Abstract
Differentiation-related DNA methylation is receiving increasing attention, partly owing to new, whole-genome analyses. These revealed that cell type-specific differential methylation in gene bodies is more frequent than in promoters. We review new insights into the functionality of DNA methylation during differentiation, with emphasis on the methylomes of myoblasts, myotubes and skeletal muscle versus non-muscle samples. Biostatistical analyses of data from reduced representation bisulfite sequencing are discussed. Lastly, a model is presented for how promoter and intragenic DNA hypermethylation affect gene expression, including increasing the efficiency of polycomb silencing at some promoters, downmodulating other promoters rather than silencing them, counteracting enhancers with heterologous specificity, altering chromatin conformation by inhibiting the binding of CTCF, modulating mRNA transcript levels by inhibiting overlapping promoters of noncoding RNA genes or by regulating the use of alternative mRNA promoters, modulating transcription termination, regulating alternative splicing and acting as barriers to the spread of activating chromatin.
Keywords: alternative splicing, CTCF, DNA methylation, enhancer, epigenetic, HOX gene, LSP1, skeletal muscle, TBX1, transcription termination
Importance of DNA methylation to mammalian differentiation
The development of methods for whole-genome methylation profiling has afforded major new insights into the functions of mammalian DNA methylation [1,2]. Especially powerful are methylome profiling techniques with single-base resolution, like reduced representation bisulfite sequencing (RRBS) [1]. Genome-wide, tissue-specific or cell type-specific DNA methylation profiling has begun to shift the focus of DNA methylation research from mostly promoters and immediate upstream enhancers to including intragenic regions and distal intergenic regions [3–7]. In parallel, chromatin epigenetic profiles have uncovered abundant differentiation-related epigenetic marks using antibodies specific for histone modifications in chromatin immunoprecipitation coupled with next-generation DNA sequencing (ChIP-seq) [8,9]. New functionalities for chromatin epigenetic changes have been derived from studies of histone modifications [8–10], minor histones in chromatin [11], the insulator- and chromatin loop-associated protein CTCF [4,12], and DNaseI hypersensitivity by DNase-sequencing [13]. Here, we provide the first overview of differentiation-associated DNA methylation in the skeletal muscle lineage that highlights genome-wide profiling of differential methylation. Using mostly myogenesis as an example, we describe new insights into DNA methylation’s role in controlling gene expression in a cell type- and development-specific manner. In addition, we summarize important features of the biostatistical analysis of differential methylation, using RRBS profiles from many different samples.
The importance of genomic 5-methylcytosine (5mC) in helping to regulate mammalian development is indicated by diverse studies demonstrating the following: the strong repressive effect of in vitro methylation at promoters [14–19]; deleterious consequences of knockout or conditional knockout of DNA methyltransferases in mice [20,21]; major changes in DNA methylation during early mammalian embryogenesis and gametogenesis [22]; tissue-specific differences in human DNA methylation [6,23]; the contribution of altered DNA methylation to certain human diseases, especially cancer, immunological and neurological diseases [24–26]; the involvement of DNA methylation in controlling the activity of some enhancers and the DNA binding of some transcription factors [3,18]; and the roles of DNA methylation in X chromosome inactivation [27] and imprinting [28]. Targeted deletion of the gene encoding the chromatin remodeling DNA helicase LSH (HELLS) is associated with both DNA demethylation in many regions of the genome and embryonic lethality accompanied by many developmental abnormalities [29]. Another line of evidence that evolution has selected 5mC as a base in all vertebrate DNAs for much more than just silencing retrotransposons and foreign DNA comes from reports in 2009 and thereafter [30–32] describing 5-hydroxymethylcytosine (5hmC), a sixth genetically programmed base in mammalian DNA. This derivative of genomic 5mC is present in highly cell type-specific levels and is localized preferentially to certain genomic and chromatin subregions.
Converting non-muscle cells to muscle cells
Skeletal muscle progenitor cells are critical for forming skeletal muscle during embryogenesis and for regenerative muscle repair after birth [33]. The differentiation of mononuclear myoblasts (Mb) to large, multinuclear myotubes (Mt) provides an in vitro model for complex differentiation-linked cellular changes. Moreover, these progenitor cells can be compared with skeletal muscle tissue, which consists largely of their fully differentiated products. The role of chromatin modifications and miRNAs in controlling the expression of specific genes during myogenesis has been reviewed recently [33,34].
Some of the earliest studies on the importance of DNA methylation to differentiation involved the skeletal muscle lineage. Constantinides et al. found that treatment of an embryonic fibroblast cell line with 5-azacytidine (5azaCR), an inhibitor of DNA methylation, induces the formation of Mt [35]. The effect of 5azaCR was demonstrated to be on the DNA itself, and not a side effect of the inhibitor. This was shown by induction of myogenesis in 10T1/2 mouse fibroblasts upon transfection with DNA from 5azaCR-treated 10T1/2 cells [36] or by transfection with an antisense DNMT1 construct [37]. However, it was proposed that the critical target for myogenesis-inducing demethylation was the abnormal, in vitro-associated methylation of the MyoD1 gene, which encodes one of the master myogenic transcription factors. While MYOD1 itself is constitutively unmethylated in normal mouse tissues [14], human tissues, and nontransformed cell cultures [Ehrlich et al., Unpublished Data], a critical MYOD1 enhancer at 20 kb upstream (−20 kb) of the transcription start site (TSS) is unmethylated in mouse and human muscle cells but highly methylated in primary human keratinocytes [38]. Furthermore, the −20 kb enhancer became demethylated during reprogramming of keratinocyte nuclei to express MYOD1 RNA by fusion with C2C12 Mt [38]. We have also observed hypomethylation in the distal upstream region of MYOD1 by comparing methylomes from Mb, Mt and non-muscle cells [Chandra & Ehrlich, Unpublished Data]. Moreover, other genes may also be involved in experimentally induced demethylation-associated myogenic conversion [37]. For example, in a study of C2C12 Mb, which have a functional (and presumably unmethylated) Myod1 promoter and can differentiate to Mt upon serum deprivation, treatment with 5azaCR upregulated muscle-associated genes at the Mb stage and gave Mt with a more mature muscle organization [39].
Restricting the differentiation potential of stem or progenitor cells
Treatment with DNA demethylating agents can not only convert non-myogenic progenitor cells to Mt, but also can induce other cell-type interconversions in progenitor cells. For example, with 5azaCR treatment, the C2C12 Mb cell line can be induced to express genes for key osteogenic transcription factors as well as adipocyte markers [40]. The outcome of DNA demethylation treatment is dependent upon the cell type as well as the treatment and growth conditions [41].
The conversion of a multipotent adult stem cell to dissimilar differentiation products by treatment with DNA demethylating agents can be explained by the hypothesis that some genomic methylation restricts the differentiation potential of progenitor cells [40]. This hypothesis is consistent with results from genome-wide profiling of promoter DNA methylation, polycomb silencing-associated H3K27me3, RNA polymerase II binding, and activation-associated H3K4me2 in mouse embryonic stem cells (ESCs; pluripotent) and their derivative neuronal progenitors (multipotent) [42]. Terminal differentiation predominantly led to increases in DNA methylation and both increases and decreases in H3K27me3, depending on the gene involved. The acquisition of H3K27me3 was associated with gains in DNA methylation, but only at a subset of sites. Similarly, another study of promoter methylation throughout the genomes of mouse ESCs, sperm, embryonic germ cells, trophoblast stem cells and primary embryonic fibroblasts led Farthing et al. to hypothesize that changing the DNA methylation status of pluripotency genes in vivo is critical to their function [43]. DNA methylation is considered a more stable repressive mark than repression-associated histone modifications [42]. The association of differentiation and the loss of pluripotency with DNA methylation at previously unmethylated sites (de novo methylation) is consistent with the inability of ESCs to differentiate when Dnmt1, the most abundantly expressed DNA methyltransferase gene, is homozygously knocked out [44].
Changes in DNA methylation upon differentiation: whole-genome profiling
RRBS made it practical to study genome-wide DNA methylation at single-base resolution from many cell and tissue types because, at a moderate cost, it covers a variety of types of sequences [1]. It detects at least some CpG sites in about 50% of the CpG islands (CGIs; CpG-rich) but only approximately 5% of total CpG sites. Although it identifies cytosine methylation status in gene bodies as well as intergenic regions, both single-copy and repeated sequences, and nonisland sequences as well as CGIs, RRBS is strongly biased toward CGIs. In a search for differentially methylated (DM) regions (DMRs) by RRBS, Meissner et al. found that approximately 8% of CpGs that were unmethylated in ESCs became methylated in ESC-derived neural progenitor cells and approximately 2% of CpGs methylated in ESCs were unmethylated in the neural progenitor derivatives. They compared methylome profiles with ChIP-seq profiles of H3K4me3, which is characteristic of active promoters but also found in some active enhancers [45]. There was a strong association between loss of H3K4me3 and the gain of DNA methylation in ESCs versus neural progenitors. This is consistent with the hypothesis that DNA hypermethylation during development could lock in repression. In addition, highly conserved methylated noncoding DNA elements generally exhibited a major decrease in DNA methylation if they overlapped subregions that acquired H3K4me2 (characteristic of active promoters or enhancers) in the ESC-to-neural progenitor comparison.
A study by Varley et al. (ENCODE/RRBS, HudsonAlpha [HudsonAlpha, AL, USA]) used 82 different cell cultures, including cancer-derived cultures and tissues, to generate RRBS profiles for analysis of the cancer-specificity of DNA methylation patterns [6]. They also looked for genome-wide relationships between tissue-specific DMRs, expression profiles and ChIP-seq profiles for H3K27me3, the H3K27me3-associated polycomb group protein EZH2, and the enhancer protein EP300. By hierarchical clustering of DNA methylation data from seven pairs of tissues and the corresponding cell cultures, including skeletal muscle plus myogenic progenitor cells, they found that tissue-specific DMRs were most closely related to those of the analogous primary cell cultures. They also examined the minor fraction of non-CpG-methylated sites and tried to reduce errors from false positives due to incomplete bisulfite modification by requiring that the site be identified as methylated in two replicate samples. ESCs had the highest non-CpG 5mC content, as previously reported [46]. Skeletal muscle had much less non-CpG 5mC. They confirmed reports [17,47–49] of general trends for the association of gene repression with hypermethylation of promoters or intragenic regions within 2 kb of the TSS. This negative relationship was observed irrespective of whether the methylated CpGs in the extended promoter regions overlapped with CGIs. Previous studies had reported generally positive relationships between gene-body methylation and gene expression [50,51] but Varley et al. inferred a more complicated relationship [6]. They found that methylated intragenic CpGs >2 kb downstream of the TSS had, in general, a positive association with expression if they were not located in CGIs and either a positive or negative relationship if they were in CGIs. By EP300 ChIP-seq, they obtained evidence for enhancers frequently overlapping intragenic CGIs whose methylation was inversely correlated with expression.
Maunakea et al. profiled CGIs in the methylome of human brain DNA by an immunoprecipitation method and by using CpG methylation-sensitive restriction endonucleases [49]. One of their overall conclusions was that understanding gene body methylation requires examining the sequence context and consideration of individual RNA isoforms. We also propose that the relationships between intragenic DNA methylation and gene expression are complex and highly context dependent, as will be illustrated below.
Muscle lineage-associated differential methylation seen in promoter or CGI methylomes
Prior to our RRBS analysis of muscle lineage-associated changes in DNA methylation, in collaboration with Myers ([7]; see below), and the abovementioned RRBS study of general differentiation and cancer from their laboratory [6], there have been only a small number of reports about genome-wide DNA methylation that compared skeletal muscle progenitor cells or tissue with non-muscle samples. Moreover, these have involved just promoter regions or CGIs. Illingworth et al. compared DNA methylation in CGIs of human skeletal muscle, brain, leukocytes and spleen using affinity purification of 5mC-enriched DNA fragments followed by hybridization to microarrays consisting of a panel of human CGIs [52]. Between 5.7 and 8.3% of the CGIs were methylated. Muscle and spleen had the highest levels of methylation and of differential methylation. An average of 5% of the CGIs in all of the studied tissues displayed statistically significant differential methylation. From the association of intragenic or intergenic CGI DMRs with H3K4me3, they propose that many of them are unannotated promoters of noncoding RNA (ncRNA) genes.
Using samples from normal second-trimester fetuses, Yuen et al. examined muscle, brain, kidney, lung and skin cells for differential methylation at 1315 CpG sites in promoters with a single-base resolution, multiplex platform for 1505 CpG loci (GoldenGate Methylation Cancer Panel I, Illumina, CA, USA) [53]. Tissue-specific DMRs were identified by analysis of variance with p-values adjusted for multiple comparisons. For autosomal loci, the Pearson’s linear correlation was used to analyze similarities in the average methylation between different tissues. They found that 195 sites (23% of the variably methylated sites) were significantly different between the different fetal tissues. Muscle cells had the second highest number of tissue-specific DMRs and brain cells had the highest number. Isagawa et al. profiled DNA methylation in murine promoters by immunoprecipitation of methylated DNA and hybridization to a promoter microarray [54]. They compared an ESC sample, cells representative of three germ layers derived from ESCs, skeletal muscle, brain, liver and sperm from a young adult mouse. The authors assigned relative methylation levels and used a false discovery rate cut-off of 4% to identify ‘candidate methylated regions’. Most of the promoters did not differ much in methylation between the three germ layer derivatives or the postnatal tissues, although each of the tissue types had a minor fraction of candidate methylated regions. Muscle had the highest number of candidate methylated regions relative to the other postnatal samples. Sperm and ESCs were hypomethylated relative to the induced derivatives from ESCs or the postnatal tissues. They also found that the ESC differentiation products had more promoter methylation than somatic tissues.
Microarray-based DNA methylation profiling (HumanMethylation27 BeadChip, Illumina) was used by Berdasco et al. to compare DNA methylation in promoter regions of human adipose-derived stem cells with or without in vitro differentiation to myogenic-type or osteogenic-type cells [55]. CpG sites for which the average change in methylation proportion was a least 0.2, with an adjusted p-value of <0.01, were considered to be differentially methylated. They found 85 and 23 differentially methylated CpG sites in the adipose stem cells versus their myogenic and osteogenic cell derivatives, respectively. One of the genes whose DNA methylation profiles were shown in detail was the germline stem cell-associated PIWIL2, which exhibited both DNA hypermethylation of its CGI promoter and repression in the myogenic and osteogenic cells relative to the adipose stem cells.
Methylome profiling by RRBS: biostatistical methods
In the above-cited studies, researchers employed traditional statistical methods, which assume that if there is no differential methylation, the p-values associated with each CpG site should be independent and identically distributed. However, for both experimental and biological reasons, these assumptions are not valid for RRBS methylation profiles. Systematic sources of variation include wide-ranging levels of read coverage, rapidly fluctuating methylation levels that are locally correlated, and differing group sample-sizes across genomic regions. As a result, the sensitivity and specificity of single-site analyses to detect DM sites in RRBS profiles will vary greatly across the genome.
For our first study of RRBS data from many different human samples, we employed mixed-effects binomial regression models to identify individual DM sites and applied the results to our previous study [7] and the analysis shown in Figure 1, below. We employed stringent selection criteria, requiring changes in the percentage of methylation levels of at least 50% and statistical significance at the 0.01 level. This ensured very high specificity of our findings and emphasized only the most salient DM sites. We then mapped DM sites to specific regions (−2 kb to TSS, first exon or intron, internal exons, internal introns, last exon, transcription termination site to 2 kb distal and other intergenic regions) using R scripts. Analysis of individual DM sites is a valuable tool in DNA regions where there is low coverage by RRBS or where there may be individual CpGs whose DNA methylation status might affect gene expression irrespective of methylation of neighboring sequences [56,57].
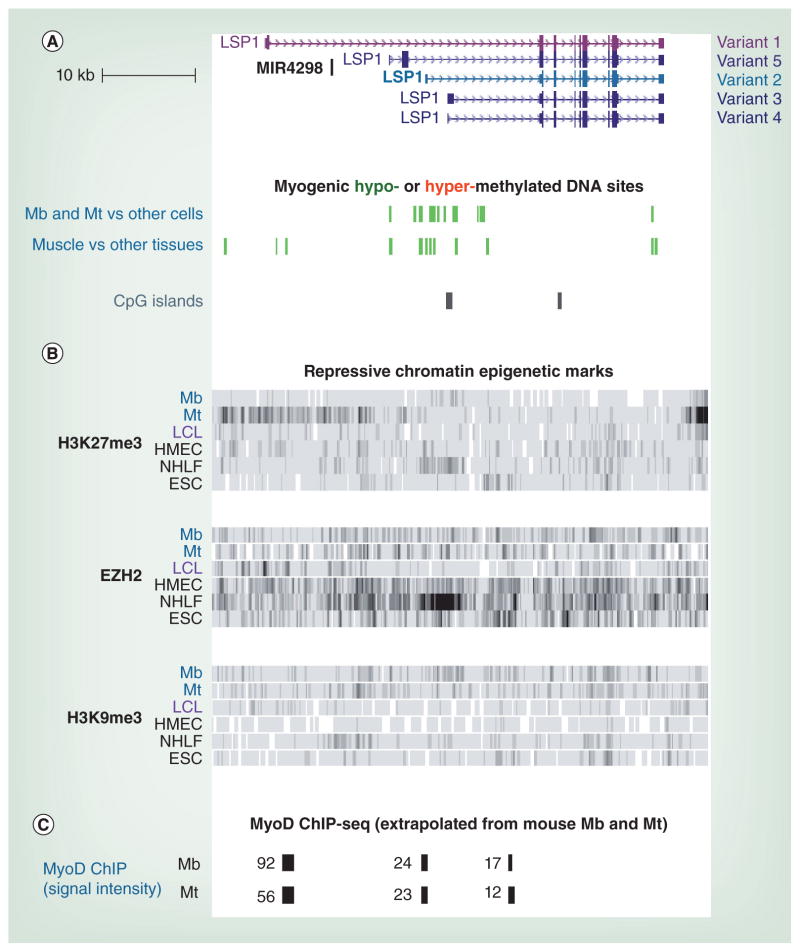
Figure 1. Myoblast-specific differential DNA methylation, but not H3K27 or H3K9 trimethylation, is implicated in myoblast-specific promoter usage for LSP1.
(A) RefSeq gene variants for LSP1 are shown. The lymphoid-specific gene variant and the most prominent Mb-specific variant are indicated in pink and blue, respectively. Underneath are custom tracks for differential methylation between myogenic and non-myogenic samples (p < 0.01 difference). Significant myogenic hypomethylation, but not myogenic hypermethylation, was observed. The depicted region in this figure and Figure 2 is chromosome 11: 1,868,993–1,917,902 (hg19). All tracks are aligned. At this scale, most individual differentially methylated sites cannot be resolved from neighboring differentially methylated sites. (B) ChIP-seq profiles of characteristically repressive chromatin marks are shown (ENCODE/histone modifications, Broad Institute [MA, USA]). The shading of the bars is proportional to the intensity of the signal. In the Mb sample, note the lack of H3K27me3, EZH2 and H3K9me3 signal at the 5′ region of LCL-specific variant 1. Reciprocally, in the LCL, there was no signal for these typically repressive chromatin marks at the 5′ region of the major Mb-specific variant 2. (C) Data from Myod ChIP-seq on murine C2C12 Mb and Mt cell cultures was extrapolated to orthologous human sequences. The numbers indicate the relative signal intensity, with >50 indicating strong binding.
ChIP-seq: Chromatin immunoprecipitation coupled with next-generation DNA sequencing; ESC: Embryonic stem cell; HMEC: Human mammary epithelial cell; LCL: Lymphoblastoid cell line; Mb: Myoblast; Mt: Myotube; NHLF: Normal human lung fibroblast.
(B) Data taken from [101].
(C) Data taken from [67].
In studies for which the primary goal is the detection of DMRs rather than DM sites, an additional statistical concern with RRBS methylation profiles is the spatial distribution of detected CpGs, characterized by clusters of sites in close proximity separated by larger lags. Due to the nature of the RRBS profiles, methods targeting the detection of focal DMRs spanning fewer than 2000 bp are likely to be more accurate than methods like BSmooth [58], which are optimized for much larger regions. Some investigators have developed ad hoc procedures that require all contiguous sites within a string to be significantly differentially methylated and to exceed a given threshold for methylation change [3,6]. While such approaches are unlikely to yield false-positive DMRs, their specificity will be exceedingly low, and many DMRs of interest will be missed. Furthermore, as closely spaced CpGs are likely to have the same experimental characteristics with respect to read coverage, sample size and underlying methylation levels, observed correlations among p-values from closely neighboring sites are more often a consequence of experimental factors than of true biological signal. The comb-p method has been recently developed to adjust single-site p-values for spatial autocorrelation [59], but such adjustments do not directly address systematic biases, and sometimes may actually lead to dampening of the signal when true differential methylation is present.
We recently developed a novel procedure to identify focal DMRs with both high sensitivity and specificity [Lacey et al., Modeling, simulation and analysis of methylation profiles from reduced representation bisulfite sequencing experiments (2013), Submitted]. We first employ a quantile-based transformation to adjust skewed p-values from single-site regression models by correcting for systematic biases in RRBS experiments associated with variable read coverage and sample size. Under the assumption of no differential methylation, the algorithm simulates samples with the average read coverage and sample sizes at each site, generates a distribution of null p-values, and assigns the adjusted p-value to be the quantile of the raw p-value with respect to the null distribution. This transformation improves the uniformity of the distribution of single-site p-values and makes them more suitable for the subsequent detection of DMRs. We then use the uniform product distribution to determine the probabilities associated with adjusted model p-values on strings of contiguous sites, with the additional requirements that all differences in methylation along the sites are strictly positive or strictly negative, that the sites are in sufficiently close proximity, and that the p-values at the boundary sites are sufficiently small. Our algorithm enables the identification of focal DMRs for which not all sites are significantly differentially methylated due to variations in sample size, read coverage or biological signal. We applied our approach in a revised analysis of differential methylation in the myogenic versus non-myogenic cell cultures and thereby identified 3233 DMRs, including 1963 that directly overlap with genes.
Muscle-associated DNA hypermethylation & hypomethylation from RRBS profiles
We recently compared RRBS DNA methylation profiles from two normal human skeletal muscle tissue samples and 14 normal non-muscle samples. In parallel, we studied nine immunocytochemically characterized myogenic progenitor cell cultures and 16 types of non-muscle cell cultures with technical duplicates ([7,101] ENCODE/HudsonAlpha). The myogenic progenitors were Mb and Mt, which were analyzed as a set (MbMt) because their methylomes are so similar. The only transformed cell cultures were Epstein–Barr virus-transformed lymphoblastoid cell lines (LCLs). Data were averaged from replicates. Using the biostatistical methods described above for individual DM sites, we determined that 12016 and 19640 CpG sites (1.0 and 1.7%, respectively, of the ~1 million CpG sites with sufficient RRBS coverage) exhibited significant differences in methylation in comparisons of muscle with non-muscle tissues or MbMt with non-muscle cell cultures. For muscle versus non-muscle tissues, 93% of the DM sites were hypomethylated. In contrast, in MbMt versus non-muscle cell cultures, almost the same numbers of DM sites were either hypermethylated or hypomethylated [7].
The large decrease in the number of CpG sites hypermethylated in Mb and Mt (9592 sites) relative to skeletal muscle (761 sites) and the many genes that were hypomethylated in skeletal muscle but not in Mb and Mt (928 genes) [7] indicate that there is active demethylation in the muscle lineage after the Mt stage. Owing to Mt and mature muscle cells being giant multinuclear, postmitotic cells, the decrease in DNA methylation in muscle versus progenitor cells could not occur as a result of a failure of maintenance methylation (methylation of newly synthesized CpGs opposite template 5mCpGs) and must be due to active demethylation. In addition, hypomethylation in myogenic progenitor cells might be partly attributable to active demethylation, as indicated by evidence from a previous study of demethylation of the α-actin gene in rat Mb [15]. TET1, TET2 and TET3 enzymes are implicated in pathways for active demethylation of mammalian DNA by catalyzing the conversion of 5mC residues to 5hmC residues, intermediates in demethylation as well as stable components of DNA [60]. Our previous exon expression microarray data for 21 cell types [61] indicated that that samples of Mb and Mt have much higher levels of TET1 and TET2 RNA than all of the analyzed nontransformed, non-muscle cell strains, with the expected exception [62] of high TET1 RNA levels for ESCs [7]. Moreover, results from an enzymatic assay for quantifying 5hmC and 5mC, demonstrated that, in comparison to Mb or Mt, skeletal muscle is twofold enriched globally in genomic 5hmC [7]. Skeletal muscle was also the only type of sample enriched in 5hmC at tested myogenic hypermethylated sites associated with PAX3 andTBX1 [7].
Mapping DM sites to a single isoform of the gene nearest to them indicated that 2407 and 1983 genes are associated with 2–178 or 2–87 MbMt or muscle DM sites, respectively [7]. More myogenic DM sites were in internal introns or exons than in the region from −2 kb through the first intron. There was considerable overlap of muscle versus MbMt DM sites. The relevance of differential methylation to the skeletal muscle lineage was seen in the overrepresentation of DNA-binding sequence motifs for the myogenic MYOD and MYOG transcription factors in genes associated with myogenic DM sites. There was an extremely strong enrichment for homeobox genes among genes associated with MbMt hypermethylation and overrepresentation of muscle structural protein genes among those exhibiting muscle hypomethylation. However, no overall correlation was observed between gene expression and myogenic hypomethylation or hypermethylation, although some functional classes of genes did show general trends. For example, among the 44 contractile fiber genes displaying muscle hypomethylation, 32 were significantly upregulated in Mt versus non-muscle cells and only three were downregulated [7,61].
Illustration of analysis of a gene from RRBS profiles: LSP1, a gene with a little-recognized association with myogenesis
Locus-specific analyses have revealed changes in methylation of promoters for the myogenic Myod1, Myog and α-actin genes and have been reviewed previously [31,63,64]. In this section, we describe a gene that exhibited remarkable myogenic differential methylation in various subregions according to analysis of DM sites from RRBS methylomes of skeletal muscle lineage samples and non-muscle samples [7]. This analysis reveals various epigenetic associations and also illustrates the power of mining ENCODE epigenetic and transcription data from the UCSC Genome Browser [101].
LSP1 displayed 40 MbMt-hypomethylated sites and 20 muscle-hypomethylated sites, most of which overlap with at least one variant of the gene (Figure 1A; [Ehrlich et al., Unpublished Data] from a methylome database [7]). A 1993 report mentioned that LSP1 is expressed in rat Mb cultures to give an RNA of a different length from that in lymphoid cells [65]. However, there have been no other reports about LSP1 in the skeletal muscle lineage despite its very high and specific expression in myogenic progenitor cells (Figure 2B & C). It encodes an intracellular F-actin binding protein implicated in cell migration, cell signaling and adhesion to fibrinogen matrix proteins in leukocyte subpopulations [66], and so is an excellent candidate for participating in myogenesis. The myogenesis association is consistent with the binding of the myogenic transcription factor Myod to the murine Lsp1 gene region (Figure 1C), as we determined by data-mining Myod ChIP-seq profiles of mouse C2C12 Mb and Mt [67]. In addition, MIR4298, a miRNA gene of unknown function that overlaps one of the LSP1 variants (Figure 1), was identified as being near a binding site for MYOG in differentiating Mb cultures in a genome-wide study of myogenic cis-regulatory DNA modules [68].
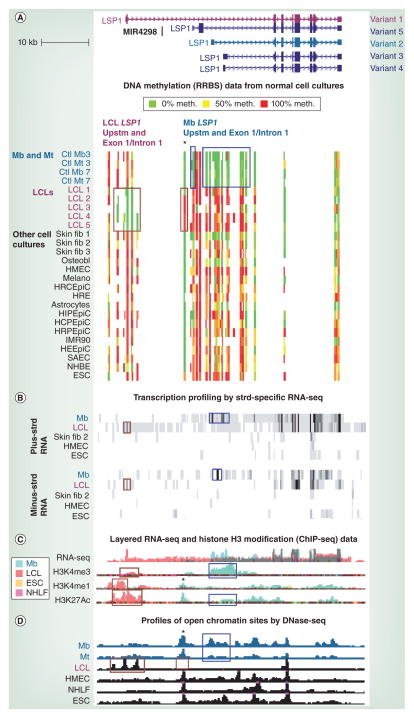
Figure 2. Opposite distributions of DNA methylation and open chromatin in myoblast and lymphoblastoid cell line samples were correlated with LSP1 alternative promoter usage (facing page).
(A) RRBS data tracks (ENCODE/DNA methylation by reduced representation bisulfite sequencing; HudsonAlpha Institute for Biotechnology, AL, USA) for the region in Figure 1 are illustrated. Each culture is from a different individual except for Mb and Mt samples with the same number. The average methylation level of each detected CpG is shown according to the indicated color scheme; intermediate values are indicated by intermediate colors. At this scale, almost all the signal seen in the figure is from clusters of CpGs rather than individual CpGs. Blue boxes show myogenic DNA hypomethylation at the Mb-specific variant 2 upstm region, exon 1 and part of intron 1. Wide or narrow brown boxes show LCL-specific hypomethylation or hypermethylation, respectively. (B) Strd-specific RNA-seq profiles (ENCODE/long RNA-seq, polyA+, Cold Spring Harbor [NY, USA]) are shown for the indicated cell types (vertical viewing range 1–100 for the plus strd and 1–10 for the minus strd). Blue and brown boxes denote Mb- and LCL-specific signal, respectively, for exon 1 sense RNA or nearby antisense RNA signal. The multiple exons in the blue boxed region for Mb are from the first exons of variant 2 and other, less prevalent, Mb-specific RNAs that are similar, but not identical to, variants 3 and 4 (Cufflinks analysis [69]). (C) RNA-seq (not strd-specific; ENCODE/ CalTech [CA, USA]) and modified-H3 ChIP-seq (ENCODE/histone modification, Broad Institute [MA, USA]) are shown with results for four cell types superimposed, as indicated by the color key. Blue boxes denote Mb-specific H3 modifications characteristic of active promoters and brown boxes show LCL-specific H3 modifications typical of active promoters or enhancers. (D) DNaseI hypersensitivity mapping (ENCODE/DNase sequencing, Duke University [NC, USA]) using Mb and Mt samples that overlapped those of (A). Combined results from two to three biological replicates are shown. Boxes show the Mb-, Mt- or LCL-specific DNase sequencing peaks in promoter or enhancer regions. The only LCL shown is LCL1, but the other four LCL samples gave similar results. Asterisks mark the subregion with LCL-associated hypermethylation and a DNase-seq peak seen in all cell types other than in LCLs. ChIP-seq: Chromatin immunoprecipitation coupled with next-generation DNA sequencing; Ctl: Control; ESC: Embryonic stem cell; HCPEpiC: Choroid plexus epithelial cell; HEEpiC: Esophageal epithelial cell; HIPEpiC: Iris pigment epithelial cell; HMEC: Human mammary epithelial cell; HRCEpiC: Retinal pigment epithelial cell; HRE: Renal epithelial cell; HRPEpiC: Retinal pigment epithelial cell; IMR90: Fetal lung fibroblast; LCL: Lymphoblastoid cell line; Melano: Melanocyte; Mb: Myoblast; Mt: Myotube; NHBE: Bronchial epithelial cell; NHLF: Normal human lung fibroblast; Osteobl: Osteoblast; RNA-seq: RNA sequencing; RRBS: Reduced representation bisulfite sequencing; SAEC: Small airway epithelial cell; Skin fib 1: Fibroblast cell strain from a child; Skin fib 2: A neonatal foreskin fibroblast cell strain; Skin fib 3: A different neonatal foreskin fibroblast cell strain; Strd: Strand; Upstm: Upstream.
Data taken from [101].
LSP1 is located between the muscle-specific troponin TNNT3 and TNNI2 genes. However, RRBS revealed that LSP1 has a much higher concentration of MbMt-hypomethylated sites than these neighboring genes (data not shown), indicating that its hypomethylation is not just a nonfunctional association with a myogenic gene neighborhood. Using a program (Cufflinks; [69]), that separately quantifies different RNA isoforms from RNA sequencing (RNA-seq) data (ENCODE/RNA-seq, CalTech, CA, USA), we found many isoforms of LSP1 RNA in Mb, one of which was the most plentiful in those cells (Figure 2; variant 2). In the LCL sample, a different RNA isoform predominated (variant 1) from a far-upstream TSS. These RNAs are predicted to encode proteins with different N termini. The combined RNA-seq signal from Mb LSP1 RNA isoforms was high and almost as high as that of the very strongly expressed LCL LSP1 RNA isoform.
From the separate RRBS DNA methylation tracks (ENCODE/HudsonAlpha) for Mb, Mt and 16 different types of non-myogenic cell cultures, prominent differences were seen in the methylation patterns of LCLs (Figure 2A, two brown boxes) and Mb or Mt (Figure 2A, two blue boxes) relative to other cell cultures and each other. MbMt-hypomethylated sites overlapped the promoter and exon 1 regions of the Mb-specific LSP1 variants (Figure 2A & B, RNA-seq, plus strand, blue box) and a Mb-specific antisense RNA signal (Figure 2B, minus strand, blue box). They also overlapped Mb- and Mt-associated peaks of H3K4me3, H3K27Ac (both positively associated with transcription), and Mb- and Mt-associated DNaseI hypersensitivity (Figure 2C & D, blue boxes and data not shown). LCL-specific hypomethylation was observed at the LCL-specific promoter/enhancer/exon 1 region, which displayed LCL-specific peaks of H3K27Ac and H3K4me1 (active enhancer), DNaseI hypersensitivity (open chromatin), an antisense RNA signal (sometimes indicative of active transcription-promoting elements), and a small peak of H3K4me3 (often, active promoter; Figure 2A–D, brown boxes on the left). B-cell lymphocytes shared this DNA hypomethylation with LCLs (data not shown). Corresponding to the cell type-specific epigenetics, differential promoter usage at LSP1 in the Mb and LCL samples was extraordinarily specific with no detectable signal of the predominant LCL RNA variant in Mb and vice versa (RNA-seq, Figure 2B & C).
The five LCL samples were the only non-ESC samples that displayed strong methylation and no DNaseI-hypersensitivity in the LSP1 Mb promoter region at the position indicated by an asterisk in Figure 2A & C & D. This subregion also displayed strong hypermethylation and no DNaseI hypersensitivity in leukocytes versus nonlymphoid tissues (data not shown). Importantly, this lymphoid-hypermethylated subregion overlaps Mb-specific peaks of H3K4me1 and H3K27Ac, chromatin marks indicative of an active enhancer (Figure 2C, asterisk). In LCLs, one might have expected repressive chromatin marks in addition to LCL hypermethylation in this gene subregion. To the contrary, as shown in Figure 1B, LCLs exhibited negligible H3K27me3 and H3K9me3 signal, and EZH2 binding in this LSP1 subregion.
These findings suggest that repression of LSP1 transcription from myogenic promoters in lymphoid cells involves myogenic enhancer hypermethylation as an early event in these cells divorced from H3K27me3, H3K9me3, or EZH2 binding. These results also implicate lymphoid-specific DNA hypomethylation at the lymphoid promoter/enhancer region in the very strong transcription of LSP1 in lymphoid cells. Conversely, the complete silencing of the LCL promoter/enhancer region in Mb was correlated with DNA methylation in this region in myogenic cells (and other nonlymphoid cells) but was not accompanied by local H3K27me3, EZH2 binding, or H3K9me3 in Mb. Only at the later Mt stage in development was some H3K27me3 signal seen in this region (Figure 1B). In summary, the DNA methylation status of LSP1 may be critical for alternative promoter usage, as has been previously described for other genes [49,51].
A model for multiple functions of differentiation-linked intragenic DNA hypermethylation
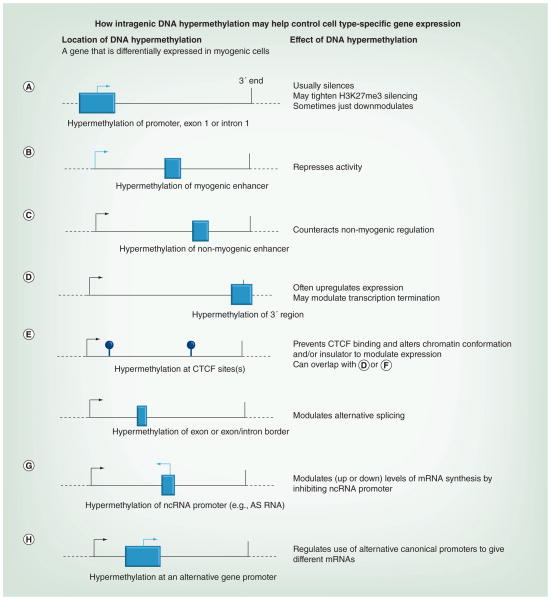
Figure 3 presents a model for diverse functions of differentiation-linked, intragenic DNA hypermethylation. Underlying the model are associations of DNA methylation with inhibition of binding of many sequence-specific transcription factors (directly or indirectly by favoring closed chromatin conformations) [16,17] and positive associations of DNA methylation with binding of methyl-DNA-preferring proteins and/or recruitment or inhibition of activity of histone modifying enzymes [18,70,71]. DNA methylation changes may usually play a stabilizing role after binding of transcription factors. DNA and chromatin epigenetic changes can influence or reinforce each other depending on the genomic sequence and cellular context. Many of the consequences of DNA hypomethylation could oppose (Figure 3A, B, E & G) or emulate (Figure 3F & H) those proposed for DNA hypermethylation. Some examples of genes with myogenic differential methylation that support this model are given below.
Figure 3. Model for the differentiation-associated regulation of gene expression by DNA hypermethylation.
Some known or proposed types of DNA methylation-dependent control of gene expression using the example of myogenesis-associated DNA hypermethylation are shown. The model depicts just cis-acting regulation from within the gene or from its canonical promoter. DNA methylation changes may be stabilizing and/or initiating the depicted changes in transcription. Boxes show hypermethylated differentially methylated regions. Light arrows indicate inhibited transcription start sites and black arrows indicate active transcription start sites that were not silenced by DNA methylation. (A) Hypermethylation of the promoter region, exon 1 or intron 1. (B) Hypermethylation of a myogenic enhancer in nonmyogenic cells. (C) Hypermethylation of a non-myogenic enhancer in myogenic cells. (D) Hypermethylation of the 3′ region. (E) Hypermethylation at CTCF sites (lollipops) somewhere in the gene. (F) Hypermethylation in an exon or at an exon/ intron border. (G) Hypermethylation of an intragenic ncRNA promoter. (H) Hypermethylation of an alternative promoter for a gene.
AS: Antisense.
Hypermethylation around the 5′ end of the gene
Methylation of the promoter region of Myog is implicated in its silencing before activation of muscle satellite cells and in non-muscle tissues [63,64,72]. CCDC140, which encodes a protein of unknown function and is expressed preferentially, but only very weakly in Mb, displays a Mb- and Mt-associated hypermethylated site approximately 90 bp downstream of the TSS that might downmodulate this gene’s expression rather than silence it. HOXD4 also has myogenic hypermethylation in the exon 1/intron 1 region [73]. The average methylation of seven myogenic HOXD4 DM sites in Mb and Mt was 78% and for human mammary epithelial cells (HMEC) and an LCL was only 9.5%. There is H3K27me3 and EZH2 binding (ChIP-seq, ENCODE/Broad [Broad Institute, MA, USA]) throughout the HOXD4-containing half of the HOXD gene cluster, as well as negligible RNA-seq signal in this region in most examined cell types, including Mb, Mt, HMEC and the LCL. Nonetheless, the skeletal muscle lineage displays distinctive DNA hypermethylation in this region. This suggests that H3K27me3 and the associated polycomb group silencing, which play a major role in HOX gene repression [74], do not suffice for repression of HOXD4 in Mb and Mt, and that DNA methylation provides an additional level of repression required specifically in the skeletal muscle lineage (Figure 3A).
Hypermethylation of a myogenic intragenic enhancer
As discussed above, DNA hypermethylation is implicated in repressing an intragenic myogenic enhancer of LSP1 in LCLs. OBSCN illustrates the association of enhancer-type chromatin and DNA hypomethylation in the skeletal muscle lineage (Figure 3B). It encodes a muscle structural protein and is expressed mostly after the Mb stage. This gene has three clusters of intragenic muscle-hypomethylated sites that overlap small peaks of enhancer-type histone modifications (H3K4me1 and H3K27Ac) in Mt, which were barely detectable in Mb [7]. Two of these muscle-hypomethylated CpG clusters do not display hypomethylation in Mb or Mt. The third one exhibits DNA hypomethylation in Mb, Mt and skeletal muscle tissue. These results suggest three intragenic muscle lineage-associated enhancer regions, one undergoing DNA demethylation before enhancer formation and the others undergoing DNA demethylation afterwards.
Hypermethylation of a non-myogenic enhancer
The 3′ end of TBX1, an early development T-box transcription factor gene that was very strongly upregulated in Mb and Mt, has a cluster of 79 MbMt hypermethylated sites [7]. Many of these persist in skeletal muscle. Because they overlap a predicted region of enhancer activity in HMEC, one of the functions of this hypermethylation may be to prevent the formation of an interfering heterologous enhancer in myogenic cells (Figure 3C). TBX1 haploinsufficiency has been linked to most of the symptoms of the DiGeorge 22q11.2 deletion syndrome [75]. Therefore, DMRs may be needed for precise regulation of this cell type-specific and development stage-specific gene.
Hypermethylation of the 3′ region
Yu et al. reported that gene expression was strongly and positively associated with CGIs at the 3′ end of genes that gained methylation upon in vitro differentiation of human ESCs [76]. Moreover, almost half of these 3′ CGIs displayed large reductions in ChIP-seq CTCF binding in differentiated cells relative to undifferentiated ESCs. In addition, several of these 3′ CGIs were shown to exhibit insulator activity upon transient transfection. This suggests that DNA hypermethylation is often regulating CTCF-dependent insulator activity at 3′ CGIs (Figure 3E). Exactly paralleling the observations of Yu et al., we found that the MbMt-hypermethylated 3′ region of TBX1, which coincides with a CGI, lacked CTCF binding detectable by ChIP-seq (ENCODE/Broad) in Mb and Mt, while other cell types without this methylation exhibited CTCF binding near the terminus of this 3′ CGI [7]. Downstream of it there is a predicted myogenic enhancer. Therefore, the 3′ CGI methylation of TBX1 in Mb and Mt is likely to have as one of its functions the suppression of CTCF insulator activity in cis and thereby allowing myogenic enhancer activity. The proposed effect of DNA methylation on CTCF binding to the 3′ CGI of TBX1 could be due to a predicted CTCF binding motif centered on the myogenic ChIP-seq CTCF peak [77], which contains a CpG dyad. Such CTCF recognition sites are subject to downregulated binding of CTCF by DNA methylation [4]. The 3′ terminal hypermethylation of TBX1 might also help direct the choice of transcription termination sites (Figure 3D) because downstream are transcription termination sites for two other gene isoforms of TBX1 that are not expressed in myogenic cells.
Hypermethylation at CTCF sites
Associations between CTCF and DMRs are not limited to the 3′ ends of genes. At the 5′ end of TBX1 there is CTCF binding in nonexpressing cell types, which is missing from expressing Mb and Mt [7]. This CTCF-binding region partially overlaps with a cluster of three myogenic hypermethylated sites approximately 0.1 kb downstream of the TSS. CTCF is generally implicated in chromatin looping [78]. Therefore, this may be an example of an intragenic DMR (not in the 3′ end of the gene) that alters the higher order chromatin structure by affecting CTCF binding and thereby affects gene expression independent of insulator activity (Figure 3E).
Hypermethylation of an exon or exon/intron border
MYH7B/MIR499 has a cluster of DM sites that might function to direct alternative splicing. There is mounting evidence that DNA methylation, especially at exons, exon–intron borders or retained introns, helps regulate RNA splicing by affecting the rate of transcription elongation, repressing binding of CTCF, or modulating the binding of splicing regulatory factors to CpG-containing exonic splicing enhancers (Figure 3F) [5,46,79–81]. MHY7B/MIR499 encodes a muscle-associated structural protein and a miRNA that is important for myogenesis [82]. MYH7 is a myosin protein found in low amounts in skeletal muscle, cardiac muscle, testis and brain [82]. Brain DNA exhibited much methylation at alternative exon 10 of MYH7B/MIR499 while skeletal and heart muscle displayed focal hypomethylation of this exon versus 14 other tissue types [7]. The methylation of exon 10 in brain DNA might explain the increase in the retention of this exon in this tissue versus skeletal and heart muscle. This would be consistent with evidence that exon DNA methylation can promote retention of alternative exons [81]. Skipping of this exon is highly consequential because it results in nonsense-mediated decay of the mRNA while still allowing the intragenic MIR499 to be generated [82].
Hypermethylation of an ncRNA promoter
The extended promoter region of the antisense HOXB-AS3 variant 3 contains 20 MbMt-hypermethylated sites from approximately 40 to 400 bp downstream of the TSS and overlaps the second and last exon of HOXB5 [73]. There was much more methylation in this region and much lower levels of expression of these sense and antisense genes in Mb than in lung fibroblasts. These findings suggest that Mb DNA hypermethylation close to the TSS of HOXB-AS3 variant 3 is downmodulating its transcription, and that HOXB-AS3 transcription favors HOXB5 expression in cis (Figure 3G). Positive sense–antisense gene relationships for overlapping genes were seen in other studies of HOX genes [83,84].
Regulation of the use of alternative gene promoters
LSP1 vividly illustrates that tissue-specific alternative promoter usage correlates with DMRs overlapping with alternative promoters (Figure 2 & 3H).
Other functions
The types of associations of intragenic DMRs with gene expression in Figure 3 are not meant to be inclusive. For example, we recently found large regions of myogenic hypermethylation that include intragenic locations in the HOXA and HOXC gene clusters. These myogenic DMRs were located at boundaries of multigenic regions consisting mostly of chromatin with the typical characteristics of active promoters and enhancers (P/E-like domains) in Mb and Mt [73]. Expression levels of genes in this vicinity suggest that the large hypermethylated DMRs help to counteract the spread of transcription upregulatory P/E domains in myogenic cells. Another function deduced for large regions of intragenic DNA methylation is to decrease the rate of transcription elongation [85] in a context-dependent manner, even to the extent of causing repression by stalling of RNA polymerase II [19]. In short, DMRs are likely to play many diverse roles in regulating gene expression during differentiation.
Future perspective
In the near future, chromatin epigenetic and RNA-seq, and chromatin–chromatin interaction profiles are likely to be used much more frequently to supplement methylome analyses in studies of transcription control. Studies of DM sites and focal DMRs will aid the discovery of transcription factors and transcription regulatory elements involved in controlling the expression of specific genes in vivo. More experiments in model systems will be done to directly test the functionality of DMRs or individual DM sites [17] identified in epigenomic profiles. These studies will help to distinguish between bystander changes in DNA methylation in vivo and those DMRs or DM sites that do exert a biological effect. Bystander changes refer to demethylation or de novo methylation at sites simply as a result of imprecise targeting in vivo of biologically important DNA methylation changes. However, a caveat in assays of the functionality of differential methylation in model systems is that they usually involve removing the DNA sequence in question from its normal genomic and chromatin neighborhood, which could alter the effects of changing the methylation status of the DNA sequence. Alternatively, these assays utilize treatments that change DNA methylation throughout the genome, which can have indirect effects.
In the years ahead, advances in understanding differentiation-specific DNA methylation changes will help elucidate age-related, physiology-associated and disease-linked alterations in DNA methylation, and their biological significance. It is likely that intragenic and distant intergenic changes in DNA methylation will be studied much more than at present for their contribution to diseases involving epigenetic deregulation, especially cancer, immunological diseases and neurological diseases. Lastly, the role of sequence-specific DNA-binding proteins, ncRNAs, DNA repair, active demethylation, passive demethylation and de novo methylation in generating differentiation-specific patterns of DNA methylation will soon be much better understood.
Executive summary.
Importance of DNA methylation to mammalian differentiation
DNA demethylating treatments can generate myotubes from certain non-muscle progenitor cells.
De novo DNA methylation is implicated in restricting the differentiation potential of stem or progenitor cells.
The loss of pluripotency upon differentiation probably partly involves the acquisition of DNA methylation at specific genetic loci.
Changes in DNA methylation upon differentiation: whole-genome profiling
Muscle lineage-associated DNA hypomethylation or hypermethylation has been seen in studies of promoter or CpG islands methylomes.
Limited numbers of promoters display muscle-associated DNA methylation patterns.
Nontraditional biostatistical methods are needed to deal with systematic sources of experimental and biological variation in methylome profiling by reduced representation bisulfite sequencing (RRBS).
Sources of experimental and biological variation include fluctuations in read coverage, sample sizes, regional methylation levels and the spatial distribution of detected CpGs. RRBS profiles are better suited for the identification of individual differentially methylated sites and focal differentially methylated regions than for identifying differential methylation over extended regions.
RRBS profiling on myoblasts, myotubes, skeletal muscle and many non-muscle samples revealed extensive and specific muscle lineage hypermethylation and hypomethylation in many gene regions. There was no overall correlation between myogenic differential methylation and myogenesis-associated levels of expression. This is consistent with there being complex relationships between them.
Example of the analysis of a specific gene from whole-genome RRBS profiles
LSP1 is expressed from different promoters in myoblasts and lymphoblasts. RRBS, histone modification, and RNA-sequencing profiles show that DNA methylation at promoter and enhancer regions, rather than repressive chromatin marks, was correlated with cell type-specific silencing of alternative LSP1 promoters.
A model for multiple functions of differentiation-linked intragenic DNA hypermethylation
A model for the effects of differentiation-associated intragenic DNA hypermethylation is illustrated for genes with differential expression in the skeletal muscle lineage.
Examples of genes that fit the model are given.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure:
This work was supported in part by NIH grant NS048859. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ndlovu MN, Denis H, Fuks F. Exposing the DNA methylome iceberg. Trends Biochem Sci. 2011;36(7):381–387. doi: 10.1016/j.tibs.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Maurano MT, Qu H, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22(9):1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, Lu Y, Tian W. Epigenetic features are significantly associated with alternative splicing. BMC Genome. 2012;13:123. doi: 10.1186/1471-2164-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪.Varley KE, Gertz J, Bowling KM, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23(3):555–567. doi: 10.1101/gr.147942.112. Comparisons of methylomes (reduced representation bisulfite sequencing profiles) of many normal samples and cancer cell lines provide evidence of a context-dependent relationship of DNA methylation to gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7▪▪.Tsumagari K, Baribault C, Terragni J, et al. Early de novo DNA methylation and prolonged demethylation in the muscle lineage. Epigenetics. 2013;8(3):317–332. doi: 10.4161/epi.23989. Differentially methylated CpG sites were mapped by reduced representation bisulfite sequencing for myogenic progenitor cells and skeletal muscle tissue versus a large variety of non-muscle samples and provided evidence for active demethylation in the skeletal muscle lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8▪▪.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. Genome-wide histone modifications and CTCF binding were mapped from nine cell types to predict cis-regulatory elements for transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19(6):541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Hu G, Cui K, Northrup D, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12(2):180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le May N, Fradin D, Iltis I, Bougneres P, Egly JM. XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Mol Cell. 2012;47(4):622–632. doi: 10.1016/j.molcel.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 13.Xi H, Shulha HP, Lin JM, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3(8):e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA, Wolkowicz MJ, Harrington MA, Gonzales F. Methylation and expression of the Myo D1 determination gene. Philos Trans R Soc Lond B Biol Sci. 1990;326(1235):277–284. doi: 10.1098/rstb.1990.0011. [DOI] [PubMed] [Google Scholar]
- 15.Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: involvement of RNA. Cell. 1996;86(5):709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich M, Ehrlich K. Effects of DNA methylation on the binding of vertebrate and plant proteins to DNA. In: Jost JP, Saluz HP, editors. DNA Methylation: Biological Significance. Birkhauser Verlag; MA, USA: 1993. pp. 145–168. [DOI] [PubMed] [Google Scholar]
- 17.Rishi V, Bhattacharya P, Chatterjee R, et al. CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proc Natl Acad Sci USA. 2010;107(47):20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oikawa Y, Omori R, Nishii T, Ishida Y, Kawaichi M, Matsuda E. The methyl-CpG-binding protein CIBZ suppresses myogenic differentiation by directly inhibiting myogenin expression. Cell Res. 2011;21(11):1578–1590. doi: 10.1038/cr.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao Y, Xi S, Briones V, Muegge K. Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS One. 2011;5(2):e9163. doi: 10.1371/journal.pone.0009163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):S245–S254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 21.Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 22.Rivera RM, Ross JW. Epigenetics in fertilization and preimplantation embryo development. Prog Biophys Mol Biol. 2013 doi: 10.1016/j.pbiomolbio.2013.02.001. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 23.Ehrlich M, Gama-Sosa M, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 25.Patel DR, Richardson BC. Dissecting complex epigenetic alterations in human lupus. Arthritis Res Ther. 2013;15(1):201. doi: 10.1186/ar4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huidobro C, Fernandez AF, Fraga MF. The role of genetics in the establishment and maintenance of the epigenome. Cell Mol Life Sci. 2013;70(9):1543–1573. doi: 10.1007/s00018-013-1296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153(4):773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weaver JR, Sarkisian G, Krapp C, Mager J, Mann MR, Bartolomei MS. Domain-specific response of imprinted genes to reduced DNMT1. Mol Cell Biol. 2010;30(16):3916–3928. doi: 10.1128/MCB.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao Y, Xi S, Shan J, et al. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc Natl Acad Sci USA. 2011;108(14):5626–5631. doi: 10.1073/pnas.1017000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, Pan Q, Lin L, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21(26):5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146(6):866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sartorelli V, Juan AH. Sculpting chromatin beyond the double helix: epigenetic control of skeletal myogenesis. Curr Top Dev Biol. 2011;96:57–83. doi: 10.1016/B978-0-12-385940-2.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sousa-Victor P, Munoz-Canoves P, Perdiguero E. Regulation of skeletal muscle stem cells through epigenetic mechanisms. Toxicol Mech Methods. 2011;21(4):334–342. doi: 10.3109/15376516.2011.557873. [DOI] [PubMed] [Google Scholar]
- 35.Constantinides PG, Jones PA, Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267(5609):364–366. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- 36.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47(5):649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 37.Szyf M, Rouleau J, Theberge J, Bozovic V. Induction of myogenic differentiation by an expression vector encoding the DNA methyltransferase cDNA sequence in the antisense orientation. J Biol Chem. 1992;267(18):12831–12836. [PubMed] [Google Scholar]
- 38.Zhang F, Pomerantz JH, Sen G, Palermo T, Blau HM. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc Natl Acad Sci USA. 2007;104(11):4395–4400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hupkes M, Jonsson MK, Scheenen WJ, et al. Epigenetics: DNA demethylation promotes skeletal myotube maturation. FASEB J. 2011;25(11):3861–3872. doi: 10.1096/fj.11-186122. [DOI] [PubMed] [Google Scholar]
- 40.Hupkes M, van Someren EP, Middelkamp SH, Piek E, van Zoelen EJ, Dechering KJ. DNA methylation restricts spontaneous multi-lineage differentiation of mesenchymal progenitor cells, but is stable during growth factor-induced terminal differentiation. Biochim Biophys Acta. 2011;1813(5):839–849. doi: 10.1016/j.bbamcr.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Rosca AM, Burlacu A. Effect of 5-azacytidine: evidence for alteration of the multipotent ability of mesenchymal stem cells. Stem Cells Dev. 2011;20(7):1213–1221. doi: 10.1089/scd.2010.0433. [DOI] [PubMed] [Google Scholar]
- 42.Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Farthing CR, Ficz G, Ng RK, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4(6):e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 45.Pekowska A, Benoukraf T, Zacarias-Cabeza J, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30(20):4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurent L, Wong E, Li G, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20(3):320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toth M, Lichtenberg U, Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci USA. 1989;86(10):3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Appanah R, Dickerson DR, Goyal P, Groudine M, Lorincz MC. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet. 2007;3(2):e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50▪.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–368. doi: 10.1038/nbt.1533. Methylome analyses on human brain cortex DNA provide evidence that DNA methylation regulates intragenic promoter activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci USA. 2009;106(3):671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Illingworth R, Kerr A, Desousa D, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6(1):e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuen RK, Neumann SM, Fok AK, et al. Extensive epigenetic reprogramming in human somatic tissues between fetus and adult. Epigenet Chromatin. 2011;4:7. doi: 10.1186/1756-8935-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isagawa T, Nagae G, Shiraki N, et al. DNA methylation profiling of embryonic stem cell differentiation into the three germ layers. PLoS One. 2011;6(10):e26052. doi: 10.1371/journal.pone.0026052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berdasco M, Melguizo C, Prados J, et al. DNA methylation plasticity of human adipose-derived stem cells in lineage commitment. Am J Pathol. 2012;181(6):2079–2093. doi: 10.1016/j.ajpath.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 56.Wang T, Chen M, Liu L, et al. Nicotine induced CpG methylation of Pax6 binding motif in StAR promoter reduces the gene expression and cortisol production. Toxicol Appl Pharmacol. 2011;257(3):328–337. doi: 10.1016/j.taap.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venza I, Visalli M, Fortunato C, et al. PGE2 induces interleukin-8 derepression in human astrocytoma through coordinated DNA demethylation and histone hyperacetylation. Epigenetics. 2012;7(11):1315–1330. doi: 10.4161/epi.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13(10):R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28(22):2986–2988. doi: 10.1093/bioinformatics/bts545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsumagari K, Chang SC, Lacey M, et al. Gene expression during normal and FSHD myogenesis. BMC Med Genomics. 2011;4:67. doi: 10.1186/1755-8794-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 63.Palacios D, Summerbell D, Rigby PW, Boyes J. Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse Myogenin gene. Mol Cell Biol. 2010;30(15):3805–3815. doi: 10.1128/MCB.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faralli H, Dilworth FJ. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp Funct Genomics. 2012;2012:836374. doi: 10.1155/2012/836374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gimble JM, Dorheim MA, Youkhana K, et al. Alternatively spliced pp52 mRNA in nonlymphoid stromal cells. J Immunol. 1993;150(1):115–121. [PubMed] [Google Scholar]
- 66.Jongstra-Bilen J, Jongstra J. Leukocyte-specific protein 1 (LSP1): a regulator of leukocyte emigration in inflammation. Immunol Res. 2006;35(1–2):65–74. doi: 10.1385/IR:35:1:65. [DOI] [PubMed] [Google Scholar]
- 67▪.Cao Y, Yao Z, Sarkar D, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18(4):662–674. doi: 10.1016/j.devcel.2010.02.014. Shows that in mouse C2C12 myoblasts and myotubes, Myod binds to approximately 60,000 sites throughout the genome, including many nonpromoter regions of genes that do not show differential expression in myogenic progenitor cells, implying a major reshaping of the epigenome in myoblasts and myotubes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs’ target genes and functions. Nat Methods. 2008;5(4):347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang LH, Wang R, Gama-Sosa MA, Shenoy S, Ehrlich M. A protein from human placental nuclei binds preferentially to 5-methylcytosine-rich DNA. Nature. 1984;308:293–295. doi: 10.1038/308293a0. [DOI] [PubMed] [Google Scholar]
- 71.Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20(4):1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lucarelli M, Fuso A, Strom R, Scarpa S. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J Biol Chem. 2001;276(10):7500–7506. doi: 10.1074/jbc.M008234200. [DOI] [PubMed] [Google Scholar]
- 73▪▪.Tsumagari K, Baribault C, Terragni J, et al. DNA methylation and differentiation: HOX genes in muscle cells. Epigenet Chromatin. 2013;6:25. doi: 10.1186/1756-8935-6-25. Describes several of the examples of myogenic differentially methylated regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soshnikova N, Duboule D. Epigenetic regulation of vertebrate Hox genes: a dynamic equilibrium. Epigenetics. 2009;4(8):537–540. doi: 10.4161/epi.4.8.10132. [DOI] [PubMed] [Google Scholar]
- 75.Simrick S, Szumska D, Gardiner JR, et al. Biallelic expression of Tbx1 protects the embryo from developmental defects caused by increased receptor tyrosine kinase signaling. Dev Dyn. 2012;8:1310–1324. doi: 10.1002/dvdy.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76▪▪.Yu DH, Ware C, Waterland RA, et al. Developmentally programmed 3′ CpG island methylation confers tissue- and cell-type specific transcriptional activation. Mol Cell Biol. 2013;33(9):1845–1858. doi: 10.1128/MCB.01124-12. Over-representation of differentially methylated CpG islands at the 3′ ends of genes, many of which overlap with chromatin immunoprecipitation sequencing detected CTCF binding sites only in pluripotent embryonic stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ziebarth JD, Bhattacharya A, Cui Y. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res. 2013;41(Database issue):D188–D194. doi: 10.1093/nar/gks1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA. 2010;107(8):3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gelfman S, Cohen N, Yearim A, Ast G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013;23(5):789–799. doi: 10.1101/gr.143503.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anastasiadou C, Malousi A, Maglaveras N, Kouidou S. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol. 2011;30(5):267–275. doi: 10.1089/dna.2010.1094. [DOI] [PubMed] [Google Scholar]
- 81.Oberdoerffer S. A conserved role for intragenic DNA methylation in alternative pre-mRNA splicing. Transcription. 2012;3(3):106–109. doi: 10.4161/trns.19816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82▪.Bell ML, Buvoli M, Leinwand LA. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol Cell Biol. 2010;30(8):1937–1945. doi: 10.1128/MCB.01370-09. Describes evidence for muscle-associated alternative splicing generating predominantly an intragenically encoded miRNA rather than a protein-coding mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sasaki YT, Sano M, Kin T, Asai K, Hirose T. Coordinated expression of ncRNAs and HOX mRNAs in the human HOXA locus. Biochem Biophys Res Commun. 2007;357(3):724–730. doi: 10.1016/j.bbrc.2007.03.200. [DOI] [PubMed] [Google Scholar]
- 84.Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA. 2007;13(2):223–239. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11(11):1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
Website
- 101.UCSC Genome Bioinformatics. http://genome.ucsc.edu.