The process by which organisms evolved elaborate signaling networks is a fascinating mystery in the fields of evolutionary and cellular biology. Emergence, diversity, and evolution of signaling networks are being rapidly understood with improved knowledge of various genomes. However, continued progress must not solely depend on analyses of the primary gene and protein sequences, rather, we must incorporate analyses of the evolution of the physical, dynamic, and biochemical properties of the encoded signaling elements. For example, compare the Gα subunits of the animal and plant heterotrimeric G-protein complex. These 2 molecules are about 30% identical at the primary sequence level, yet they have nearly the exact 3D crystal structure.1 However, despite having the same 3D structure, these 2 molecules have different dynamic properties and share a different evolutionary history (discussed below). Here, we address the evolution of G signaling using a highly divergent system of G signaling in the protist Trichomonas vaginalis and propose a regulatory mechanism for G proteins that do not follow the animal paradigm.
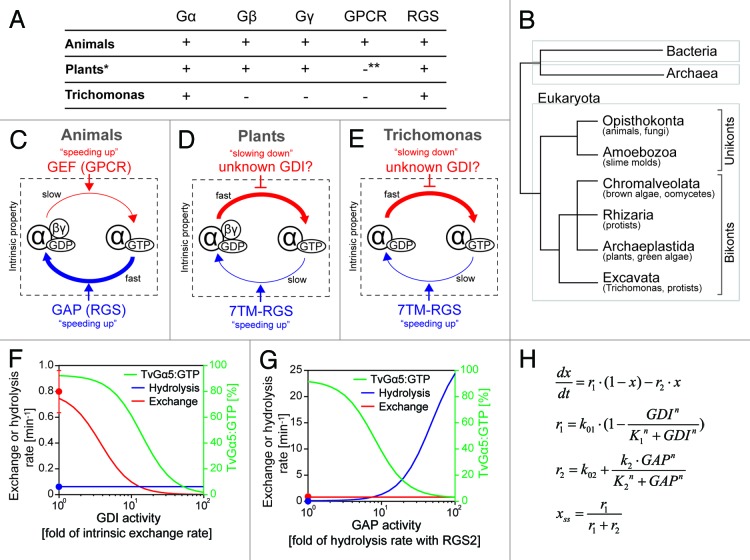
As is well known in animals and fungi, G-proteins are activated by ligand-bound G protein-coupled receptors (GPCRs) and consequently modulate activity of downstream effectors (Fig. 1C).2,3 However, GPCR-dependent activation is limited to Unikonts, defined below,4-6 despite the presence of canonical G protein subunits (Gα, Gβ, and Gγ) broadly throughout eukaryotes (Fig. 1A).6 For example, in the model plant, Arabidopsis thaliana, the G-protein can form the active guanosine trisphosphate (GTP)-bound state without GPCRs (Fig. 1D).1,4 How is this unusual Gα regulated in vivo? Arabidopsis has a 7-transmembrane protein tethered to the GTPase-accelerating protein (GAP) called regulator of G signaling (RGS) protein RGS domain (7TM-RGS), which constitutively promotes GTP hydrolysis by Gα on plasma membrane.7 Therefore, animal GPCRs modulate the activation level by speeding up the exchange rates of G protein, while plant 7TM-RGSs determine the level by modulating the inactivation step (Fig. 1C and D). Eukaryotes are separated into 2 basal clades, unkonta, and bikonta (Fig. 1B). The unikonta includes animals, yeasts, and slime molds, while bikonta includes plants, algae, and some protists. Recently, we analyzed comprehensively the intrinsic property of Gα from several unikonts and bikonts,6 and found that bikonta Gα rapidly exchange guanosine diphosphate (GDP) for GTP, while hydrolyzed GTP slowly, indicating that the rate-limiting step determining the steady-state of G cycle is GTP hydrolysis, not nucleotide exchange. This implies that bikonta G proteins do not require guanine nucleotide exchange factors (GEFs), like GPCRs, for activation, and that the primary regulators in bikonta may be GAPs or guanine nucleotide dissociation inhibitors (GDIs), which slow down GDP dissociation from Gα.
Figure 1. G-protein components and intrinsic properties seen in animals, plants, and Trichomonas. (A) G-protein-related genes encoded in the animal, plant, and Trichomonas genomes. *Some plants (e.g., cereals) lack RGS genes. **Plants have one 7TM protein that has weak similarity to the slime mold cAMP receptor, but the original function of this protein is not a GPCR. (B) Classification of eukaryotes by molecular phylogenetics. Eukaryotes are classified into 2 basal clades, unikonta and bikonta, subclassified into 6 monophyletic supergroups, Opisthokonta, Amoebozoa, Chromalveolata, Rhizaria, Archaeplastida and Excavata. Unikonta includes animals, fungi, and slime molds, while bikonta includes plants, algae, and protists. Note: G-protein genes (Gα, Gβ, and Gγ) are only found in eukaryotes. (C–E) G-protein machinery in animals, plants, and Trichomonas. (C) Animal G-protein slowly exchanges GDP for GTP and rapidly hydrolyzes GTP. GPCRs and other GEFs increase the rate of nucleotide exchange, therefore shifts the Gα state onto active “GTP-bound” form. RGSs and other GAPs speed up the hydrolysis reaction. (D) Plant G protein rapidly exchanges nucleotide and slowly hydrolyses GTP. 7TM-RGSs promote GTP hydrolysis by Gα and keep the inactive GDP-bound state. Regulators controlling the exchange rate are not found in plants, except the Gβγ dimer, which has a weak GDI activity. (E) Trichomonas G-proteins rapidly exchange nucleotide and slowly hydrolyze GTP. 7TM-RGSs enhance the GAP activity of Gα, but the hydrolysis rate with RGS is not enough to keep the GDP-bound state of Gα. Note: Trichomonas lacks Gβγ dimer. (F–H) Simple qualitative model was developed to simulate the steady-state fraction of GTP-bound TvGα5 (% of TvGα5:GTP) in response to the variation of GDI or GAP activities. (F) The model shows that as GDI activity increased, the exchange rate and the % of TvGα5:GTP decreased, while the hydrolysis rate did not vary. The red or blue line (with y-axis on the left) shows rates for nucleotide exchange or GTP hydrolysis over increased activity of GDI. The red or blue dots indicate intrinsic exchange rate of TvGα5 or the hydrolysis rate promoted by TvRGS2, as reported [Bradford W, 2013]. The green line (with y-axis label on the right) shows the percentage of the % of TvGα5:GTP. (G) The model shows an enhanced hydrolysis rate as GAP activity increased, resulting in a significant decrease in the % of TvGα5:GTP. The nucleotide exchange rate did not vary. (H) Mathematical equations used in the model. x = % of TvGα5:GTP in the total pool of TvGα5, which is assumed here to remain at a constant level. The steady-state of x is denoted as xss. r1 or r2 is the rate of nucleotide exchange or hydrolysis, respectively. Their computed values are shown in panels (F and G) (with y-axis on the right) with respect to changes in GDI and GAP activity. Rate constant k01 or k02 is the rate of intrinsic nucleotide exchange or steady-state hydrolysis rate with the cognate GAP, TvRGS2; k01 = 0.8 min−1, k02 = 0.062 min−1. K1 and K2 represents the functional threshold of GDI and GAP activity, K1 = K2 = 3 min−1. k2 denotes the maximum enhance in the hydrolysis as the GAP activity increases, k2 = 30 min−1. n is the Hill coefficient, n = 2.
In addition to the unusual intrinsic property of these “self-activating” G proteins, some bikonts do not possess genes for canonical G protein regulators, such as GPCR, RGS, and/or Gβγ. Take as an example the simplest G protein system seen in the protist, Trichomonas vaginalis, which has several isotypes of Gα (TvGα) and 7- or 5-transmembrane RGSs (TvRGS), but no Gβ, Gγ and GPCRs (Fig. 1A and E). The intrinsic exchange (0.80 min−1) or hydrolysis (6.2 x 10−3 min−1) rate of TvGα5 indicated that the rate-limiting step is GTP hydrolysis, like other bikonts. The hydrolysis rate was increased by TvRGS2 (0.062 min−1), although this rate is still too low to shift the TvGα5 pool predominantly to the GDP-bound state.
In order to investigate potential mechanisms controlling the activation dynamics of the Trichomonas G protein, a simple qualitative mathematical model was developed to predict the steady-state fraction of active TvGα with respect to changes in GDI or GAP activity (Fig. 1F–H). Specifically, we modeled the nucleotide exchange and GTP hydrolysis reactions between GDP-bound (TvGα5:GDP) and GTP-bound (TvGα5:GTP) status, with rates being affected by either an unknown GDI (Fig. 1F) or an unknown stimulator/ligand enhancing TvRGS2 activity (Fig. 1G), respectively. As shown in Figure 1F, the exchange rate decreased as GDI activity increased, resulting in a continuous drop in the fraction of TvGα5:GTP. A similar result was achieved by enhancing the GAP activity alone. The model showed that the hydrolysis rate exceeded the intrinsic exchange rate with a ~10-fold increase in the GAP activity (Fig. 1G). Since both models succeeded to control the fraction of GDP- or GTP-bound TvGα5, GDIs or GAPs, but not GEFs, regulate the G cycle in Trichomonas and thus likely in other bikonta. Therefore, based on these results, we propose that either a GDI or GAP controls the nucleotide-binding state of bikont eukaryotes (Fig. 1F–H).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26350
References
- 1.Jones JC, et al. Sci Signal. 2011;4:ra8. doi: 10.1126/scisignal.2001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilman AG. Annu Rev Biochem. 1987;56:615–49. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 3.Dohlman HG. Annu Rev Physiol. 2002;64:129–52. doi: 10.1146/annurev.physiol.64.081701.133448. [DOI] [PubMed] [Google Scholar]
- 4.Johnston CA, et al. Proc Natl Acad Sci U S A. 2007;104:17317–22. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urano D, et al. PLoS Genet. 2012;8:e1002756. doi: 10.1371/journal.pgen.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford W, et al. Sci Signal. 2013;6:ra37. doi: 10.1126/scisignal.2003768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JG, et al. Science. 2003;301:1728–31. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]