Abstract
Throughout the developing world, urban centers with sprawling slum settlements are rapidly expanding and invading previously forested ecosystems. Slum communities are characterized by untended refuse, open sewers, and overgrown vegetation, which promote rodent infestation. Norway rats (Rattus norvegicus), are reservoirs for epidemic transmission of many zoonotic pathogens of public health importance. Understanding the population ecology of R. norvegicus is essential to formulate effective rodent control strategies, as this knowledge aids estimation of the temporal stability and spatial connectivity of populations. We screened for genetic variation, characterized the population genetic structure, and evaluated the extent and patterns of gene flow in the urban landscape using 17 microsatellite loci in 146 rats from 9 sites in the city of Salvador, Brazil. These sites were divided between three neighborhoods within the city spaced an average of 2.7 km apart. Surprisingly, we detected very little relatedness among animals trapped at the same site and found high levels of genetic diversity, as well as structuring across small geographic distances. Most FST comparisons among sites were statistically significant, including sites <400 m apart. Bayesian analyses grouped the samples in three genetic clusters, each associated with distinct sampling sites from different neighborhoods or valleys within neighborhoods. These data indicate the existence of complex genetic structure in R. norvegicus in Salvador, linked to the heterogeneous urban landscape. Future rodent control measures need to take into account the spatial and temporal linkage of rat populations in Salvador, as revealed by genetic data, to develop informed eradication strategies.
Keywords: population genetics, urban ecology, Rattus norvegicus, rodent control
INTRODUCTION
Norway rats (Rattus norvegicus) are geographically widespread, reaching maximal population densities in urban centers where abundant food resources and harborage are present (Glass et al. 1989). This commensal species is responsible for damage to the environment, infrastructure, and agriculture (Macdonald et al. 1999; Singleton et al. 1999; Howald et al. 2007), causing significant financial costs across different ecological settings. In addition to their impact as a pest, Norway rats are reservoir hosts for the transmission many infectious diseases (Webster & Macdonald 1995; Himsworth et al. 2013), including leptospirosis (Ko et al. 2009), bartonellosis (Kosoy et al. 2010), bubonic plague (Chanteau et al. 1998), murine typhus (Civen & Ngo 2008), and salmonellosis (Nkogwe et al. 2011).
The dramatic growth of urban slum settlements, most of which has occurred in tropical regions of the world in resource poor countries over the past 50 years, has transformed the urban ecology creating new habitats for Norway rats. To date more than one billion of the worlds’s inhabitants reside in slum settlements or shantytowns. This population will double by 2025 (UN-HABITAT, 2010). In slum communities lack of access to services and poor housing and sanitation, promote infestation by R. norvegicus (Glass et al. 1989, 2005; Childs et al. 1991), promoting conditions for epidemic transmission of infectious diseases in humans (Ko et al. 1999). Leptospirosis, a spirochetal infection, which causes severe life-threatening disease in humans, has emerged as a cause of large epidemics in urban slum communities throughout the developing world (Ko et al. 1999, 2009; Sarkar et al. 2002; Amilasan et al. 2012; Dechet et al.2012). In these outbreaks, R. norvegicus, as well as R. rattus, are the main reservoirs for human transmission (Pereira et al. 2000; Faria et al. 2008; Matthias et al. 2008).
Despite the ecological and public health impacts of Norway rats in shantytowns worldwide, there is a paucity of data describing the population dynamics of Norway rats in urban landscapes. Ecological studies evaluating rodent demography have been limited to temperate urban environments of the Northern Hemisphere, most notably in Baltimore, Maryland, USA, where rodent ecological research has been continually conducted for over a half-century (Davis, Emlen, & Stokes 1948; Davis 1953; Orgain & Schein 1953; Calhoun 1962; Farhang-Azad & Southwick 1979; Glass et al. 1989; Childs et al. 1991; Glass et al. 2005). To date only one study has used a population genetic approach on urban R. norvegicus populations (Gardner-Santana et al. 2009), while a handful of others have looked at island invasion dynamics, mostly in R. rattus (Calmet et al. 2001; Abdelkrim et al. 2005; Russell et al. 2009b; Russell et al. 2010). It is unclear how applicable results from studies of rat population dynamics in temperate cities in developed countries will be to populations inhabiting urban slums, especially those situated in tropical environments.
Microsatellite loci have been used to study disease vector movements and help define eradication units consisting of discrete populations (Slotman et al. 2006; Gilabert et al. 2007; McCoy 2008; Solano et al.2010; Beadell et al. 2010; Brown et al. 2011) for vectors such as insect and rodent reservoirs of zoonotic pathogens. Previous studies involving R. norvegicus in temperate environments indicate that microsatellite loci provide a valuable tool for assessing population genetic structure at spatial scales of several kilometers (Robertson & Gemmell 2004; Russell et al. 2009b; Adelkrim et al. 2007, 2010). However, microsatellite-based approaches have not been applied to investigate R. norvegicus populations at the finer spatial resolution required to elucidate genetic structure and gene flow to inform delineation of eradication units for this reservoir host.
Herein we present the findings of a study, which investigated the population genetic structure of R. norvegicus from slum settlements (favelas) within Salvador, situated in Northeast Brazil. This urban center has undergone recent rapid growth and expansion which has led to a ~280% increase in its population since 1970 (Brazilian Institute of Geography and Statistics) and establishment of densely populated slum settlements in formerly-uninhabited, Atlantic forested regions of the city (Reis et al. 2008). At present, 58% of the population of Salvador resides in favelas (Moreira & Pereira 2008). Large outbreaks of leptospirosis occur in the same favela communities each year during seasonal periods of heavy rainfall (Ko et al. 1999; Sarkar et al. 2002; Reis et al. 2008). In Salvador R. norvegicus is the principal reservoir for leptospirosis (Ko et al. 1999; Pereira et al. 2000; Sarkar et al. 2002; Reis et al. 2008; Faria et al. 2008), as in all major urban centers in Brazil (Pereira & Andrade 1988; Pereira et al. 2000).
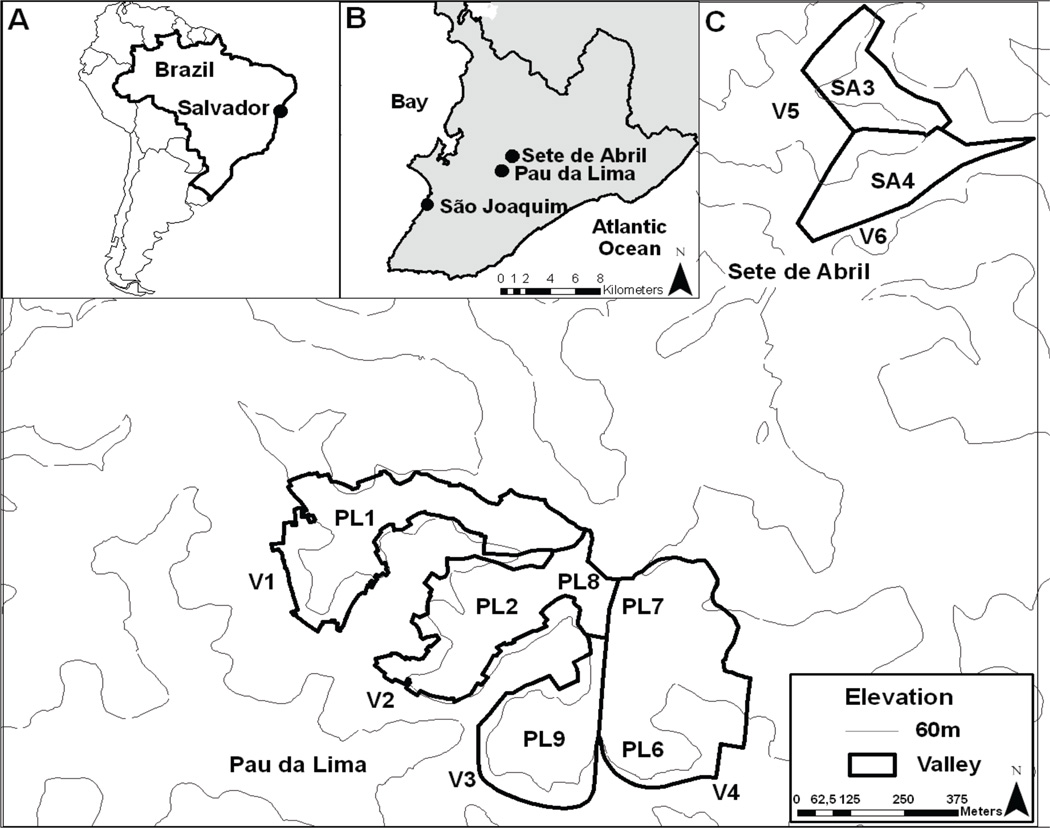
We captured Norway rats from nine sites located at the base of the valleys of three different neighborhoods, each of which had a documented history of ongoing rodent infestations and moderate to high annual incidence of leptospirosis (Fig. 1). Since most sites were only a few hundred meters apart, we had the opportunity to examine the patterns of population genetic structure at a very fine spatial resolution. Our goal was to identify a panel of microsatellite loci, which would generate robust inferences at micro-spatial scales, and use them to quantify the genetic variation at each site, evaluate the patterns of genetic differentiation within and between sites, and assess the extent of genetic admixture between sites. Because R. norvegicus dispersal and re-colonization following eradication helps sustain population abundance over time, understanding these dynamics will inform rodent control efforts by elucidating the spatial and temporal scope of eradication units and identifying potential barriers or corridors for dispersal.
Figure 1.
Map of Salvador, Brazil showing rat sampling locations. A. Map of South America identifying Brazil and the city of Salvador. B. Boundaries of Salvador city (dark outline) and neighborhoods where rodent capture was performed (dark dots). C. Topographic map of Sete de Abril and Pau da Lima valleys (black outlines, named as V1, V2, V3, V4 and V5) and rodent trapping sites (PL1, PL2, PL8, PL6-9, SA3 and SA4) in 2010 and 2011.
MATERIALS AND METHODS
Study Area and Sampling
Salvador is a Brazilian city of about 2.7 million residents (Brazilian Institute of Geography and Statistics 2011) spread over 313 km2 (12° 55' 34" southern latitude and 38° 31' 12" western longitude; Fig. 1). The city has a subtropical climate characterized by average annual temperatures (the mean of the 365 days of each year) ranging from 21– 37°C (mean 25.3°C), and heavy rainfall during winter. The three neighborhoods selected for study included two residential slum areas, Pau da Lima and Sete de Abril, with documented high annual incidence of leptospirosis cases (in 2010, 10.4 and 23.3 per 100,000 people, respectively, (Costa, personal communication). The third neighborhood, São Joaquim, although experiencing lower incidence of leptospirosis, was included to broaden the spatial extent of the study (Fig. 1), and because of its location near the city port offers the potential sampling of rats originating from different geographic regions. The two residential slum communities include a series of valleys (mean valley depth 60 m), where the highest human population density and worst slum conditions are concentrated below 30 m from the valley floor.
Six sampling sites (PL1, PL2, PL6, PL7, PL8, PL9) were located at the base of four valleys (V1-V4) in Pau de Lima and two additional sites (SA3 and SA4) in two valleys (V5-V6) were located in Sete de April (Table 1, Fig. 1). The distances between sampling sites averaged 2.7 km, although most were only 300 m-1.7 km apart (Table S1). Inter-site distances fall within the documented range of R. norvegicus migration/dispersal distances of for both urban (25–150 m, Davis et al. 1948, Glass et al. 1989, Traweger et al. 2006) and rural settings (260–2000 m, Taylor 1978, Taylor & Quy 1979, Macdonald & Fenn 1995). The ninth sampling site (SJ5) was located at a local market area in a valley (V7) located in São Joaquim.
Table 1.
Number of individuals (N) sampled per year at each collection site. Refer to Figure 1 for site locations. N refers to sample size for each site (year of collection in parenthesis), The last two columns list for each sample the number of males and female and their age class. (1= juveniles,2= sexually mature young adult; and 3 = large adult).
| Neighborhood | Site | Valley | N (2010) | N (2011) | N (Total) | Male/Female | Age Class Composition (1/2/3) |
|---|---|---|---|---|---|---|---|
| Pau da Lima | PL1 | V1 | 15 | 12 | 27 | 14/13 | 8/14/5 |
| Pau da Lima | PL2 | V2 | 15 | 15 | 30 | 16/14 | 7/20/3 |
| Pau da Lima | PL6 | V4 | 3 | 15 | 18 | 10/8 | 9/7/2 |
| Pau da Lima | PL7 | V4 | 3 | - | 3 | 2/1 | -/3/- |
| Pau da Lima | PL8 | V2 | 7 | 16 | 23 | 13/10 | 7/9/7 |
| Pau da Lima | PL9 | V3 | - | 11 | 11 | 7/4 | 7/4/- |
| Sete de Abril | SA3 | V5 | 32 | - | 32 | 14/18 | 10/17/5 |
| Sete de Abril | SA4 | V6 | 3 | - | 3 | 2/1 | 1/-/2 |
| São Joaquim | SJ5 | V7 | 3 | - | 3 | 2/* | -/2/1 |
| Total | - | - | 81 | 69 | 150 | - | - |
The sex of one individual from SJ5(V7) is unknown.
In both 2009 and 2010 between January and April, the months immediately preceding the wet season (May-July), the annual peak of leptospirosis cases, rodenticide control was implemented in the four Pau de Lima valleys (V1-V4), which include six of the sampling sites in this study (PL1, PL2, PL6, PL7, PL8, PL9). In these sites R. norvegicus was by far the predominant rodent, infesting 85% of rodent-infested households. Approximately 70,000 houses were treated with both chemical intervention (second generation anticoagulants as brodifacoum, bromadiolone and coumatetralyl) and environmental and educational activities to eliminate possible rat habitats and make the resident community aware of how to reduce exposure to the pathogen. Following this rodenticide control there was a decrease in the proportion of rodent-infested houses in both years, from 25.3% to 7.8% (2009) and from 23.2% to 14.6% (2010; Costa, personal communication).
Sampling was carried out in June, July, and August of 2010 and 2011, using either live Tomahawk traps (2010) or snap traps (2011). Three to five traps were placed at each of six to eight contiguous households defining an individual site. Each site contained multiple, separate burrow systems, presumably containing colonies of rats of varying kinship (Calhoun 1962). Traps were set before sundown and collected at sunrise the following morning. Because rats are known carriers of Seoul virus (genus Hantavirus), all procedures were conducted outdoors using approved biosafety measures (Mills et al. 1995). After double bagging and transport to an outdoor processing area live-trapped rats were euthanized with thiopental solution then weighed, sexed, and 1cm of tail tissue was removed. In total, 150 tail tips were collected and stored at −80°C until tested. Table 1 lists the samples obtained by year and site together with capture effort, sex, and age class. Body mass was used as a proxy for age and rats were classified into in 3 groups (1 = <200g, juveniles, non breeding; 2 = 201–400g, young adults, breeding; 3 = >400g, adults, breeding and senescent). Body mass has been shown to be a reliable indicator of age and reproductive maturity (Farhang-Azad & Southwick 1979; Glass et al. 1989; Iskajer et al. 1989; Porter et al., in preparation).
Genetic Methods
DNA was extracted from 1–3 mm of tail tips using the prepGEM Tissue kit (ZyGEM Corp Ltd, Hamilton New Zealand) according to the manufacturer’s protocol.
Following a literature review of studies utilizing microsatellites in Rattus, 30 microsatellite loci were selected based on high allelic richness, absence of null alleles, lack of deviation from Hardy-Weinberg equilibrium (HWE), and no evidence of linkage to other loci in the set. All microsatellite primer sequences and references are given in the supplementary material (Table S2). A touchdown PCR (5 min. of initial denaturation at 95°C; 10 cycles of denaturing at 95°C, annealing at temperatures from 62°C to 53°C, decreasing by 1°C per cycle, and extension at 72°C; 40 cycles of 95°C, 52°C and 72°C; and a final 20 min. extension at 72°C) in 13.5 µL reactions containing 0.4 µM fluorescently labeled forward primer (6-FAM and HEX, Sigma-Aldrich), 0.4 µM reverse primer (Eurofins Mwg/Operon), 1X buffer (Promega), 0.8 mM dNTPs (NEB), 2.0 mM MgCl2 (Promega), 0.2 mg/mL BSA (NEB), and 0.5 U GoTaq polymerase (Promega) was used on a subset of individuals (12–14) to optimize amplification and assess the individual locus contribution out of the full complement of 30 loci to population genetic variation. PCR amplicons were analyzed on an ABI 3730xl automated sequencer. PCR products were loaded in groups of two loci with non-overlapping size ranges with a GS600 LIZ size standard. Alleles were scored using the program GeneMarker v 1.95 (SoftGenetics, State College, PA) with manual editing of the automatically scored peaks.
Of the 30 microsatellite loci tested, 17 were selected to screen all individuals based on high variability (4–7 alleles/locus) and reliable amplification. All 17 microsatellite loci chosen for analysis were tested for within-population deviations from HWE and all pairs of loci across populations were tested for linkage disequilibrium (LD) using the software Arlequin 3.5 (Excoffier & Lischer 2010). Markov chain parameters were set at 10,000 dememorizations, 1,000 batches, and 10,000 iterations per batch for both HWE and LD. Significance values were adjusted using the sequential Bonferroni method (Holm 1979).
Genetic Diversity and Population Structure
Arlequin 3.5 (Excoffier & Lischer 2010) was used to calculate summary statistics including observed (Ho) and expected (He) heterozygosities and allele frequencies for all loci in all populations. Allelic richness (Ar) was calculated with rarefaction correction for sample size in the program HP-Rare v 1.0 (Kalinowski 2004, 2005), and multilocus averages of allelic richness and private (unique to a population) allelic richness were used to examine genotypic diversity differences between sites.
Genetic differentiation among populations was assessed by pairwise FST in the program Arlequin 3.5 (Excoffier & Lischer 2010). The significance of these FST values was tested with 10,000 permutations. To determine which hierarchical level of population structure (neighborhood, valley, site, and nested combinations thereof) explained the largest significant proportion of variance in allele frequencies, we performed an analysis of molecular variance (AMOVA) in Arlequin 3.5 (Excoffier & Lischer 2010). The AMOVA was run using 4 different groupings of individuals: (a) by site, (b) by valley, (c) by site within neighborhood, and (d) by valley within a neighborhood. Prior to grouping samples from the same site from two different years we used AMOVA and pairwise FST to test for significant temporal differentiation between samples originating from rats captured at the same site. Although little is known about the generation times of Norway rats in tropical climates, as many as 4–8 generations could have passed between the two samplings in 2010 and 2011, as Norway rats have several litters per year (Davis 1953).
Using the Bayesian clustering method implemented in STRUCTURE v2.3 (Pritchard et al. 2000), patterns of population structure and individual assignment to sampling localities were tested by identifying genetic clusters without using a priori sampling information (details in supplementary material). To identify the most likely number of clusters (K), three independent runs for each K = 1–15 were carried out. For all runs, the admixture model with independent allele frequencies was used with a burn-in value of 10,000 steps followed by 100,000 replications. The most likely K was determined by following the guidelines of Prichard et al.2000, and using the second order rate of change in the likelihood distribution (ΔK, Evanno et al. 2005). Discriminant analysis of principal components (DAPC, Jombart et al. 2010) was also used to assess genetic clustering. DAPC is a two-step multivariate ordination analysis that makes no assumptions about deviations from HWE and LD, unlike the Bayesian approach implemented in STRUCTURE, which assumes HWE and linkage equilibrium (details in supplementary material).
Furthermore, because many trap sites were separated by distances compatible with Norway rat migration capabilities, patterns of genetic differentiation were examined using a Mantel test of isolation by distance (IBD) implemented in Genepop (Raymond & Rousset 1995; Rousset 2008), comparing the linearized pairwise FST values with the Euclidean distances between capture sites using 10,000 permutations. Due to the noncontiguous nature of our sampling, analyses were run using only the 6 sites within the Pau da Lima neighborhood.
Relatedness, Dispersal, and Migration
To determine if relatedness of individuals within sampling sites influenced genetic divergence patterns between those sites, we estimated relatedness through maximum likelihood methods and examined relationships for all individual pairs using ML-Relate (Kalinowski et al. 2006), running the analyses only for sites with six or more individuals. Additionally, Bayesian approaches implemented in STRUCTURE (Prichard et al. 2000) and GENECLASS 2.0 (Piry et al. 2004) were used to identify first generation migrants (individuals putatively born at a site different from their capture site). We reran STRUCTURE (Prichard et al. 2000) with the USEPOPINFO option (i.e. prior sample site information) for each individual included in the analysis. Burn-in and run lengths were the same as above, and migration rate was set at 0.05. STRUCTURE detects migrants by quantifying individual membership to a site and cluster by calculating Q values, the proportion of that individual's ancestry from a population, for each site in each individual. If an individual is a putative migrant, the Q value for its site of origin/capture (Qo) is significantly lower than the Q value for its site of assignment (Qa). In Geneclass 2.0 we utilized the ‘detect migrants’ function, which, in combination with Bayesian resampling methods, calculates likelihood-based statistics, including Lh, the likelihood of finding an individual in the population in which it was trapped, and Lmax, the maximum likelihood of assignment to any sampled population. We used both Lh and Lh/Lmax to detect first generation migrants. Lh is the most appropriate statistic to use when all potential source populations have not been sampled. Lh lacks power, however, compared to other estimators, so we also used the Lh/Lmax ratio, despite it being more informative when all source populations have been sampled (Paetkau et al. 2004). Because migrants from unsampled populations can be misclassified as residents, we selected the Rannala & Mountain (1997) criterion with the resampling method of Paetkau et al. (2004) to determine the critical value of Lh and Lh/Lmax, using 1,000 simulated individuals and the default α = 0.01.
We used Fstat 2.9.3 (Goudet 1995) to test whether the observed population structure could be attributed to differences in dispersal between the sexes and calculated four different statistics and their significance values: FST, mAIc and vAIc (the corrected mean Assignment Index and its variance, Favre et al. 1997), and mPr (mean pairwise relatedness). The estimates were obtained for the full dataset excluding the poorly sampled localities (N<4 rats) of PL7(V4), SA4(V6), SJ5(V7) (details in supplementary material).
Population Size (NE and NC)
We calculated both effective (NE) and census (NC) population sizes for the three genetic clusters identified by the clustering analysis, under the assumption that they form units within which random mating occurs. NE was computed using Waples & Do’s linkage disequilibrium method implemented in LDNe (Waples & Do 2008). Since linkage disequilibrium is affected by migration, the low migration detected between genetic clusters should reduce the bias in the NE estimation. NC was computed based on NE estimates and the ratio of the realized (observed genotypes) and possible (expected genotypes) diploid combinations of all sampled alleles (Hyseni et al. 2012). This analysis used a Bayesian method (Petit & Valiere 2006) adapted from Gazey & Staley (1986; details in supplementary material)
RESULTS
Marker Validation, Genetic Diversity, and Relatedness
We successfully genotyped 146 out of 150 samples across 17 microsatellite loci. After sequential Bonferroni corrections for multiple tests, only one locus, D8Mgh7, from one sampling site, PL1(V1), showed a significant probability of deviation from HWE, and no pairwise comparisons of loci across sampling sites showed significant signatures of linkage disequilibrium (LD). This reaffirmed our choice of loci from previous studies based on HWE and absence of linkage (data available on request).
Both the AMOVA (Table S3) and the FST (Table S4) results on the temporally separated samples obtained from the same sites suggested that the 2010 and 2011 collections were not significantly different with the exception of the PL8(V2) site (FST = 0.13, p < 0.05). Thus, diversity statistics were calculated by site, with temporal collections combined for each site, except for the PL8(V2) site. Genetic variation was high, despite the small spatial scale of the sampling effort and the small sample sizes from a few sites (Table 2). Corrected allelic richness (Ar) ranged from 2.71–3.29, while private (unique to a population) allelic richness (Ap) ranged from 0.06–0.30, and He ranged from 0.57–0.72 (Table 2).
Table 2.
Genetic diversity by site for 146 individuals. The first two columns report the sampling sites with the respective valley in parentheses and samples size (N). Measures include allelic richness (Ar) and private allelic richness (Ap) averaged over all loci, and mean observed (Ho) and expected (He) heterozygosity.
| Site (Valley) | N | Ar | Ap | Ho | He |
|---|---|---|---|---|---|
| PL1(V1) | 25 | 3.29 | 0.17 | 0.67 | 0.70 |
| PL2(V2) | 30 | 3.05 | 0.10 | 0.67 | 0.68 |
| PL6(V4) | 18 | 3.12 | 0.06 | 0.65 | 0.69 |
| PL7(V4) | 3 | 3.00 | 0.12 | 0.62 | 0.72 |
| PL8_2010(V2) | 7 | 3.07 | 0.22 | 0.61 | 0.67 |
| PL8_2011(V2) | 16 | 2.99 | 0.11 | 0.70 | 0.67 |
| PL9(V3) | 11 | 3.07 | 0.11 | 0.67 | 0.69 |
| SA3(V5) | 30 | 3.00 | 0.14 | 0.63 | 0.64 |
| SA4(V6) | 3 | 2.71 | 0.21 | 0.55 | 0.57 |
| SJ5(V7) | 3 | 2.71 | 0.30 | 0.71 | 0.59 |
Results of the relatedness analyses within each site with at least 11 samples per site, indicated that most of the samples (87%) came from unrelated individuals with approximately 12% sharing a half-sibling relationship and ~1% being either full siblings or parent-offspring (Table 3). To further explore this result we repeated the analysis on a subset of samples from 4 households in Pau da Lima with samples size > 6 where traps were placed close to a single burrow (Table 3). Even for this subset of samples that was more likely to include family groups than the whole data set, most (> 90%) of trapped individuals, comprised of adult and young rats of both sexes, were unrelated (Table 3).
Table 3.
Percent of pairwise comparisons of individuals that fell into each relatedness category (e.g. unrelated, half sibling, full sibling) as calculated in ML-Relate (Kalinowski et al. 2006). Comparisons were made among all individuals, and among individuals within sites and households with N > 6.
| All Sites |
PL1 (V1) |
PL2 (V2) |
PL6 (V4) |
PL8 (V2) |
PL9 (V3) |
SA3 (V5) |
House 5 (PL1) |
House 15 (PL2) |
House 16 (PL9) |
House 19 (PL8) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Unrelated | 87.1% | 92.0% | 87.6% | 92.2% | 85.4% | 89.1% | 82.8% | 100% | 90.5% | 95.2% | 90.9% |
| Half Sibling | 11.6% | 7.3% | 9.2% | 6.5% | 9.1% | 7.3% | 12.4% | 0.0% | 4.8% | 4.8% | 6.1% |
| Full Sibling | 1.1% | 0.7% | 2.1% | 0.0% | 3.9% | 3.6% | 2.5% | 0.0% | 4.8% | 0.0% | 1.5% |
|
Parent- Offspring |
0.2% | 0.0% | 1.1% | 1.3% | 1.6% | 0.0% | 2.3% | 0.0% | 0.0% | 0.0% | 1.5% |
Genetic Differentiation, Sex-biased Dispersal, and Population Sizes
The AMOVA analyses show that most of the variation was attributable to intra-site differences (89.4%) with inter-site variation contributing 8.2% (Table 4). The majority of the pairwise FST values for inter-site comparisons were also significantly different and ranged from 0.06 to 0.28 (Table 5S), even though the geographic distance between sampling sites was small, ranging from 79 m to a few hundred meters (Fig. 1). The four non-significant FST values were all associated with comparisons that included sites with small samples sizes (PL7(V4), SA4(V6), and SJ5(V7), N=3, Table 1, Table S5).
Table 4.
Analysis of molecular variance (AMOVA) results by neighborhood and collection site for all rat samples (geographic structure).
| Source of variation | Variance component | % of variation | p-value |
|---|---|---|---|
| Among groups (neighborhoods) | Va = 0.11 | 2.43 | 0.05 |
| Among sites within groups (sites) | Vb = 0.37 | 8.18 | 0.00 |
| Within sites (individuals) | Vc = 4.03 | 89.39 | 0.00 |
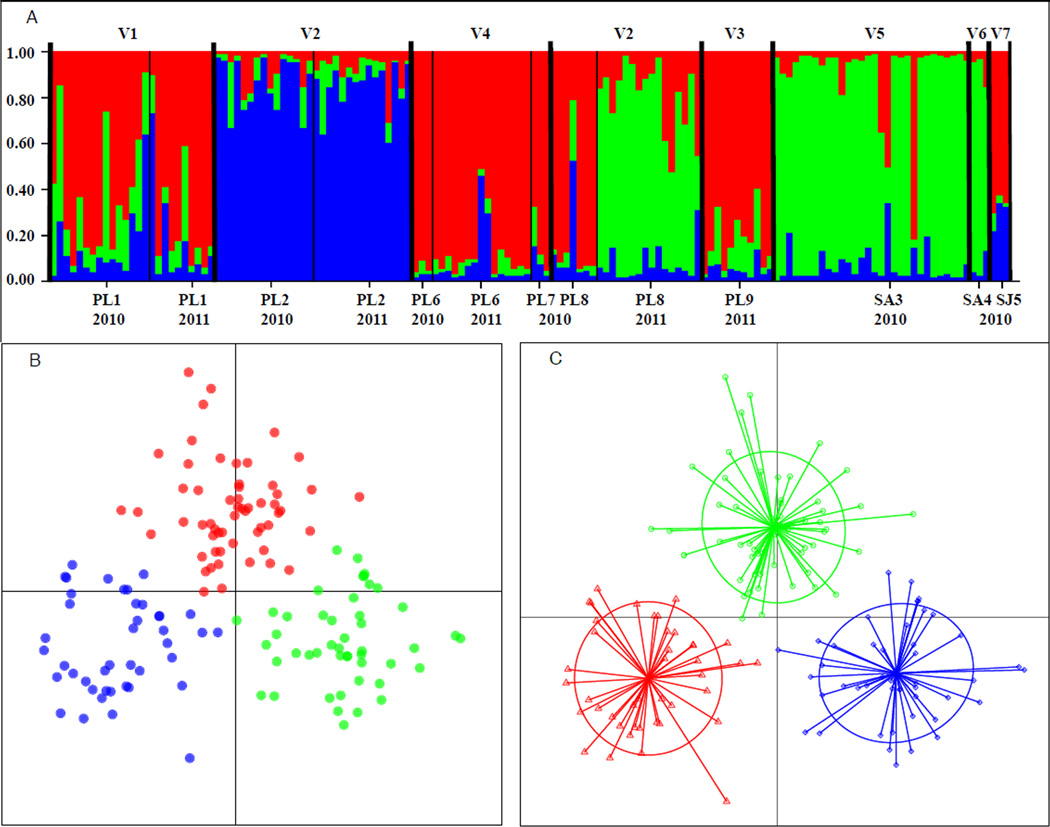
Bayesian clustering analyses evaluating ΔK (Evanno et al., 2005) identified three genetic clusters (Fig. 2A). Cluster 1 grouped several of the Pau da Lima sites (PL1(V1), PL6(V4), PL7(V4), PL8-2010(V2), and PL9(V3)) with the São Joaquim site, SL5(V7). Cluster 2 included only the two sets of samples obtained a year apart from Pau da Lima site 2 (PL2-2010(V2) and PL2-2011(V2)). Cluster 3 grouped the samples from PL8 obtained in 2011 with the two Sete de Abril sites (SA3(V5) and SA4(V6)) (Fig. 2A). The multivariate analyses (DAPC) also detected three genetic clusters with most individuals assigned with greater than 95% posterior probability to the same three groups identified by the clustering analysis (Fig. 2B, 2C). The Mantel test between pairwise Fst values and geographic distance sites did not detect any isolation by distance for the contiguous sites in Pau de Lima (p = 0. 957 > 0.05; Fig. S1).
Figure 2.
Genetic clusters and population structure. In all panels, Cluster 1 (red) = Pau da Lima sites 1, 6, 7, 8 (2010 only), and 9, Cluster 2 (blue) = Pau da Lima site 2, Cluster 3 (green) = Sete de Abril sites 3 and 4, Pau da Lima site 8 (2011 only), A) STRUCTURE bar plot (K=3) with individuals organized by site, year, and valley. Each vertical bar represents a single individual. The height of each color represents the probability of assignment to that genetic cluster, B) Principal Components Analysis, dots of different colors identify individuals from different genetic clusters, colors as in A, C) Discriminant Analysis of Principal Components with 3 K-means clusters. Circles indicate 95% confidence interval of assignment, dots identify individuals, colors of the individuals are as in A.
Table 5 shows the results from the migrant analyses. Although most individuals (93.2%) were correctly assigned to their site of capture, the two GENECLASS methods identified a total of ten migrants with the majority (9/10) being adults and all but one male. The Lh method detected six migrants, while the other GENECLASS method (Lh/Lmax) detected four of the same individuals plus 3 additional individuals (Table 5). Four of the migrants identified by the GENECLASS methods were also identified as migrants on the estimated membership coefficients for the capture site and assigned site (Qo and Qa, respectively) calculated using STRUCTURE (Table 5). These analyses also suggested that these four individuals have mixed ancestry (Qa from 0.358 to 0.875), possibly deriving from ancestral matings across multiple clusters.
Table 5.
First generation migrant analyses for all the sites and sampling times, except PL8(V2) which is separated by year. The first three columns report the individual identification number followed by its age class and sex. The following two colums show the origin and the assigned sites. The next two columns report the results of the GENECLASS 2.0 analyses, listing p-values for Lh and Lh/Lmax. The last two columns show the assignment values (Q) using STRUCTURE for the site of origin (Qo) and the assigned site (Qa). Individuals in bold were detected as first generation migrants by two or more methods.
| Sample | Age | Sex | Site (origin) | Site (assigned) | Lh p-value | Lh/Lmax p-value | Qo | Qa |
|---|---|---|---|---|---|---|---|---|
| 5 | 3 | M | PL8_2010(V2) | PL1(V1) | 0.005 | 0.003 | 0.110 | 0.358 |
| 62 | 3 | M | SA4(V6) | SA3(V5) | 0.001 | 0.002 | - | - |
| 47 | 3 | M | SA3(V5) | PL1(V1) | 0.000 | 0.000 | 0.000 | 0.875 |
| 9 | 3 | M | PL8_2010(V2) | PL9(V3) | 0.006 | 0.003 | 0.092 | 0.708 |
| 101 | 2 | M | SJ5(V7) | SJ5(V7) | 0.004 | - | - | - |
| 13 | 2 | M | PL7(V4) | SA4(V6) | 0.000 | 0.000 | - | - |
| 131 | 1 | M | PL8_2011(V2) | SA3(V5) | - | 0.004 | - | - |
| 134 | 3 | F | PL8_2011(V2) | PL1(V1) | - | 0.004 | - | - |
| 146 | 2 | M | PL2(V2) | PL1(V1) | - | 0.008 | - | - |
| 42 | 2 | M | SA3(V5) | PL1(V1) | - | 0.007 | 0.157 | 0.554 |
Male-biased dispersal was inferred by three (Fst, vAIc, and mPr) and one (mPr) estimate in the two-sided and one-sided tests, respectively (Table 6). Neither a one-sided nor two-sided test showed evidence for female-biased dispersal. These results complement the finding from the assignment tests (Table 5) that indicate that the majority of migrants are males.
Table 6.
Results from tests for sex-biased dispersal using four different statistics reported for both the two-and the one-sided tests for females and males. Number of individuals for each sex is listed in parenthesis. The second columns shows Fst values followed by the correct mean assignment index (mAIc) and its variance (vAIc), using Favre et al 1997 method. The last column reports the mean pairwise relatedness (mPr). Significance values (p-values) for these measures are reported in the last row.
| Two sided | FST | mAIc | vAIc | mPr |
|---|---|---|---|---|
| F (67) | 0.104 | 0.585 | 16.03 | 0.191 |
| M (70) | 0.076 | -0.560 | 27.38 | 0.135 |
| p-value | 0.059 | 0.152 | 0.192 | 0.034 |
| One sided | FST | mAIc | vAIc | mPr |
| F (67) | 0.104 | 0.585 | 16.03 | 0.191 |
| M (70) | 0.076 | -0.560 | 27.38 | 0.135 |
| p-value | 0.046 | 0.081 | 0.026 | 0.027 |
Computations based on linkage disequilibrium (LDNe, a measure of Ne based on non-random associations of linked alleles, Waples & Do 2008) for the three genetic clusters identified by Structure and multivariate analyses (Fig 2) resulted in mean estimates of NE ranging from 34 to 94 with corresponding NC estimates ranging from 273 to 897 (Table S6). Cluster 2, which only included the two temporally separated samples from PL2, had the smallest NE and NC estimates (NE: 34, with 95% CI ranging from 10–40; NC=273, 95% HPD= 167–396). Cluster 1, which grouped together five sites from 3 valleys in Pau da Lima (PL1, PL6, PL7, PL8-2010, and PL9) and the São Joaquim site, SL5, had the highest population size estimates (NE: 94, with 95% CI ranging from 75–125; NC=897, 95% HPD= 357–1608). Cluster 3, comprising one of the two PL8 temporally distinct samples and the two Sete de Abril sites (SA3 and SA4), had intermediate values (NE: 54, with 95% CI ranging from 35–85; NC=454, 95% HPD= 233–722).
DISCUSSION
Patterns of Genetic Diversity, Differentiation, and Relatedness
Given the relatively recent colonization of Salvador by Norway rats (coinciding with the construction of slum settlements in the last half-century, Moreira & Pereira 2008) and the small spatial scale of this study (around 300 m-1.8 km between most sites), the finding of relatively high levels of genetic variation within sampling sites and significant genetic differentiation between sites was unexpected. We found high genetic diversity (He ranging from 0.57 to 0.72, Tab. 2), even for sites represented by small samples sizes (N=3, Tab. 2); and no difference in diversity between samples (Pau de Lima sites) that had experienced recent (2009–2010) rodenticide control and those (Sete de April and São Joaquim sites) that did not. The levels of genetic diversity detected are comparable to the those reported for rat samples from urban Baltimore (Maryland, USA), where He values ranged from 0.67–0.78 for 277 Norway rats trapped from 11 sampling sites (Gardner-Santana et al. 2009), as well as from rural populations on a much larger spatial scale (He = 0.42 Abdelkrim et al. 2005; He = 0.59 Abdelkrim et al. 2009; He = 0.72–0.82 Abdelkrim et al. 2010; He = 0.55 Russell et al. 2009). In neither case had these populations been subjected to rodenticide treatment in times just prior to sampling.
Our results highlight the existence of complex population genetic structure among Norway rats in Salvador with moderate genetic admixture (Fig. 2A, Table 6). There was a considerable amount of genetic differentiation between geographically proximate sites, and in one case, even between samples obtained at the same site but in two consecutive years (PL8(V2), Fig. 2). These observations indicate that rapid population turnover, although uncommon, does happen.
The results of the clustering analyses (Fig. 2) suggest the existence of three genetic groups, which differentiate rats captured from sampling sites between the two neighborhoods of Pau da Lima and Sete de Abril, as well as sites within Pau da Lima. The three clusters correspond to the sites with the highest sample size (~ 30 individuals). For instance, sites with low sample sizes, including SJ5(V7), grouped with PL1(V1), a site located in a different neighborhood more than 8 km away, in cluster 1. This result suggests that inadequate sampling might be responsible for our inability to discriminate between some of the sites, highlighting the need for further studies with increased sample sizes.
The results of the Mantel test and FST analyses show moderate but significant pairwise genetic differentiation without a pattern of isolation by distance (examined for the more densely sampled Pau da Lima neighborhood). This parallels the results of the clustering analyses, and suggests little correlation (p = 0. 957, R2 = 0.342) between genetic and geographic distance. In addition to the non-significant p-value in the Mantel test, the plotted distribution in Fig.S1 shows widespread, clumped points, connected by a negatively-sloped trend line, rather than a scatter of points, suggesting a negative correlation of genetic and geographic distance. While the FST value for PL6 and PL7, located 317 m apart in V4, is not statistically significant, the FST values for localities in different valleys (PL7(V4) and PL8(V2)), separated by just 79 m are significant, as are FST values for sites separated by geographic distances comparable to the one between PL7(V4) and PL8(V2) (393-263 m; Tables 5, Fig. S1).
More comprehensive sampling, involving placing more traps at sites already included in this study, may improve the resolution of the genetic differentiation between valleys. In the only study directly comparable to ours, Gardner-Santana et al. 2009 analyzed rats captured from11 sites in Baltimore, separated by an average of 4.7 km, and detected three to four genetic clusters across the city. In this study we found genetic differentiation between sites separated by less than 100 m. This discrepancy could be influenced by the topographic complexity of Salvador, as evidenced by the presence of numerous semi-isolated valleys, compared to the relatively flat and uniform topography of Baltimore. Interestingly, in a three year-long capture-mark-recapture study in Denmark, there was no movement of Norway rats between adjacent, but separated, sewer systems; rats were genetically clustered within the same sewer system, moving an average maximal distance of ~200 m, suggesting the existence of separate population units with only limited mixing between them (Heiberg et al. 2012).
Since dispersal patterns and social structures have been shown to influence the distribution of genetic variation in populations (Chesser 1991a, 1991b; Matocq et al. 2000), we initially hypothesized that the genetic differentiation observed between these sites could be partly due to the social behavior and ecology of this species, as related extended families of Norway rats tend to inhabit burrow systems proximal to one another (Calhoun 1962). However, the observed patterns of genetic divergence among geographically close groups of sampling sites cannot be attributed to high relatedness among individuals, since our sampling overwhelmingly (~90% pairwise relationships) consisted of unrelated individuals, even from traps located around the same household (Table 3). No information is available for tropical urban settings, but temporal and seasonal fluctuations, i.e. colony instability that only increased sampling efforts will elucidate, at local scales may rapidly change allele frequencies (Balloux & Lugon-Moulin 2002; Gileva et al. 2006). The presence of unrelated juveniles and adults captured from the same household could reflect colony instability that for the Pau de Lima sites, could have been aggravated by rodenticide poisoning in the two years before the samples were collected and could have permitted burrow re-colonization by unrelated rats from other neighborhoods.
Sex-biased Dispersal
The relatively high level of genetic structuring in our data could also be influenced by Norway rat behavioral traits (Brouat et al. 2007). R. norvegicus, like the majority of rodents, tend to exhibit female philopatry and male dispersal (Calhoun 1962), although social aggression (Davis 1951) and environmental disturbances can precipitate long distance movement by both sexes (Taylor 1978). Despite using similar methods, genetic studies in Baltimore (Gardner-Santanta 2009) failed to find evidence for philopatry and sex-biased dispersal among urban Norway rats, suggesting that both sexes might move in high-density urban populations because adult males may not be able to establish and hold breeding territories (Childs et al. 1991; Macdonald & Fenn 1995). It is possible that these rat populations, or at least those from the Pau de Lima sites, were not subject to these density-dependent effects on social hierarchies and dispersal since populations were below carrying capacity due to the previous rodent control efforts. Gardner-Santana et al. did not mention any rodent control activities during their study. Population reductions could result in conditions favorable for re-colonization of burrow systems by migrating or dispersing animals from adjacent populations, especially if adult male dominated breeding territories were disrupted in populations destabilized by rodenticide control interventions (Calhoun 1962).
Male-biased migration among study sites in Salvador, as assessed by Fstat, STRUCTURE, and Geneclass, provides support for the hypothesis that population disruption increases genetic diversity and admixture between sites. Out of the ten individuals detected as migrants, all but one was male, and among the males, nine were sexually mature adults or subadults (>200g). Moderate genetic structuring could result as subordinate males migrate/disperse and, in areas where highly structured dominant-male based territories are less common, could have greater access to females (Macdonald et al. 1999; Hinson et al. 2006). The influx of new genes would be amplified by the high fecundity and short generation time typical of this species.
It is also important to note the promiscuous breeding habits of the Norway rat, where numerous copulations occur nightly among receptive females and the presence of a female in estrus entices male rats from other burrow systems to travel outside their accustomed ranges (Calhoun 1962). Although dominant males may decrease access of sub-dominant males to adult females residing in the same burrow, this protection is always incomplete, and numerous mountings of dominant male-associated females with other males are assured (Calhoun 1962). Although not every mating attempt results in ejaculation, the potential for mixed parentage among litters is great and will reduce the degree of relatedness among extended families. Additionally, the ability of male rats to control the amount of ejaculate based on the presence of other males in the breeding environment (sperm allocation) may increase the probability that males lower down in the succession of males mounting females have an advantage in contributing their sperm/genes to receptive females (Pound & Gage 2004).
Our migrant analyses identified putative dispersal/migration routes both within and between valleys and neighborhoods, which are key for implementation of pest management programs targeting the eradication unit (Russell et al. 2009a). Although further investigations with a denser sampling scheme are needed to clarify “source” and “sink” population dynamics, our results suggested that PL1(V1) could be a source population, providing migrants to other sites within and beyond the same neighborhood (i.e. PL8(V2) and SA3(V5)).
The shape and adjacency of two of the valley systems sampled (Fig 1) could facilitate dispersal/migration, as no obvious impermeable barriers exist. During the rainy season, especially if flooding occurs, rats can disperse to neighboring valleys riding on flowing water (Cottam 1948). PL8, located at the bottom of valley V2 seems to represent a sink, receiving migrants from sites in other valleys (V1 and V3) and from different neighborhoods (Sete de April, SA3). The dynamic nature of the genetic variation at this site was emphasized by the finding of significant pairwise FST values between rat samples from successive years (Table 5), and their genetic assignment to different clusters (Fig. 2). This result is probably due to the geographic position of PL8 in valley 2, at the entrance to the other valleys in Pau da Lima, and the existence of potential corridors between Pau da Lima and Sete de Abril (Fig. 1). At this time we can only conjecture about the mechanisms contributing to the genetic variability present at this site, and only future efforts to acquire additional samples will provide the means to understand source-sink dynamics within and between valleys.
The São Joaquim market site and two sites in Pau da Lima (PL6(V4) and PL7(V4)), although showing signs of genetic admixture (Fig. 2), did not show strong evidence of recent migration from other sampled sites, possibly because of the greater distance between the São Joaquim site and all other sites (~8.2 – 10.1 km). São Joaquim is located near the international sea port, which could be a source of additional genetic diversity, but sites as yet unsampled could also be the source of the migrants to SJ5(V7), PL6(V4), and PL7(V4), leading to the detection of genetic admixture. The potential contributions of as yet unsampled sites is a complicating factor precluding firm conclusions regarding the processes leading to the observed genetic admixtures and thus, dictates the need for increased sampling.
The population size estimates were obtained using the genetic clusters identified by the Bayesian and multivariate analyses (Fig. 2), which include multiple sampling sites within each genetic cluster. As expected cluster 2, which included only one site with two temporal samplings, had the lowest population size estimates, while cluster 3, which groups most of the other sites, had the largest (Table S6). The sparse sampling scheme of the present study does not support robust population size estimates, as we suspect our ability to identify additional genetic clusters may have been hindered by the small number of sampling sites both within and around the valleys. A denser sampling scheme will allow a better spatial definition of the genetic units and thus lead to more accurate population estimates.
However, it is reassuring to find that the NC estimates based on genetic rather than traditional mark-recapture data are higher than the corresponding NE estimates since NC estimates are usually higher than NE estimates. In this study we report a 1/10 NE/ NC ratio, which is similar to other estimates (Frankham 1995; Luikart et al. 2010). In further studies we aim to confirm the validity of this approach and provide a solid understanding of both the stability of the NE/ NC ratio, and the factors that could impact its change in this study system. Calculating NE estimates from genetic data could provide a cost effective method to assess the number of rats in Salvador at different spatial and temporal scales with obvious benefits for designing more targeted control interventions simultaneously targeting genetically linked populations that provide rats a corridor to disperse/migrate between neighborhoods.
Implications for Control
Currently, the eradication unit for rodent control programs in Salvador, as well as many urban centers in Brazil and worldwide, is the city “block,” a unit based on man-made boundaries. The finding of genetic differentiation between sampling sites in neighboring valleys only a few hundred meters apart, coupled with evidence of genetic admixture between distant sites, and genetic discontinuities between temporal samplings at some sites suggests that eradication unit sizes should vary between sites. Our data suggest that, given the complex urban landscape, an eradication unit based on the city block might not be necessary or advisable, as geographically based eradication units can be equally efficient, resulting in less expensive in the long run and easier to sustain city-wide control and monitoring measures.
The temporal comparisons of samples from the same sites collected in successive years provide further insight on the re-emergence dynamics of these rat populations that is relevant for developing control interventions and their monitoring through time. In some sites individuals belonging to the same genetic cluster were captured in the same location in successive years, while in others we observed a population replacement. These observations suggest that colony re-establishment can happen either by in situ re-emergence or by re-colonization from surrounding areas. The detection of first generation migrants in a few sites implies that rats can move to new sites quite readily and underscores the importance of understanding the factors that enable such movements, especially in geographic areas at the base of each valley, which act at the same time as dispersal corridors between valleys and sources of migrants for slum areas located within a valley.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Centro de Controle de Zoonoses (CCZ) in Salvador for their assistance in conducting the study. This work was supported by the Secretariat of Health Surveillance and Oswaldo Cruz Foundation, Brazilian Ministry of Health; CAPES (Coordination for the Improvement of Higher Education Personnel), Brazilian Ministry of Education; National Institutes of Health (grants R01 TW009504, U01 AI088752, R01 AI052473, R24 TW007988 and D43 TW00919); and Yale Institute for Biospheric Studies, Molecular Systematics and Conservation Genetics Center and Wilbur Downs Fellowship Program. We thank Danielle Edwards, Sarah Federman, and Ryan Garrick for their assistance during data collection and analysis, as well as The Yale Institute for Biospheric Studies, Molecular Systematics and Conservation Genetics Center for use of laboratory facilities. Brittney Kajdacsi was supported by a Science Scholarship from The Graduate Center, City University of New York (CUNY) during the writing of this manuscript.
Footnotes
DATA ACCESSIBILITY
Microsatellite genotypes and analysis input files: DRYAD entry doi: 10.5061/dryad.hd300.
AUTHOR INFORMATION
This project was carried out under the direction of Dr. Adalgisa Caccone in the Yale Department of Ecology and Evolutionary Biology and Dr. Albert Ko at the Department of Epidemiology of Microbial Diseases, Yale School of Public Health. FC, FP, GR, HF, MGR, JEC, AIK, and AC designed the study. FC, FP, and GR performed sample collection and rat tissue preparation in Brazil. FC and FP also contributed to manuscript preparation. JB and CH helped with analyses, performing PCA and DAPC, and NE/NC estimates, as well as contributing to manuscript preparation. BK, currently a Biology PhD student at City University of New York, performed all laboratory work and most of the analysis, in addition to preparing the manuscript. Her current research focuses on urban land- and stream-scape genomics of two salamander species in New York City. CH is currently a graduate student at Cornell University, and his research focuses on understanding spatial and environmental correlates of genomic variation in a variety of taxa. FC is an eco-epidemiologist interested in the study of urban ecology and infectious diseases in the developing with the aim to implement and evaluate disease control programs. GR and HF are biologists interested in rodent control to prevent zoonotic diseases. FP is a parasite ecologist interested in how reservoir host ecology and population dynamics impact the spatial and temporal distribution of zoonotic disease risk. JB is an evolutionary geneticist interested in disease vectors and pathogens, and their ecological interactions with humans. MGR is a molecular epidemiologist interested in human pathology and immunpathogenesis of parasitological and infectious diseases including hepatitis C, dengue, meningites, leptospirosis, chagas disase, and schistosomisais. JEC is an infectious disease epidemiologist interested in the transmission of zoonotic pathogens to humans, including directly-transmitted zoonotic viruses, such as the hantaviruses, arenaviruses and rabies, and vector-borne bacteria, including rickettsia, bartonella and borrelia. AIK is an infectious disease epidemiologist whose research focuses on the dynamics of health problems which have emerged, due to rapid urbanization and social inequity, and the interventions that are implemented in urban slum communities. AC is an evolutionary biologist interested in understanding spatial and temporal patterns and levels of genomic diversity in organisms of conservation or epidemiological concern.
REFERENCES
- Abdelkrim J, Pascal M, Calmet C, Samadi S. Importance of Assessing Population Genetic Structure before Eradication of Invasive Species: Examples from Insular Norway Rat Populations. Conservation Biology. 2005;19(5):1509–1518. [Google Scholar]
- Abdelkrim J, Pascal M, Samadi S. Establishing Causes of Eradication Failure Based on Genetics: Case Study of Ship Rat Eradication in Ste. Anne Archipelago. 2007;21(3):719–730. doi: 10.1111/j.1523-1739.2007.00696.x. [DOI] [PubMed] [Google Scholar]
- Abdelkrim J, Byrom AE, Gemmell NJ. Fine-scale genetic structure of mainland invasive Rattus rattus populations: implications for restoration of forested conservation areas in New Zealand. Conservation Genetics. 2010;11(5):1953–1964. [Google Scholar]
- Amilasan AS, Ujiie M, Suzuki M, Salva E, Belo MC, Koizumi N, Yoshimatsu K, Schmidt WP, Marte S, Dimaano EM, Villarama JB, Ariyoshi K. Outbreak of leptospirosis after flood, the Philippines, 2009. Emerging Infectious Diseases. 2012;18(1):91–94. doi: 10.3201/eid1801.101892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson IAE. The spread of commensal species of Rattus to oceanic islands and their effects on island avifaunas. In: Moors PJ, editor. Conservation of island birds. No. 3. Cambridge: International Council for Bird Preservation Technical Publication; 1985. pp. 35–81. [Google Scholar]
- Balloux F, Lugon-Moulin N. The estimation of population differentiation with microsatellite markers. Molecular Ecology. 2002;11:155–165. doi: 10.1046/j.0962-1083.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- Beadell JS, Hyseni C, Abila PP, Azabo R, Enyaru JCK, Ouma JO, Mohammed YO, et al. Phylogeography and population structure of Glossina fuscipes fuscipes in Uganda: implications for control of tsetse. PLoS neglected tropical diseases. 2010;4(3):e636. doi: 10.1371/journal.pntd.0000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazilian Institute of Geography and Statistics. Brazilian Ministry of Health, estimated population retrospective statistical series 1960–70. [Google Scholar]
- Brazilian Institute of Geography and Statistics. Summary of the 2010 demographic census. 2011 [Google Scholar]
- Brouat C, Loiseau A, Kane M, Bâ K, Duplantier JM. Population genetic structure of two ecologically distinct multimammate rats: the commensal Mastomys natalensis and the wild Mastomys erythroleucus in southeastern Senegal. Molecular ecology. 2007;16(14):2985–2997. doi: 10.1111/j.1365-294X.2007.03353.x. [DOI] [PubMed] [Google Scholar]
- Brown JE, McBride CS, Johnson P, Ritchie S, Paupy C, Bossin H, Lutomiah J, et al. Worldwide patterns of genetic differentiation imply multiple “domestications” of Aedes aegypti, a major vector of human diseases. Proceedings Biological sciences / The Royal Society. 2011;278(1717):2446–2454. doi: 10.1098/rspb.2010.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun JB. The Ecology and Sociology of the Norway Rat. Bethesda, Maryland: US Department of Health; 1962. [Google Scholar]
- Calmet C, Pascal M, Samadi S. Is it worth eradicating the invasive pest Rattus norvegicus from Molène archipelago? Biodiversity and Conservation. 2001;10:911–928. [Google Scholar]
- Chanteau S, Ratsifasoamanana L, Rasoamanana B, Rahalison L, Randrianbelosoa J, Roux J, Rabeson D. Plague, a reemerging disease in Madagascar. Archives de l'Institut Pasteur de Madagascar. 1998;64:12–14. doi: 10.3201/eid0401.980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs JE, Glass GE, LeDuc JW. Rodent sightings and contacts in an inner-city population of Baltimore, Maryland, U.S.A. Bulletin of the Society of Vector Ecologists. 1991;16:245–255. [Google Scholar]
- Civen R, Ngo V. Murine typhus: an unrecognized suburban vectorborne disease. Clinical Infectious Diseases. 2008;46(6):913–918. doi: 10.1086/527443. [DOI] [PubMed] [Google Scholar]
- Cottam C. Aquatic habitats of the Norway rat. Journal of Mammalogy. 1948;29:299. [Google Scholar]
- Davis D, Emlen J, Stokes A. Studies on home range in the brown rat. Journal of Mammalogy. 1948;29(3):207–225. [Google Scholar]
- Davis DE. The Relation Between Level of Population and Size and Sex of Norway Rats. Ecology. 1951a;32(3):462–464. [Google Scholar]
- Davis DE. The Characteristics of Global Rat Populations. American Journal of Public Health and the Nations Health. 1951b;41(2):158–163. doi: 10.2105/ajph.41.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DE. The characteristics of rat populations. The Quarterly Review of Biology. 1953;28:373–401. doi: 10.1086/399860. [DOI] [PubMed] [Google Scholar]
- Dechet AM, Parsons M, Rambaran M, Mohamed-Rambaran P, Florendo-Cumbermack A, et al. Leptospirosis outbreak following severe flooding: a rapid assessment and mass prophylaxis campaign; Guyana, January-February 2005. PLoS ONE. 2012;7(7):e39672. doi: 10.1371/journal.pone.0039672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular ecology. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular ecology resources. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Farhang-Azad A, Southwick CH. Population ecology of Norway rats in the Baltimore Zoo and Druid Hill Park, Baltimore, Maryland. Annals of Zoology. 1979;15:1–42. [Google Scholar]
- de Faria MT, Calderwood MS, Athanazio DA, McBride AJA, Hartskeerl RA, Pereira MM, Ko AI, et al. Carriage of Leptospira interrogans among domestic rats from an urban setting highly endemic for leptospirosis in Brazil. Acta tropica. 2008;108(1):1–5. doi: 10.1016/j.actatropica.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre L, Balloux F, Goudet J, Perrin N. Female-biased dispersal in the monogamous mammal Crocidura russula: evidence from field data and microsatellite patterns. Proceedings of the Royal Society Biological sciences. 1997;264(1378):127–132. doi: 10.1098/rspb.1997.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Effective population-size: adult-population size ratios in wildlife - A review. Genetical Research. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- Gazey WJ, Staley MJ. Population estimation from mark-recapture experiments using a sequential Bayes algorithm. Ecology. 1986;67:941–951. [Google Scholar]
- Goudet J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. Journal of Heredity. 1995;86(6):485–486. [Google Scholar]
- Goudet J, Perrin N, Waser P. Tests for sex-biased dispersal using bi-parentally inherited. Molecular Ecology. 2002;11:1103–1114. doi: 10.1046/j.1365-294x.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- Gardner-Santana LC, Norris DE, Fornadel CM, Hinson ER, Klein SL, Glass GE. Commensal ecology, urban landscapes, and their influence on the genetic characteristics of city-dwelling Norway rats (Rattus norvegicus) Molecular ecology. 2009;18(13):2766–2778. doi: 10.1111/j.1365-294X.2009.04232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilabert A, Loiseau A, Duplantier JM, Rahelinirina S, Rahalison L, Chanteau S, Brouat C. Genetic structure of black rat populations in a rural plague focus in Madagascar. Canadian Journal of Zoology. 2007;85(9):965–972. [Google Scholar]
- Gileva EA, Rakitin SB, Cheprakov MI. Genomic instability in the bank vole: population-ecological aspects. Russian Journal of Ecology. 2006;37:271–277. [Google Scholar]
- Giraudeau F, Apiou F, Amarger V, Kaisaki PJ, Bihoreau M-T, Lathrop M, Vergnaud G, Gauguier D. Linkage and physical mapping of rat microsatellites derived from minisatellite loci. Mammalian Genome. 1999;10(4):405–409. doi: 10.1007/s003359901012. [DOI] [PubMed] [Google Scholar]
- Glass GE, Childs JE, Korch GW, LeDuc DE. Occasional Papers of the Museum of Natural History. Vol. 130. University of Kansas, Lawrence, Kansas; 1989. Comparative ecology and social interactions of Norway rat (Rattus norvegicus) populations in Baltimore, Maryland; pp. 1–33. [Google Scholar]
- Glass GE, Klein SL, Shields T, Easterbrook J. The Norway rat population in Baltimore, Maryland. Vector-Borne and Zoonotic Diseases. 2005;5:296–299. doi: 10.1089/vbz.2005.5.296. [DOI] [PubMed] [Google Scholar]
- Glass GE, Gardner-Santana LC, Holt RD, Chen J, Shields TM, Roy M, Schachterie S, Klein SL. Trophic garnishes: cat-rat interactions in an urban environment. PLoS ONE. 2009;4(6):e5794. doi: 10.1371/journal.pone.0005794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiberg A, Leirs H, Siegismund HR. Reproductive success of bromadiolone-resistant rats in absence of anticoagulant pressure. Pest Management Science. 2006;871:862–871. doi: 10.1002/ps.1249. [DOI] [PubMed] [Google Scholar]
- Heiberg A, Sluydts V, Leirs H. Uncovering the secret lives of sewer rats (Rattus norvegicus): movements, distribution and population dynamics revealed by a capture-mark-recapture study. Wildlife Research. 2012;39:202–219. [Google Scholar]
- Himsworth CG, Parsons KL, Jardine C, Patrick DM. Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ccology of rat-associated zoonoses in urban centers. Vector Borne and Zoonotic Diseases. 2013;13:1–11. doi: 10.1089/vbz.2012.1195. X. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6(2):65–70. [Google Scholar]
- Howald G, Donlan JC, Galván JP, Russell JC, Parkes J, Araceli S, Wang Y, Veitch D, Genovesi P, Pascal M, Saunders A, Tershy B. Invasive Rodent Eradication on Islands. Conservation Biology. 2007;21(5):1258–1268. doi: 10.1111/j.1523-1739.2007.00755.x. [DOI] [PubMed] [Google Scholar]
- Hyseni C, Kato AB, Okedi LM, Masembe C, Ouma JO, Aksoy S, Caccone A. The population structure of Glossina fuscipes fuscipes in the Lake Victoria basin in Uganda: implications for vector control. Parasites & Vectors. 2012;5(1):222. doi: 10.1186/1756-3305-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskjaer C, Slade NA, Childs JE, Glass GE, Korch GW. Body mass as a measure of body size in small mammals. Journal of Mammalogy. 1989;70:662–667. [Google Scholar]
- Jacob HJ, Brown DM, Bunker RK, Daly MJ, Dzau VJ, Goodman A, Koike G, Kren V, Kurtz T, Lernmark A. A genetic linkage map of the laboratory rat, Rattus norvegicus . Nature genetics. 1995;9(1):63–69. doi: 10.1038/ng0195-63. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST. Counting Alleles with Rarefaction: Private Alleles and Hierarchical Sampling Designs. Conservation Genetics. 2004;5(4):539–543. [Google Scholar]
- Kalinowski ST. Hp-Rare 1.0: a Computer Program for Performing Rarefaction on Measures of Allelic Richness. Molecular Ecology Notes. 2005;5(1):187–189. [Google Scholar]
- Kalinowski ST, Wagner AP, Taper ML. Ml-relate: a computer program for maximum likelihood estimation of relatedness and relationship. Molecular Ecology Notes. 2006;6(2):576–579. [Google Scholar]
- Ko AI, Reis MG, Ribeiro Dourado CM, Johnson WD, Riley LW. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet. 1999;354(9181):820–825. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nature reviews. Microbiology. 2009;7(10):736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA, Boonmar S, Bhengsri S, Dowell SF, Sitdhirasdr A, Lerdthusnee K, Richardson J, Peruski LF. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. The American Journal of Tropical Medicine and Hygiene. 2010;82(6):1140–1145. doi: 10.4269/ajtmh.2010.09-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conservation Genetics. 2010;11:355–373. [Google Scholar]
- Lowe S, Browne M, Boudjelas S, De Poorter M. 100 of the World’s Worst Invasive Alien Species: a Selection from the Global Invasive Species database. Gland, Switzerland: Invasive Species Specialist Group (ISSG) a specialist group of the Species Survival Commission (SSC) of the World Conservation Union (IUCN; 2000. [Google Scholar]
- Macdonald DW, Fenn MPG. Rat ranges in arable areas. Journal of Zoology (London) 1995;236:349–353. [Google Scholar]
- Macdonald DW, Mathews F, Berdoy M. The behaviour and ecology of Rattus norvegicus: From opportunism to kamikaze tendencies. In: Singleton, Hinds L, Leirs H, Zhang Z, editors. Ecologically-based Rodent Management. Canberra: Australian Centre for International Agricultural Research; 1999. [Google Scholar]
- Matthias MA, Ricaldi JN, Cespedes M, Diaz MM, Galloway RL, Saito M, Steigerwalt AG, Patra KP, Ore CV, Gotuzzo E, Gilman RH, Levett PN, Vinetz JM. Human leptospirosis caused by a new, antigenically unique leptospira associated with a rattus species reservoir in the peruvian Amazon. PLoS Neglected Tropical Diseases. 2008;2(4):e213. doi: 10.1371/journal.pntd.0000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JN, Childs JE, Ksiazek TG, Peters CJ, Velleca WM. Methods for trapping and sampling small mammals for virologic testing. Atlanta, GA: US Dept. of Health & Human Services, Centers for Disease Control and Prevention; 1995. [Google Scholar]
- Moreira IM, Pereira GC. Salvador and Metropolitan Region. Edufba: Salvador, Brazil; 2008. p. 228. [Google Scholar]
- Nkogwe C, Raletobana J, Stewart-Johnson A, Suepaul S. Frequency of detection of Escherichia coli, Salmonella spp., and Campylobacter spp. in the faeces of wild rats (Rattus spp.) in Trinidad and Tobago. Veterinary Medicine International. 2011;2011:1–7. doi: 10.4061/2011/686923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgain H, Schein MW. A preliminary analysis of the physical environment of the Norway rat. Ecology. 1953;34:467–473. [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Molecular Ecology. 2004;13(1):55–65. doi: 10.1046/j.1365-294x.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- Pereira MM, Andrade J. Epidemiological aspects of leptospirosis in a slum area in the city of Rio de Janeiro, Brazil. Search for leptospires and specific antibodies in rodents. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1988;82:768–770. doi: 10.1016/0035-9203(88)90231-3. [DOI] [PubMed] [Google Scholar]
- Pereira MM, Matsuo MGS, Bauab AR, Vasconcelos SA, Moraes ZM, Baranton G, Saint GI. A clonal subpopulation of Leptospira interrogans sensu stricto is the major cause of leptospirosis outbreaks in Brazil. Journal of Clinical Microbiology. 2000;38:450–452. doi: 10.1128/jcm.38.1.450-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit E, Valiere N. Estimating population size with noninvasive capturemark- recapture data. Conservation Biology. 2006;20:1062–1073. doi: 10.1111/j.1523-1739.2006.00417.x. [DOI] [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. GENECLASS2: a software for genetic assignment and first-generation migrant detection. The Journal of heredity. 2004;95(6):536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FH, Costa F, Rodrigue G, Farias H, Glass G, Mitemayer GR, Ko AI, Childs J. Demographic Differences between Tropical and Temperate Norway Rats. In preparation [Google Scholar]
- Pound N, Gage MJG. Prudent sperm allocation in Norway rats, Rattus norvegicus: a mammalian model of adaptive ejaculate adjustment. Animal Behaviour. 2004;68(4):819–823. [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. The Journal of heredity. 1995;86(3):248–249. [Google Scholar]
- Reis RB, Ribeiro GS, Felzemburgh RDM, Santana FS, Mohr S, Melendez AXTO, Queiroz A, Santos AC, Ravines RR, Tassinari WS, Carvalho MS, Reis MG, Ko AI. Impact of Environment and Social Gradient on Leptospira Infection in Urban Slums. PLoS Neglected Tropical Diseases. 2008;2(4):e228. doi: 10.1371/journal.pntd.0000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson BC, Gemmell NJ. Defining eradication units to control invasive pests. Journal of Applied Ecology. 2004;41(6):1042–1048. [Google Scholar]
- Rousset F. Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular ecology resources. 2008;8(1):103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Russell JC, Mackay JWB, Abdelkrim J. Insular pest control within a metapopulation context. Biological Conservation. 2009a;142(7):1404–1410. [Google Scholar]
- Russell JC, Abdelkrim J, Fewster RM. Early colonisation population structure of a Norway rat island invasion. Biological Invasions. 2009b;11(7):1557–1567. [Google Scholar]
- Russell JC, McMorland AJC, MacKay JWB. Exploratory behaviour of colonizing rats in novel environments. Animal Behaviour. 2010;79(1):159–164. [Google Scholar]
- Sarkar U, Nascimento SF, Barbosa R, Martins R, Nuevo H, Kalafanos I, Grunstein I, Flannery B, Dias J, Riley LW, Reis MG, Ko AI. Population-based case-control investigation of risk factors for leptospirosis during an urban epidemic. American Journal of Tropical Medicine and Hygiene. 2002;66:605–610. doi: 10.4269/ajtmh.2002.66.605. [DOI] [PubMed] [Google Scholar]
- Singleton GR, Leirs H, Hinds LA, Zhang Z, editors. Ecologically-based management of rodent pests. Canberra: Australian Centre for International Agricultural Research; 1999. pp. 49–80. [Google Scholar]
- Slotman MA, Kelly NB, Harrington LC, Kitthawee S, Jones JW, Scott TW, Caccone A, Powell JR. Polymorphic microsatellites markers for studies ofAedes aegypti (Diptera Culicidae), the vector of dengue and yellow fever. Molecular Ecology Notes. 2006;7:168–171. [Google Scholar]
- Solano P, Ravel S, de Meeûs T. How can tsetse population genetics contribute to African trypanosomiasis control? Trends in parasitology. 2010;26(5):255–263. doi: 10.1016/j.pt.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Taylor KD. Range of movement and activity of common rats (Rattus norvegicus) on agricultural land. Journal of Applied Ecology. 1978;15(3):663–677. [Google Scholar]
- Taylor KD, Quy RJ. Long distance movements of a common rat (Rattus norvegicus) revealed by radio-tracking. Mammalia. 1979;42(1):63–72. [Google Scholar]
- UN-HABITAT. Hidden cities: unmasking and overcoming health inequities in urban settings. 2010 World Health Organization, The WHO Centre for Health Development and United Nations Human Settlements Programme. [Google Scholar]
- Waples RS, Do C. LDNE: a program for estimating effective population size from data on linkage disequilibrium. Molecular Ecology Resources. 2008;8:753–756. doi: 10.1111/j.1755-0998.2007.02061.x. [DOI] [PubMed] [Google Scholar]
- Webster JP, Macdonald DW. Parasites of wild brown rats (Rattus norvegicus) on UK farms. Parasitology. 1995;111(03):247–255. doi: 10.1017/s0031182000081804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.