, Nakade H, Thibault RJ, Rotello VM. J. Am. Chem. Soc. 2004;126:14773–14777. doi: 10.1021/ja047643p. [DOI] [PubMed] [Google Scholar]
- 236.Liu Y, Lin C, Li H, Yan H. Angew. Chem., Int. Ed. 2005;44:4333–4338. doi: 10.1002/anie.200501089. [DOI] [PubMed] [Google Scholar]
- 237.Lu Y, Liu J. Curr. Opin. Biotechnol. 2006;17:580–588. doi: 10.1016/j.copbio.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 238.Cheglakov Z, Weizmann Y, Braunschweig A, Wilner O, Willner I. Angew. Chem., Int. Ed. 2008;47:126–130. doi: 10.1002/anie.200703688. [DOI] [PubMed] [Google Scholar]
- 239.Zhou J, Soontornworajit B, Martin J, Sullenger BA, Gilboa E, Wang Y. Macromol. Biosci. 2009;9:831–835. doi: 10.1002/mabi.200900046. [DOI] [PubMed] [Google Scholar]