Abstract
In this work, we present a novel, automated, registration method to fuse magnetic resonance imaging (MRI) and transrectal ultrasound (TRUS) images of the prostate. Our methodology consists of: (1) delineating the prostate on MRI, (2) building a probabilistic model of prostate location on TRUS, and (3) aligning the MRI prostate segmentation to the TRUS probabilistic model. TRUS-guided needle biopsy is the current gold standard for prostate cancer (CaP) diagnosis. Up to 40% of CaP lesions appear isoechoic on TRUS, hence TRUS-guided biopsy cannot reliably target CaP lesions and is associated with a high false negative rate. MRI is better able to distinguish CaP from benign prostatic tissue, but requires special equipment and training. MRI-TRUS fusion, whereby MRI is acquired pre-operatively and aligned to TRUS during the biopsy procedure, allows for information from both modalities to be used to help guide the biopsy. The use of MRI and TRUS in combination to guide biopsy at least doubles the yield of positive biopsies. Previous work on MRI-TRUS fusion has involved aligning manually determined fiducials or prostate surfaces to achieve image registration. The accuracy of these methods is dependent on the reader’s ability to determine fiducials or prostate surfaces with minimal error, which is a difficult and time-consuming task. Our novel, fully automated MRI-TRUS fusion method represents a significant advance over the current state-of-the-art because it does not require manual intervention after TRUS acquisition. All necessary preprocessing steps (i.e. delineation of the prostate on MRI) can be performed offline prior to the biopsy procedure. We evaluated our method on seven patient studies, with B-mode TRUS and a 1.5 T surface coil MRI. Our method has a root mean square error (RMSE) for expertly selected fiducials (consisting of the urethra, calcifications, and the centroids of CaP nodules) of 3.39 ± 0.85 mm.
2. INTRODUCTION
Prostate needle biopsy guided by transrectal ultrasound (TRUS) is the current gold standard for prostate cancer (CaP) diagnosis.1 TRUS-guided needle biopsy is typically performed using a blinded sextant procedure where the prostate is divided into 6 regions, and a biopsy is taken form each region.2 Approximately 40% of CaP lesions appear isoechoic on TRUS; hence are difficult to target on TRUS-guided needle biopsy.3, 4 TRUS-guided needle biopsy has a low detection rate of 20–25%.5 Because of a low CaP detection rate for TRUS-guided biopsy more than 1/3 of men biopsies undergo a repeat biopsy procedure.6
Comparatively, multi-parametric magnetic resonance imaging (MP-MRI) has a high positive predictive value (PPV) for CaP detection.7 T2-weighted (T2w) MRI is able to provide anatomical information about the prostate in addition to structural information about CaP.8 Other MRI protocols provide complementary functional information, such as dynamic contrast enhanced (DCE)9 and diffusion weighted imaging (DWI),10 or metabolic information, such as magnetic resonance spectroscopy (MRS).11 MRI-guided biopsies have 40–55% CaP detection rates.12, 13 However, these procedures require specialized equipment and technicians, are expensive, time-consuming, and stressful for many patients.12, 13
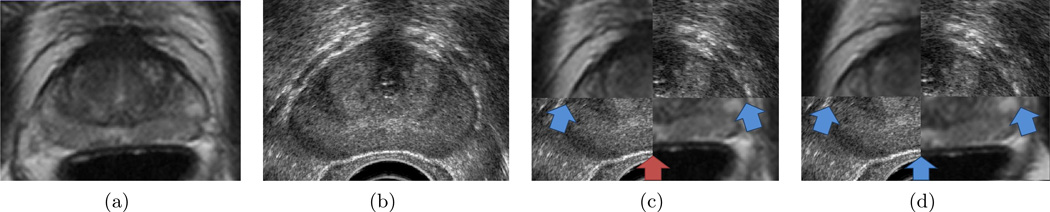
MRI-TRUS fusion, that spatially aligns MRI to TRUS, allows anatomical, structural, functional, and metabolic information obtained from MP-MRI and anatomical information obtained from TRUS to potentially be utilized to guide needle biopsy. In such protocols, MRI of the prostate is performed prior to the biopsy procedure, hence no need exists for specialized biopsy equipment. During the subsequent TRUS-guided biopsy, information from the MRI is transferred to the TRUS image. Utilizing both MRI and TRUS to guide biopsy at least doubles the positive yield of biopsy.14–17 However unique challenges exist for MRI-TRUS registration. First, intensity-based metrics are inappropriate because of the poor correlation between intensities on MRI and TRUS.18 Previous work has shown that Mutual Information alone cannot accurately align MRI to TRUS.18 Second, differences exist in prostate shape caused by the difference deformations induced by the TRUS probe and, when present, the MRI endorectal coil.19 Figure 5 illustrates an example showing fusion of prostate MRI and TRUS for one patient; no MRI endorectal coil was used resulting in pronounced differences in prostate shape.
Figure 5.
(a) T2-w MRI of the prostate. (b) TRUS of the prostate. (c) MRI-TRUS fusion using the Nakagami m-parameter for Ta displayed as a checkerboard image. Blue arrows shows boundary regions which are well aligned on MRI and TRUS, while red arrows show boundary regions which are misaligned. (d) MRI-TRUS fusion using the Nakagami m-parameter for Te displayed as a checkerboard image.
State-of-the-art MRI-TRUS fusion methods require manual intervention to establish spatial correspondence between MRI and TRUS imagery.14, 15 Labanaris et. al. performed a study that divided 260 patients into two groups: (1) an 18-core TRUS-guided biopsy with no MRI information added, and (2) a similar biopsy procedure with additional cores sampled from regions suspicious for CaP as determined by T2w MRI.14 The location of the additional cores was determined by manual inspection of the MRI and TRUS images. For the group undergoing only TRUS-guided biopsy, the CaP detection rate was 19.4%; the group with additional biopsy samples from CaP suspicious regions suspicious had a CaP detection rate of 74.9%. Hadaschik et. al. obtained a 59.4% CaP detection rate when using a semi-automated MRI-TRUS fusion system to guide the biopsy in 106 patients.15 Unfortunately, methods that require manual intervention may lead to longer examination times, and increase patient discomfort during or after TRUS acquisition.
In this work, we present a registration algorithm that is automated for all steps after TRUS acquisition. Our registration method may be well suited to MRI-TRUS fusion for guided biopsies because no time intensive manual interventions are required. In the remainder of the paper, Section 3 discusses previous work in MRI-TRUS fusion in greater detail as well as our novel contributions; Section 4 provides details of our novel MRI-TRUS fusion system; Section 5 describes our experimental design for evaluating our system and Section 6 presents our experimental results; We provide concluding remarks in Section 7.
3. PREVIOUS WORK IN MRI-TRUS FUSION AND NOVEL CONTRIBUTIONS
Table 3 briefly describes current state-of-the-art MRI-TRUS fusion methods listing the commercial system, technical work, whether the work was performed for planar (2D) or volumetric (3D) imagery, the type of manual intervention required, the number of studies, and the reported root mean squared error (RMSE). RMSE, when reported, ranges from 2 to 3 mm. Directly comparing RMSE between studies can be difficult, because RMSE is not always calculated over the same set of fiducials. For instance, Reynier et. al. only report RMSE for the prostate surface and the urethra, therefore in this study it is unclear how well other internal structures of the prostate align. Comparing between clinical patient studies (18, 20, 22, 24) and phantom studies (21, 23) is not possible. Phantom studies may not be able to appropriately model realistic deformations in the prostate between image modalities in clinical studies. Additionally, when using phantoms corresponding fiducials are easy to identify while on patient data accurately determining corresponding fiducials may be a more difficult problem. Mitra et. al. have the lowest reported RMSE of 1.60 ± 1.17 mm, however this study was performed only on corresponding 2D axial imagery. RMSE error would be expected to increase for 3D studies, as for these studies an additional dimension over which fiducials could be misaligned is introduced.
Table 3.
Description of intensity and texture features used to constructed the feature set F(c).
| Feature | Description | Formulation | |
|---|---|---|---|
| Intensity | Intensity value | f̃(c) | |
| Mean | Average intensity value within neighborhood 𝒩(c). | ||
| Median | Median intensity value within neighborhood 𝒩(c). | ||
| Range | Range of intensity values within neighborhood 𝒩(c). | ||
| Variance | Variance of a Gaussian distribution of intensity values within 𝒩(c). | ||
| Rayleigh Variance33 | Variance of the Rayleigh distribution calculated within the neighborhood 𝒩(c). | ||
| m-parameter34 | The m-parameter of the Nakagami distribution, which controls the shape of the distribution, calculated within the neighborhood 𝒩(c). | We use the method of Greenwood and Durand 35 to estimate m. |
MRI-TRUS fusion methods require manual intervention after TRUS acquisition to determine the location of the prostate.16, 18, 20–25 Manual intervention requires either selecting fiducials or delineating the prostate surface. Overall, MRI-TRUS fusion methods can be divided into (a) fiducial-based,18, 20, 23, 25 (b) surface-based,16, 21, 22 and (c) model-based methods according to the underlying structures being aligned between the MRI and TRUS imagery.24
Fiducial-based methods attempt to find a transformation that minimizes the distance between corresponding fiducials on MRI and TRUS.18, 20, 23, 25 Early work by Kaplan et. al.25 used manually selected fiducials to determine a rigid transformation between MRI and TRUS imagery. In this study only qualitative alignment was assessed between 2 MRI-TRUS studies. Mitra et. al.18 extracted the prostate surface and internal fiducials from a manual segmentation of the prostate; fiducials were used to determine a diffeomorphic transformation between MRI and TRUS on manually identified corresponding 2D axial images.
Xu et. al.23 used fiducials extracted from an automatic segmentation of the prostate to determine an affine transformation; however, the authors manually refined this segmentation as necessary to improve registration accuracy. Pinto et. al.26 found MRI-TRUS biopsy guidance using this method resulted in CaP detection in 55 out of 101 patients. Furthermore Pinto et. al.26 found 19 regions determined to be highly suspicious for CaP on MRI, 17 of which were confirmed as containing CaP on biopsy.
Reynier et. al.20 used fiducials extracted from a manual segmentation of the prostate to calculate an elastic transformation. They reported RMSE of 1.11±0.54 mm between prostate surfaces and 2.07±1.57 mm between the urethra.20 The very low RMSE between prostate surfaces is to be expected as this method is aligning fiducials extracted from the prostate surface. The increase in RMSE for the urethra suggests other internal anatomical structure may have a greater misalignment than the prostate surface. Further work has used this methodology to guide brachytherapy27 and biopsy.17 Rud et. al.17 studied 80 patients with suspicious MRI; MRI-TRUS-guided biopsy found CaP in 54 cases.
Surface-based methods eliminate the need to select fiducials by finding a transformation that minimizes the distance between prostate surfaces on MRI and TRUS.16, 21, 22 Narayanan et. al.21 aligned prostate surfaces, obtained using a semi-automated segmentation requiring manual selection of 4 or more fiducials on the prostate surface, using a deformable adaptive focus model. Karnik et. al.22 used Thin Plate Splines (TPS) to align prostate surfaces, obtained using a semi-automated segmentation requiring manual selection of 10 or more fiducials on the prostate surface. Natarajan et. al.16 extended this approach to (a) require only 4 − 6 fiducials on the prostate surface as well as (b) incorporate elastic interpolation when aligning MRI and TRUS.16 They used this system to guide biopsy in 56 patients and achieved a CaP detection rate of 23%, compared to 7% for systematic, nontargeted biopsies.16 Sonn et. al.28 found a CaP detection rate of 53% in 171 men; for patients with highly suspicious MRI findings 15 out of 16 patients were found to have a positive CaP biopsy. The difference in CaP detection rates between Natarajan et. al.16 and Sonn et. al.28 possibly reflects the different patient populations considered in each study; Sonn et. al.28 considered patients with persistently increased PSA, hence this study selected patients at an increased risk for CaP.
Hu et. al.24 performed MRI-TRUS fusion using a model-based method, where a patient-specific finite element model (FEM) of the prostate on MRI was constructed and used to determine a deformable transformation. Model initialization on the TRUS image required specifying two fiducials identifying the base and apex of the prostate. In addition, manual adjustment of filter parameters was required.
A significant limitation of the current state-of-the-art MRI-TRUS fusion methods is the need for manual intervention after TRUS acquisition. All of the presented methods rely on user interaction to identify the location of the prostate either by selecting fiducials or delineating the prostate.16, 18, 20–25 TRUS acquisition occurs just prior to needle biopsy, hence, the need for manual intervention represents a significant disadvantage, in terms of time and efficiency. Inaccuracies in defining landmarks or prostate boundaries manually may also introduce error into the registration. For instance, manual prostate delineation, on either MRI or TRUS, has been documented to have inter-observer variability of 1.7 to 2.5 mm.19
3.1 Novel Contribution
The primary novel contribution of this work is a method for MRI-TRUS fusion that is automated for all steps after TRUS acquisition. Unlike current state-of-the-art methods, our method does not require identifying landmarks or segmenting the prostate on TRUS. The registration workflow consists of the following modules: (1) delineating the prostate on MRI (to be performed prior to TRUS acquisition), (2) constructing a probabilistic model of the prostate location on TRUS using intensity and texture features, and (3) aligning the MRI prostate segmentation to the TRUS probabilistic model via a novel image metric. The only required manual pre-processing is prostate segmentation on MRI, which can be performed offline prior to TRUS acquisition.
To construct the probabilistic model of the prostate location on TRUS we introduce two novel methods (1) a novel application of attenuation correction for improved registration accuracy on TRUS and (2) a novel method to determine the location of the prostate on TRUS by calculating a probabilistic model using texture and spatial features. Finally a novel registration metric to align a mask to a probabilistic model is introduced to allow for registration of the T2w MRI segmentation to the probabilistic model of prostate locations on TRUS.
4. METHODOLOGY
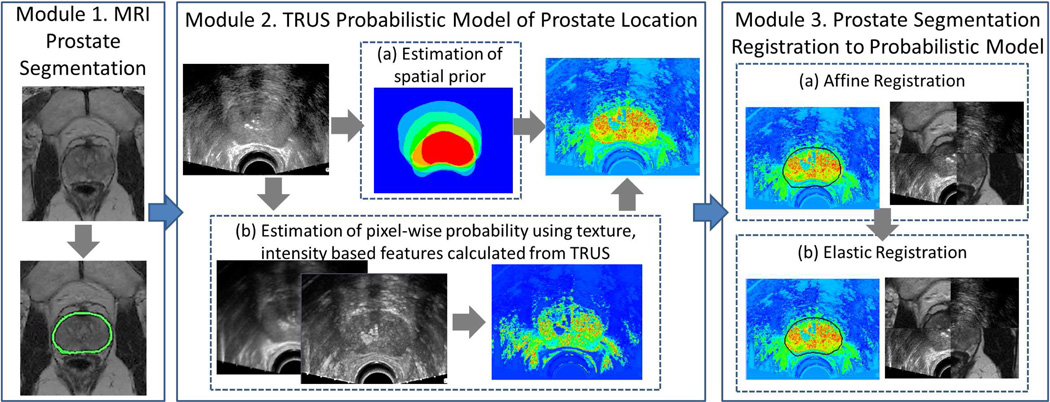
Table 2 lists the notation used throughout this paper. Figure 1 displays a flowchart of our methodology which consists of the following three modules:
Module 1: Determine prostate location on MRI. Prior to TRUS acquisition an expert manually delineated the prostate on T2w MRI.
Module 2: Create a probabilistic model of prostate location on TRUS. As an initial step attenuation correction is performed on the TRUS imagery. The probabilistic model is created by, (a) estimated the likely location of the prostate on TRUS, (b) extracting texture and intensity features, and (c) estimating the probability of each pixel belonging to the prostate using the features extracted in steps a and b.
Module 3: Register MRI prostate segmentation and TRUS probabilistic model. Registration is performed by (a) affine registration to account for translation, rotation, and scale differences between images followed by (b) elastic registration to account for differences in prostate deformation.
Table 2.
Description of notation used throughout this paper.
| Notation | Description | Notation | Description |
|---|---|---|---|
| 𝒞M | 3D MRI image scene. | Pi(FT(c)) | Probability of FT (c) belonging to class i. |
| CM | 3D grid of pixels on 𝒞M. | ℳT | 3D TRUS prostate segmentation. |
| fM(c) | MRI image intensity function for c ∈ CM. | gT(c) | TRUS segmentation function for c ∈ CT. |
| ℳM | 3D MRI prostate segmentation. | ΩT,i | Collection of pixels in CT that belong to class i. |
| Pi(c) | TRUS segmentation function for c ∈ CT. | μF,i | Mean vector of FT (c) for ΩT,i. |
| gM(c) | MRI segmentation function for c ∈ CM. | ΣF,i | Covariance matrix of FT (c) for ΩT,i. |
| 𝒞T | 3D TRUS image scene. | ℳ̂T | Estimated TRUS prostate segmentation. |
| CT | 3D grid of pixels on 𝒞T. | gT(c) | Estimated TRUS segmentation function for c ∈ CT. |
| fT(c) | TRUS image intensity function for c ∈ CT. | T | Transformation function. |
| 𝒞P,i | 3D probabilistic model for class i. | S(T(ℳM), 𝒞T) | Similarity metric for T(ℳM) and CT. |
| Pi(c) | Probability of belonging to class i for c ∈ CT. | R(T) | Regularization metric for T. |
| FT(c) | Set of intensity and texture based features for c ∈ CT. | ΩM,i | Collection of pixels in CM that belong to class i. |
| (r,θ, z) | Corresponding polar coordinates for c ∈ CT. | p | Control point location defined on CT. |
| f̃T(c) | TRUS image intensity function with out signal attenuation for c ∈ CT. | E[p] | Expected location of control point p. |
| Pi(c) | Spatial prior for c ∈ CT. | 𝒩(p) | Set of control points with neighbor p. |
Figure 1.
Flowchart for our registration algorithm consisting of the following modules: (1) Prostate delineation on MRI (prostate boundary shown in green); (2) Construction of probabilistic model of prostate location on TRUS where blue corresponds to pixels most unlikely to belong to the prostate, red corresponds to pixels most likely to belong to the prostate. The model consists of estimating (a) likely location of prostate on TRUS (spatial prior) and (b) likely appearance of prostate on TRUS (texture and intensity features); (3) Registration of MRI segmentation to TRUS model by (a) affine (translation, rotation, scale) registration and (b) elastic registration.
4.1 Module 1: Prostate Segmentation on MRI
A 3D MRI volume 𝒞M = (CM, fM) is defined by a set of 3D Cartesian coordinates CM and the image intensity function fM(c) : c ∈ CM. Modules 1 determines a 3D prostate segmentation ℳM = (CM, gM) such that gM(c) = i for a pixel c belonging to class i where i = 1 represents prostate and i = 0 represents background. In this work, the prostate is manually delineated on 𝒞M to obtain ℳM.
4.2 Module 2: Probabilistic Model of Prostate Location on TRUS
A 3D TRUS volume 𝒞T = (CT, fT) is defined in a similar way as 𝒞M (Section 4.1). From 𝒞T a model 𝒞P,i = (CT, Pi(c)) is calculated, where Pi(c) : c ∈ CT is the probability of the pixel c belonging to class i. As an initial step attenuation correction29 is performed on 𝒞T to account for spatial variations in image intensity. Pi(c) is then calculated by (i) extraction of texture and intensity features defined as FT (c), and (ii) construction of Pi(c).
4.2.1 Attenuation Correction
Ultrasound imagery may have attenuation artifacts, where pixels closer to the ultrasound probe appear brighter than pixels far away. Attenuation is caused by signal loss as the ultrasound waves propagate through tissue.29, 30 As the TRUS probe is circular, variations in image intensity will be along radial lines from the probe. To account for these changes attenuation correction methods similar to MRI bias field correction have been presented.29 Attenuation correction of TRUS imagery has been demonstrated to be important for segmentation.29, 31 To the best of our knowledge, attenuation correction has not been applied in the context of facilitating and improving registration.
Attenuation correction is performed as follows. For each pixel c ∈ CT with a set of 3D Cartesian coordinates expressed as (x, y, z) such that the probe center is defined as x = 0, y = 0, z = 0, we calculate a set of corresponding polar coordinates as follows,
| (1) |
Image attenuation is modeled within the polar coordinated reference frame as,
| (2) |
where f̃(r, θ, z) is the true, unknown TRUS signal associated with the location (r, θ, z). η(r, θ, z) is additive white Gaussian noise assumed to be independent of f̃(r, θ, z). Similar to Cohen et. al.,32 β(r, θ, z) may be estimated via convolution of a smoothing Gaussian kernel with the image, i.e. a low-pass filtering of the signal. The true underlying signal may then be recovered using the equation,
| (3) |
where lpf is a low-pass filter. f̃(r, θ, z) is then converted back into 3D Cartesian coordinates, f̃(c) : c ∈ CT.
4.2.2 Feature Extraction
For each pixel f̃(c) : c ∈ CT a set of texture and intensity features FT (c) are calculated. FT (c) is calculated using each feature in Table 3 independently. The features chosen describe (a) intensity for a pixel or a region (mean, median), (b) intensity variation in a region (range), (c) intensity variation assuming either a Gaussian (variance), Rayleigh (variance), or Nakagami (m-parameter) distribution. Rayleigh and Nakagami distributions were considered because of their utility in describing the statistics of ultrasound imagery.33, 34
4.2.3 Probabilistic Model Calculation
The probability of pixel c belong to class i defined as Pi(c) is dependent on the location of c and the feature set FT (c). We define the probability of a location c belonging to tissue class i is Pi(c). Similarly the probability of a set of features FT (c) belonging to tissue class i is Pi(FT (c)). We assume Pi(c) and Pi(FT (c)) are independent, and hence the final probability Pi(c) may be expressed as
| (4) |
For the remainder of this section we describe the calculation of Pi(c) and Pi(FT (c)).
Spatial Prior: Pi(c), referred to as the spatial prior, is the likelihood of pixel c belonging to class i based on its location. Pi(c) is calculated from a set of J training studies 𝒞T,j : j ∈ {1, …, J}, where for each study the prostate has been delineated by an expert, giving the 3D prostate segmentation ℳT,j. The prostate segmentation is defined ℳT,j = (CT, gT,j) such that gT,j(c) = i for a pixel c belonging to class i where i = 1 represents prostate and i = 0 represents background. The origin for each study is set as the center of the TRUS probe, so that the location of pixel c has a consistent position relative to the TRUS probe across all studies. Pi(c) is defined as,
| (5) |
Hence Pi(c) is the frequency of pixel c being located in the prostate across J training studies.
Feature Set Probability: The probability Pi(FT (c)) is the likelihood of a set of features FT (c) associated with pixel c belonging to class i. In this work, we assume FT (c) may be accurately modeled as a multivariate Gaussian distribution with a mean vector of μF,i and a covariance matrix ΣF,i for the ith class. Given the Gaussian distribution parameters μF,i and ΣF,i, the probability Pi(FT (c)) is calculated as,
| (6) |
where k is the number of features in FT (c). However μF,i and ΣF,i are unknown therefore these parameters must be estimated.
We assume an initial rigid transformation Tr (Section 4.3) which provides an estimate of the alignment between MRI and TRUS. An estimated prostate segmentation may then be defined as ℳ̂T = Tr(ℳM) where ℳ̂T = (CT, ĝT) and ĝT (c) = i for a pixel c estimated to belong in class i. μF,i and ΣF,i are then calculated as, , where ΩT,i is the collection of pixels in CT belonging to class i according to ĝT (c) and | · | is the cardinality of a pixel set. ΣF,i is similarly defined for a covariance matrix of FT (d) for ΩT,i.
4.3 Module 3: Registration of MRI Segmentation and TRUS Probabilistic Model
The goal of registration in this work is to find a transformation T to spatially map 𝒞M onto 𝒞T. In this work T is calculated to align ℳM and 𝒞T. T is calculated via the equation,
| (7) |
where S(T (ℳM), 𝒞T) is a similarity metric between T (ℳM) and 𝒞T. R(T) is a regularization function which penalized T which are not smoothly varying and α determines the weight of R relative to S(·, ·).
The similarity metric S(·, ·) is calculated as,
| (8) |
where ΩM,i is the collection of pixels in CM belonging to class i. T is initialized with a rigid transformation Tr such that overlap between ℳM and P1(c) is maximized calculated as
| (9) |
Given the initial alignment Tr, an affine registration Ta followed by an elastic registration Te was used to align the MRI and TRUS images.
4.3.1 Affine Registration
An affine transformation (translation, rotation, and scale) defined as Ta was used to align the MRI and TRUS. No regularization R(T) was used for affine registration, as this transformation by definition is smoothly varying. Not defining R(T) is equivalent to setting α = 0.
4.3.2 Elastic Registration
An elastic B-spline-based transformation Te was used to recover differences in prostate deformation between MRI and TRUS.36 Te is defined by a set of control points which determine the transformation Te for all c ∈ CM. Each control point, defined by its location p ∈ CM, is allowed to move independently.
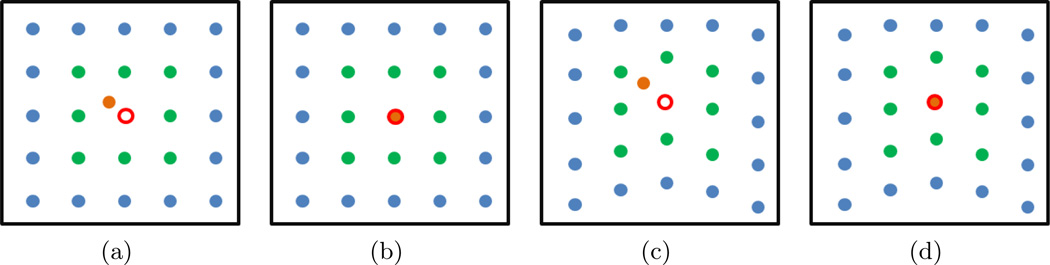
The term R(T) is added to constrain Te to only those transformations which are likely to occur. R(T) is calculated as , where p is the location of a B-Spline control point and E[p] is the maximum likelihood estimate of where p should be located. In this work E[p] was estimated as where 𝒩(p) is the set of control points which neighbor p and | · | is the cardinality of a set. Thus E[p] is the average over the set of knots which neighbor the knot at location p. Figure 2 gives a 2D pictorial representation of our regularization approach.
Figure 2.
A graphical example of B-Spline regularization. The control point of interest p is highlighted in orange. Control points corresponding to 𝒩(p) are highlighted in green. According to our formulation for E[p], the expected location of p is shown by an open red circle. (a), (c) Example where R(T) would have a high value because p is far from E[p]. (b) and (d) would both give a low R(T) value because p is near E[p]. For (c), (d) the deformation not local to p is not taken into account when considering E[p], however other control points may result in a higher R(T) compared to (b).
R(T) is defined such that if p = E[p] then the knot p will contribute 0 to the value of R(T). As p moves farther from E[p], the value of (1 − exp−∥(p − E[p])∥) increases, and contributes more to the value of R(T). Hence R(T) is lower for evenly spaced, smoothly varying control points than for randomly spaced, erratically varying control points. Deformations that are not evenly spaced and smoothly varying will only occur if they improve the similarity metric S(·, ·).
5. EXPERIMENTAL DESIGN AND RESULTS
5.1 Dataset Description
T2-weighted MRI imagery was acquired using a Siemens 1.5 T scanner and a pelvic phased-array coil for 7 patients under IRB approval. 3D TRUS imagery was acquired using a bi-planar side-firing transrectal probe for each patient. For each pair of images an expert radiologist manually selected corresponding fiducials. Corresponding fiducials included, the urethra, the center of regions suspicious for CaP, and the center of small calcifications. In addition, an expert radiologist manually delineated the prostate boundary on MRI and TRUS.
5.2 Evaluation Measures
RMSE is a measure of how well two corresponding sets of fiducials align; a RMSE of 0 represents perfect alignment. A manually selected set of fiducials on MRI is defined as . Similarly, a set of fiducials on TRUS is defined as , such that the corresponds to . RMSE is calculated as .
5.3 Implementation Details
All methods described in this paper were implemented using the Insight Segmentation and Registration Toolkit (ITK) version 4.3.37 All texture features were calculated using a 𝒩(c) of a spherical neighbourhood of size 1 mm3. Both affine and elastic transformations were optimized at a single resolution using a Powell optimization scheme.38 The regularization weight was set as α = 1, such that the similarity metric and regularization function were given equal weights.
5.4 Experiment 1: Effect of Attenuation Correction
It is our hypothesis that subtle differences in intensity characteristics across the TRUS image can lead to a probabilistic model Pi(c) that does not accurately model the prostate location. Incorrect estimation of Pi(c) can result in sub-optimal image registration. In this experiment we evaluate the effect of attenuation correction as described in Section 4.2.1 on registration accuracy, in terms of RMSE. We evaluate RMSE for MRI alignment to TRUS imagery with and without attenuation correction.
5.5 Experiment 2: Selection of TRUS Model Feature Set
A set of intensity and texture features F(c) are used to calculate the probabilistic model Pi(c) that guides registration as described in Sections 4.2.2 and 4.2.3. The accuracy of Pi(c) depends on the choice of features in F(c); those features which are best able to distinguish prostate tissue from non-prostate tissue will lead to a more accurate Pi(c) and will lead to a more accurate image registration. In this paper we evaluated 7 features described in Table 3 in terms of RMSE.
6. EXPERIMENTAL RESULTS
6.1 Experiment 1: Effect of Attenuation Correction
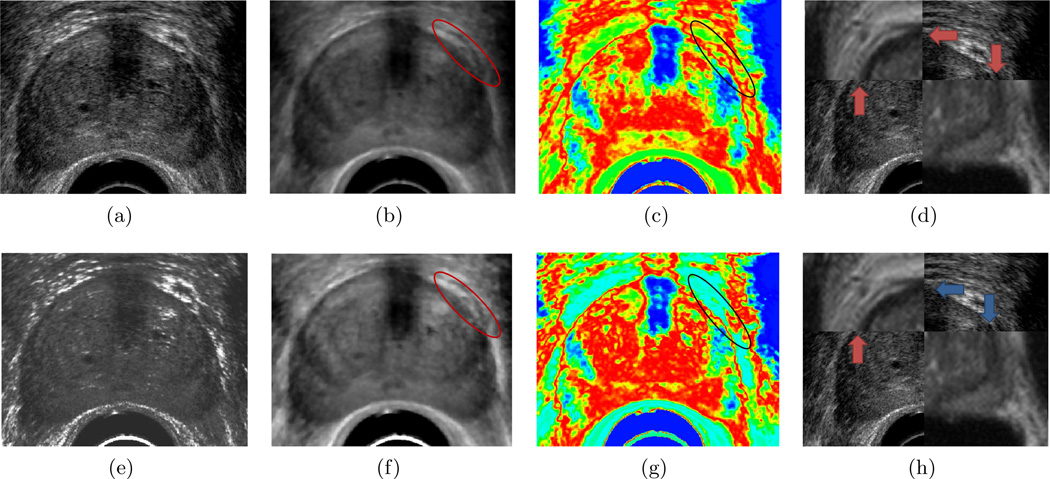
Table 4 presents quantitative results for Ta and Te, with and without attenuation correction for all 7 features. Figure 3 shows an study where attenuation correction improved the results by over 1 mm, qualitatively the attenuation corrected TRUS (h) more accurately aligns to the MRI to the non-attenuation corrected TRUS (d).
Table 4.
RMSE for Ta and Te evaluated using 7 feature sets. The symbol * denotes a spurious registration, where the two images had no alignment. The best registration results, obtained from median and Nakagami m-parameter features are bolded.
| Feature | Attenuation Correction |
Transformation | ||
|---|---|---|---|---|
| Tr | Ta | Te | ||
| Initial Alignment | 5.50 ± 1.49 mm | - | - | |
| Intensity | No | - | 4.86 ± 1.58 mm | 4.52 ± 1.62 mm |
| Yes | - | 4.15 ± 1.06 mm | 3.99 ± 1.09 mm | |
| Mean | No | - | 4.88 ± 2.03 mm | 4.61 ± 2.05 mm |
| Yes | - | 3.73 ± 1.25 mm | 3.54 ± 1.17 mm | |
| Median | No | - | 4.75 ± 1.82 mm | 4.46 ± 1.65 mm |
| Yes | - | 3.79 ± 1.29 mm | 3.32 ± 1.05 mm | |
| Range | No | - | 4.42 ± 1.27 mm | 4.13 ± 1.34 mm |
| Yes | - | 4.60 ± 1.18 mm | 4.25 ± 1.22 mm | |
| Variance | No | - | 4.23 ± 0.98 mm | 3.92 ± 1.06 mm |
| Yes | - | 4.56 ± 1.38 mm | 4.36 ± 1.39 mm | |
| Rayleigh Variance33 |
No | - | 4.54 ± 1.76 mm | 4.30 ± 1.76 mm |
| Yes | - | 3.72 ± 1.16 mm | 3.50 ± 0.90 mm | |
| m-parameter34 | No | - | * | * |
| Yes | - | 3.79 ± 1.15 mm | 3.39 ± 0.85 mm | |
Figure 3.
Example of a TRUS image (a) without and (e) with attenuation correction; the corresponding median feature (b), (f); the corresponding probability models (c), (g), where blue corresponds to those pixels most unlikely to belong to the prostate, red corresponds to those pixels most likely to belong to the prostate; and the final registration results (d), (h). Blue arrows in (d) and (h) show boundary regions which are well aligned on MRI and TRUS, while red arrows show boundary regions which are misaligned. The region highlighted by the red circle in (b) and (f) show regions where attenuation correction improved the feature contrast between the prostate and background pixels. The corresponding region on the probability models is highlighted by the black circle (c), (g). Note the image with attenuation correction (g) is better able to distinguish between pixels belonging to the prostate from the background, resulting in a more accurate registration.
In all cases, except range and standard deviation features, attenuation correction prior to feature extraction resulted in improvement of the registration accuracy, as evaluated by RMSE. For many cases, attenuation correction improved RMSE by 1 mm. The m-parameter is a special case, where without attenuation correction the registration failed to align the images. This feature is particularly sensitive to noise, and the presence of attenuation artifacts results in Pi(FT (c)) evaluated for the m-parameter feature being unable to distinguish between pixels belonging to the prostate and the background.
6.2 Experiment 2: Selection of TRUS Model Feature Set
Table 4 presents quantitative results for Ta and Te for all 7 features evaluated. The best performing features are derived from two features types (a) descriptions of the expected intensity value (mean and median) and (b) ultrasound specific descriptions of the intensity variance (Rayleigh variance, Nakagami m-parameter). The median feature performed slightly better than the mean feature, most likely because the median is more robust to pixels with outlier intensities compared to the mean.39 Similarly, the Nakagami m-parameter performed better than the Rayleigh variance, as the Nakagami distribution is a generalized form of the Rayleigh distribution therefore the Nakagami m-parameter should be better able to model the underlying distribution of pixel intensities.34 In comparison, image features which describe the variance in the data which are not ultrasound specific (range, standard deviation), performed poorly in terms of registration accuracy. This is most likely because these measures are not able to accurately model the distribution of pixel intensities in TRUS imagery.
Figure 5(c) displays an example of a qualitative registration result using the Nakagami m-parameter. Clear misalignment of the prostate near the rectum (red arrow) was apparent after Ta because of the differences in prostate deformation caused by no MRI endorectal coil and the TRUS probe. Hence, a need exists for Te to account for the differences in prostate deformation between MRI and TRUS. Figure 5(d) displays MRI and TRUS images after Te was applied; the prostate boundary near the rectum is better aligned between MRI and TRUS.
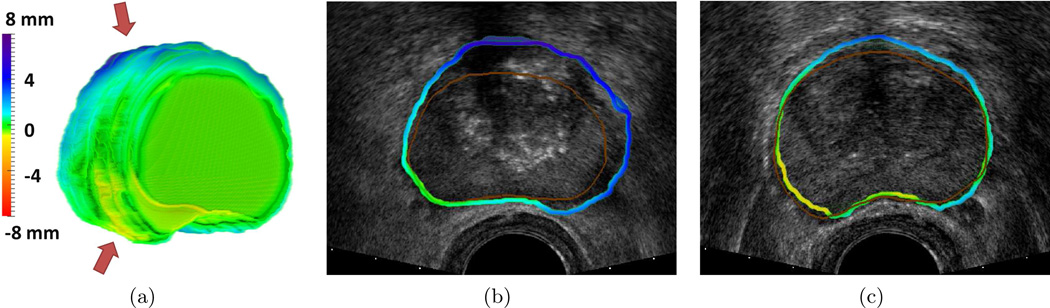
To further evaluate our registration accuracy we created surface renderings of the prostate surface as shown in Figure 4, such that blue represents regions where the MRI image was misaligned external to the prostate surface on TRUS and red represents regions where the MRI image was misaligned internal to the prostate surface on TRUS. In the qualitative example shown there are two regions of misalignment, near the rectal wall (yellow) and near the bladder (blue). In figure 4(b) an axial plane of the TRUS is displayed with two boundaries overlaid, (1) the axial cross section of the surface rendering shown in figure 4 and the true prostate boundary (brown line). The hyperechoic region distal to the TRUS probe results in Pi(c) being unable to appropriately model the location of the prostate, hence the large registration error of ≈ 4 mm. Similarly Figure 4(c) a different axial plane of the TRUS is displayed with the cross section of the surface rendering shown in figure 4 and the true prostate boundary (brown line). First note that this misalignment is much less pronounced, representing a registration error of ≈ 1 mm. Near the rectal wall the error is primarily because Te is unable to fully account for the subtle differences in prostate deformation.
Figure 4.
(a) Prostate surface rendering such that the prostate base is facing toward the right, where blue represents regions where the MRI image was misaligned external to the prostate surface on TRUS and red represents regions where the MRI image was misaligned internal to the prostate surface on TRUS. (b) 2D axial image on TRUS displaying a region of large misalignment distal to the TRUS probe, where brown represents the expert delineation of the prostate on TRUS. (c) 2D axial image on TRUS displaying a region of misalignment near the TRUS probe, where brown represents the expert delineation of the prostate on TRUS.
7. CONCLUDING REMARKS
This paper describes a novel method to spatially align MRI and TRUS images of the prostate so that no manual intervention is required after TRUS acquisition. RMSE for our method in 7 patient studies was 3.39±0.85 mm. Unlike previously described methods,16, 18, 20–25 our method requires no manual intervention.
The biggest limitation of the current implementation is the requirement to segment the MRI prior to TRUS acquisition. Therefore, planned future work will incorporate an automated prostate segmentation scheme that has been developed by our group40 to delineate the prostate in Module 1 (Section 4.1). An additional limitation of this work is the use of the B-Spline transformations in Module 3 (Section 4.3), which recover non-linear deformations with few additional constraints, to account for the difference in deformation of the prostate between MRI and TRUS imagery. In this work we imposed an additional regularization constraint to ensure the underlying deformation in the prostate was smoothly varying. However, other transformations such as Finite Element Models (FEM), which allow for explicit modeling of tissue physics, could also potentially be used to drive the MRI-TRUS fusion.24 In future work we will consider other deformation transformations and regularization constraints to model the differences in deformation of the prostate between MRI and TRUS imagery.
Finally an extended analysis of the presented method is planned to include further evaluation of, (a) optimization schemes such as evolutionary,41 and gradient descent38 optimization methods may be better able to find the transformation maximum. (b) additional features such as Haralick texture features42 and additional ultrasound specific features43 may provide greater ability to distinguish between prostate and non prostate tissues, (c) combinations of multiple features to build the probability model may lead to a more accurate and robust estimate of the prostate location on TRUS.
Table 1.
State-of-the-art MRI-TRUS fusion methods indicating the commercial system (where available), technical work, whether each method requires volumetric (3D) or planar (2D) imagery, the type of manual intervention required, the number of studies, and the reported root mean square error (RMSE). Direct comparison between RMSE for different systems is not possible as different internal fiducials (or in some cases phantoms) were used to evaluate each method.
| Commercial System |
Technical Work |
2D/3D | Intervention | Studies | Root Mean Squared Error (RMSE) |
|---|---|---|---|---|---|
| Urostation | Reynier et. al.20 | 3D | Manual prostate segmentation on TRUS | 11 | 2.07 ± 1.57 mm (urethra) 1.11 ± 0.54 mm (prostate 1.11 ± 0.54 mm (prostate |
| ProFuse | Narayanan et. al.21 | 3D | Semi-automated prostate segmentation on TRUS | 2 | 3.06±1.41 mm (phantom) |
| Karnik et. al.22 | 3D | Semi-automated prostate segmentation on TRUS | 16 | 2.13 ± 0.80 mm | |
| UroNav | Xu et.al.23 | 3D | Manual refinement of prostate segmentation on TRUS | 20 | 2.3 ± 0.9 mm (phantom) |
| - | Hu et.al.24 | 3D | Manual identification of prostate apex and base on TRUS, adjustment of filter weights | 8 | 2.40 mm (median) |
| - | Mitra et.al.18 | 2D | Manual prostate segmentation on TRUS | 20 | 1.60 ± 1.17 mm |
ACKNOWLEDGEMENTS
This work was made possible by grants from the National Institute of Health (R01CA136535, R01CA140772, R43EB015199, R21CA167811), National Science Foundation (IIP-1248316), Department of Defense (W81XWH-11-1-0179), and the QED award from the University City Science Center and Rutgers University.
REFERENCES
- 1.Wolf AMD, Wender RC, Etzioni RB, Thompson IM, D’Amico AV, Volk RJ, Brooks DD, Dash C, Guessous I, Andrews K, DeSantis C, Smith RA. American cancer society guideline for the early detection of prostate cancer: Update 2010. CA: A Cancer Journal for Clinicians. 2010;60(2):70–98. doi: 10.3322/caac.20066. [DOI] [PubMed] [Google Scholar]
- 2.Hodge KK, McNeal JE, Terris MK, Stamey TA. Random systematic versus directed ultrasound guided transrectal core biopsies of the prostate. The Journal of urology. 1989;142(1):71–75. doi: 10.1016/s0022-5347(17)38664-0. [DOI] [PubMed] [Google Scholar]
- 3.Halpern EJ, Strup SE. Using gray-scale and color and power doppler sonography to detect prostatic cancer. American Journal of Roentgenology. 2000;174(3):623–627. doi: 10.2214/ajr.174.3.1740623. [DOI] [PubMed] [Google Scholar]
- 4.Spajic B, Eupic H, Tomas D, Stimac G, Kruslin B, Kraus O. The incidence of hyperechoic prostate cancer in transrectal ultrasound-guided biopsy specimens. Urology. 2007;70(4):734–737. doi: 10.1016/j.urology.2007.06.1092. [DOI] [PubMed] [Google Scholar]
- 5.Djavan B, Ravery V, Zlotta A, Dobronski P, Dobrovits M, Fakhari M, Seitz C, Susani M, Borkowski A, Boccon-Gibod L, Schulman CC, Marberger M. Prospective evaluation of prostate cancer detected on biopsies 1, 2, 3, and 4: When should we stop? Journal of Urology. 2001;166(5):1679–1683. [PubMed] [Google Scholar]
- 6.Babaian RJ, Toi A, Kamoi K, Troncoso P, Sweet J, Evans R, Johnston D, Chen M. A comparative analysis of sextant and an extrended 11-core multisite directed biopsy strategy. The Journal of Urology. 2000;163(1):152–157. [PubMed] [Google Scholar]
- 7.Tiwari P, Viswanath S, Kurhanewicz J, Sridhar A, Madabhushi A. Multimodal wavelet embedding representation for data combination (MaWERiC): integrating magnetic resonance imaging and spectroscopy for prostate cancer detection. NMR in Biomedicine. 2012;25(4):607–619. doi: 10.1002/nbm.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng D, Tempany CM. MR imaging of the prostate and bladder. Seminars in Ultrasound, CT, MRI. 1998;19(1):67–89. doi: 10.1016/s0887-2171(98)90025-7. [DOI] [PubMed] [Google Scholar]
- 9.Parker GJM, Suckling J, Tanner SF, Padhani AR, Revell PB, Husband JE, Leach MO. Probing tumor microvascularity by measurement, analysis and display of contrast agent uptake kinetics. Journal of Magnetic Resonance Imaging. 1997;7(3):564–574. doi: 10.1002/jmri.1880070318. [DOI] [PubMed] [Google Scholar]
- 10.Charles-Edwards EM, deSouza NM. Diffusion-weighted magnetic resonance imaging and its application to cancer. Cancer Imaging. 2006;6:135–143. doi: 10.1102/1470-7330.2006.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurhanewicz J, Vigneron DB. Advances in MR spectroscopy of the prostate. Magnetic Resonance Imaging Clinics of North America. 2008;16(4):697–710. doi: 10.1016/j.mric.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anastasiadis A, Lichy M, Nagele U, Kuczyk MA, Merseburger AS, Hennenlotter J, Corvin S, Sievert K, Claussen CD, Stenzl A, Schlemmer HP. Mri-guided biopsy of the prostate increases diagnostic performance in men with elevated or increasing psa levels after previous negative trus biopsies. European Urology. 2006;50:738–749. doi: 10.1016/j.eururo.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Pondman KM, Fütterer JJ, ten Haken B, Kool LJS, Witjes JA, Hambrock T, Macura KJ, Barentsz JO. MR-guided biopsy of the prostate: an overview of techniques and a systemic review. European Urology. 2008;54:517–527. doi: 10.1016/j.eururo.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Labanaris AP, Engelhard K, Zugor V, Nutzel R, Kuhn R. Prostate cancer detection using an extended prostate biopsy schema in combination with additional targeted cores from suspicious images in conventional and functional endorectal magnetic resonance imaging of the prostate. Prostate Cancer and Prostatic Diseases. 2010;13(1):65–70. doi: 10.1038/pcan.2009.41. [DOI] [PubMed] [Google Scholar]
- 15.Hadaschik BA, Kuru TH, Tulea C, Rieker P, Popeneciu IV, Simpfendörfer T, Huber J, Zogal P, Teber D, Pahernik S, Roethke M, Zamecnik P, Roth W, Sakas G, Schlemmer HP, Hohenfellner M. A novel stereotactic prostate biopsy system integrating pre-interventional magnetic resonance imaging and live ultrasound fusion. The Journal of Urology. 2011;186(6):2214–2220. doi: 10.1016/j.juro.2011.07.102. [DOI] [PubMed] [Google Scholar]
- 16.Natarajan S, Marks LS, Margolis DJ, Huang J, Macairan ML, Lieu P, Fenster A. Clinical application of a 3d ultrasound-guided prostate biopsy system. Urologic Oncology: Seminars and Original Investigations. 2011;29(3):334–342. doi: 10.1016/j.urolonc.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rud E, Baco E, Eggesbo HB. Mri and ultrasound-guided prostate biopsy using soft image fusion. Anticancer Research. 2012;32(8):3383–3389. [PubMed] [Google Scholar]
- 18.Mitra J, Kato Z, Mart R, Oliver A, Llad X, Sidib D, Ghose S, Vilanova JC, Comet J, Meriaudeau F. A spline-based non-linear diffeomorphism for multimodal prostate registration. Medical Image Analysis. 2012;16(6):1259–1279. doi: 10.1016/j.media.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Usmani N, Ghosh S, Kamal W, Pedersen J, Pervez N, Yee D, Danielson B, Murtha A, Amanie J, Sloboda RS. Comparison of prostate volume, shape, and contouring variability determined from preimplant magnetic resonance and transrectal ultrasound images. Brachytherapy. 2012;11(4):284–291. doi: 10.1016/j.brachy.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Reynier C, Troccaz J, Fourneret P, Dusserre A, Gay-Jeune C, Descotes J-L, Bolla M, Giraud J-Y. Mri/trus data fusion for prostate brachytherapy. preliminary results. Medical Physics. 2004;31(6):1568–1575. doi: 10.1118/1.1739003. [DOI] [PubMed] [Google Scholar]
- 21.Narayanan R, Kurhanewicz J, Shinohara K, Crawford E, Simoneau A, Suri J. MRI-ultrasound registration for targeted prostate biopsy. ISBI. 2009:991–994. [Google Scholar]
- 22.Karnik VV, Fenster A, Bax J, Cool DW, Gardi L, Gyacskov I, Romagnoli C, Ward AD. Assessment of image registration accuracy in three-dimensional transrectal ultrasound guided prostate biopsy. Medical Physics. 2010;37(2):802–813. doi: 10.1118/1.3298010. [DOI] [PubMed] [Google Scholar]
- 23.Xu S, Kruecker J, Turkbey B, Glossop N, Singh AK, Choyke P, Pinto P, Wood BJ. Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies. Computer Aided Surgery. 2008;13(5):255–264. doi: 10.1080/10929080802364645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Ahmed H, Taylor Z, Allen C, Emberton M, Hawkes D, Barratt D. Mr to ultrasound registration for image-guided prostate interventions. Medical Image Analysis. 2012;16:687–703. doi: 10.1016/j.media.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan I, Oldenburg N, Meskell P, Blake M, Church P, Holupka EJ. Real time MRIultrasound image guided stereotactic prostate biopsy. Magnetic Resonance Imaging. 2002;20:295–299. doi: 10.1016/s0730-725x(02)00490-3. [DOI] [PubMed] [Google Scholar]
- 26.Pinto PA, Chung PH, Rastinehad AR, Jr, AAB, Kruecker J, Benjamin CJ, Xu S, Yan P, Kadoury S, Chua C, Locklin JK, Turkbey B, Shih JH, Gates SP, Buckner C, Bratslavsky G, Linehan WM, Glossop ND, Choyke PL, Wood BJ. Magnetic resonance imaging/ultrasound fusion guided prostate biopsy improves cancer detection following transrectal ultrasound biopsy and correlates with multiparametric magnetic resonance imaging. The Journal of Urology. 2011;186(4):1281–1285. doi: 10.1016/j.juro.2011.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daanen V, Gastaldo J, Giraud J, Fourneret P, Descotes J, Bolla M, Collomb D, Troccaz J. Mri/trus data fusion for brachytherapy. International Journal of Medical Robotics and Computer Assisted Surgery. 2006;2:256–261. doi: 10.1002/rcs.95. [DOI] [PubMed] [Google Scholar]
- 28.Targeted biopsy in the detection of prostate cancer using an office based magnetic resonance ultrasound fusion device. The Journal of Urology. 2013;189(1):86–92. doi: 10.1016/j.juro.2012.08.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao G, Brady M, Noble J, Zhang Y. Segmentation of ultrasound b-mode images with intensity inhomogeneity correction. IEEE Transactions on Medical Imaging. 2002;21(1):48–57. doi: 10.1109/42.981233. [DOI] [PubMed] [Google Scholar]
- 30.Pye S, Wild S, McDicken W. Adaptive time gain compensation for ultrasonic imaging. Ultrasound in Medicine & Biology. 1992;18(2):205–212. doi: 10.1016/0301-5629(92)90131-s. [DOI] [PubMed] [Google Scholar]
- 31.Ashton EA, Parker KJ. Multiple resolution bayesian segmentation of ultrasound images. Ultra- sonic Imaging. 1995;17(4):291–304. doi: 10.1177/016173469501700403. [DOI] [PubMed] [Google Scholar]
- 32.Cohen M, Dubois R, Zeineh M. Rapid and effective correction of rf inhomogeneity for high field magnetic resonance imaging. Human Brain Mapping. 2000;10(4):204–211. doi: 10.1002/1097-0193(200008)10:4<204::AID-HBM60>3.0.CO;2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burckhardt CB. Speckle in ultrasound B-mode scans. IEEE Transactions on Sonics and Ultrasonics. 1978;25(1):1–6. [Google Scholar]
- 34.Yacoub MD, Bautista JEV, Guerra de Rezende Guedes L. On higher order statistics of the Nakagami-m distribution. IEEE Transactions on Vehicular Technology. 1999;48(3):790–794. [Google Scholar]
- 35.Greenwood JA, Durand D. Aids for fitting the gamma distribution by maximum likelihood. Technometrics. 1960;2(1):55–65. [Google Scholar]
- 36.Rueckert D, Sonoda L, Hayes C, Hill D, Leach M, Hawkes D. Nonrigid registration using free-form deformations: application to breast mr images. IEEE Transactions on Medical Imaging. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 37.Yoo T, Ackerman M, Lorensen W, Schroeder W, Chalana V, A S, Metaxas D, Whitaker R. Proc. of Medicine Meets Virtual Reality. Westwood: IOS Press Amsterdam; 2002. Jan, Engineering and algorithm design for an image processing api: A technical report on itk - the insight toolkit; pp. 586–592. e., ed. [PubMed] [Google Scholar]
- 38.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes 3rd Edition: The Art of Scientific Computing. 3 ed. New York, NY, USA: Cambridge University Press; 2007. [Google Scholar]
- 39.Devore J, Farnum N. Applied Statistics for Engineers and Scientists. 2 ed. Belmont, CA, USA: Thomas Brooks/Cole; 2005. [Google Scholar]
- 40.Toth R, Madabhushi A. Multi-feature landmark free active appearance models: Application to prostate MRI segmentation. IEEE Transactions on Medical Imaging. 2012 Aug;38:1638–1650. doi: 10.1109/TMI.2012.2201498. [DOI] [PubMed] [Google Scholar]
- 41.Jacq J, Roux C. Registration of 3-D images by genetic optimization. Pattern Recognition Letters. 1995;16(8):823–841. [Google Scholar]
- 42.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Transactions on Systems Man, and Cybernetics. 1973;3(6):610–621. [Google Scholar]
- 43.Eltoft T. Modeling the amplitude statistics of ultrasonic images. IEEE Transactions on Medical Imaging. 2006;25(2):229–240. doi: 10.1109/TMI.2005.862664. [DOI] [PubMed] [Google Scholar]