Abstract
The RAD9A-RAD1-HUS1 (9-1-1) complex is a PCNA-like heterotrimeric clamp that binds damaged DNA to promote cell cycle checkpoint signaling and DNA repair. While various 9-1-1 functions in mammalian somatic cells have been established, mounting evidence from lower eukaryotes predicts critical roles in meiotic germ cells as well. This was investigated in 2 recent studies in which the 9-1-1 complex was disrupted specifically in the mouse male germline through conditional deletion of Rad9a or Hus1. Loss of these clamp subunits led to severely impaired fertility and meiotic defects, including faulty DNA double-strand break repair. While 9-1-1 is critical for ATR kinase activation in somatic cells, these studies did not reveal major defects in ATR checkpoint pathway signaling in meiotic cells. Intriguingly, this new work identified separable roles for 9-1-1 subunits, namely RAD9A- and HUS1-independent roles for RAD1. Based on these studies and the high-level expression of the paralogous proteins RAD9B and HUS1B in testis, we propose a model in which multiple alternative 9-1-1 clamps function during mammalian meiosis to ensure genome maintenance in the germline.
Keywords: RAD9A-RAD1-HUS1 complex, meiosis, double-strand break repair, synapsis, ATR, TOPBP1, HUS1B, RAD9B, pachytene checkpoint
Introduction
The 9-1-1 complex is an evolutionarily conserved, heterotrimeric PCNA-like clamp composed of the RAD9A (initially referred to in the literature as RAD9), RAD1, and HUS1 proteins.1,2 The 9-1-1 complex is a first responder that binds to sites of DNA damage and is thought to function in both DNA damage checkpoint signaling and DNA repair. During DNA replication stress, 9-1-1 recognizes DNA damage and promotes cell cycle checkpoint signaling through the ATR kinase. Roles for 9-1-1 in DNA repair are less well understood; however, physical and functional interactions with many DNA repair proteins have been reported, including a variety of DNA polymerases, glycosylases, nucleases, and the RAD51 recombinase.1-3 These interactions implicate 9-1-1 function in DNA repair pathways as diverse as base excision repair, nucleotide excision repair, mismatch repair, and homologous recombination, most likely in recognizing DNA damage and coordinating recruitment of critical repair factors.
While the activities of 9-1-1 in mammalian somatic cells have been studied extensively, little is known about functions for this complex in the mammalian germline. Studies in lower eukaryotes suggest that the 9-1-1 complex may have critical roles in meiotic recombination and homologous chromosome synapsis as well as meiotic checkpoint signaling.4-13 The germline functions of mammalian 9-1-1 were investigated in 2 recent studies in which conditional mutations in Rad9a14 and Hus115 were targeted to undifferentiated spermatogonia using mice that express CRE recombinase under the control of the Stra8 promoter.16 The results revealed insights into the functions of DNA damage checkpoint proteins during meiosis, including key roles for components of the 9-1-1 complex in meiotic double-strand break (DSB) repair as well as the intriguing finding that RAD1 may have RAD9A- and HUS1-independent activities during meiosis.
The 9-1-1 Complex Functions in ATR Activation and DNA Repair in Somatic Cells
Perhaps the best characterized role for the mammalian 9-1-1 complex in somatic cells is in promoting DNA damage signaling and cell cycle arrest through the ATR pathway in response to single-stranded DNA accumulation during replicative stress and DNA lesion processing. In the traditional view of ATR kinase activation, the 9-1-1 complex is loaded onto 5′ recessed ends at DNA damage sites, and subsequent interaction between RAD9A and TOPBP1 enables the latter to stimulate ATR kinase activity.17-23 An additional protein, RHINO, interacts with both 9-1-1 and TOPBP1,24 and is also necessary for ATR activation. While it has been proposed that a major role of 9-1-1 is to recruit TOPBP1 to sites of DNA damage, recent work indicates that TOPBP1 can also be recruited to particular DNA structures by the MRN (MRE11-RAD50-NBS1) complex,25-27 and that 9-1-1 plays a role in an ensuing step that enables TOPBP1 to stimulate ATR.25,27 Such evidence is consistent with previous studies in Xenopus indicating that TOPBP1 mediates recruitment of the 9-1-1 complex to damage sites for ATR activation.28,29 Alternative modes of ATR activation have been described in budding yeast, where the 9-1-1 complex (DDC1-RAD17-MEC3) or DNA2 can directly activate ATR (MEC1) independently of TOPBP1 (DPB11).30-32 In mammalian germ cells, ATR, TOPBP1, and RAD1 proteins have been reported to localize along meiotic chromosomes.33-35 However, many details remain to be resolved regarding the mode of ATR activation in germ cells, and to the best of our knowledge, the Rad9a14 and Hus1 studies are the first to suggest that meiotic ATR activation is, at least in part, 9-1-1-independent.15
A variety of studies in somatic cells indicate that the 9-1-1 complex not only contributes to ATR activation but also participates directly in DNA repair. Notably, 9-1-1 interacts with, and in many cases stimulates the activity of, multiple factors involved in base excision repair, including flap endonuclease 1 (FEN1),36 human 8-oxoguanine DNA glycosylase (hOGG1),37 MutY glycosylase (MYH),38 DNA polymerase β,39 and DNA ligase I.40 Multiple interactions have also been reported between 9-1-1 subunits and the DNA mismatch repair factors MutS homolog 2 (MSH2), MSH3, MSH6, and MutL homolog 1 (MLH1),41,42 and mismatch recognition by the MSH proteins is stimulated by the presence of 9-1-1.41 Furthermore, human RAD9A and MSH6 colocalize in nuclear foci following DNA methylating agent treatment, and RAD9A focus formation is dependent upon the presence of MSH6.41 Li and coworkers also demonstrated that RAD9A is important for nucleotide excision repair by maintaining, among others, the protein level of the excision factor DDB2 in human cells,43 and RAD9A-deficient mouse embryonic stem cells are slow to repair UV-induced DNA damage.44 These repair-related functions of 9-1-1 are likely to be relevant to meiotic chromosome biology, as many of the DNA repair factors mentioned here have known or predicted roles in meiotic cells. For example, while DNA polymerase β functions during base excision repair in somatic cells, it is also critical for synapsis of homologous chromosomes during mouse meiosis.45,46
Several lines of evidence indicate that the vertebrate 9-1-1 complex functions in repair of DSBs as well. For example, siRNA knockdown of Hus1 expression was shown to decrease the efficiency of homologous recombination in response to ionizing radiation (IR)-induced DNA damage, and HUS1-deficient mouse cells are hypersensitive to IR despite intact nonhomologous end joining (NHEJ).47 Mammalian RAD9A interacts with the RAD51 recombinase,3 and Rad9a inactivation leads to decreased repair of DSBs by homologous recombination and inefficient exit from G2 phase following IR exposure.3,48 Similarly, RAD9A-deficient mouse embryonic stem cells initiate but do not maintain IR-induced G2 delay.44 Immunoglobulin gene conversion in Rad9a−/− chicken DT40 cells is substantially reduced in a manner similar to Rad51 or Brca2 mutants with known defects in homologous recombination.49 Rad9a is also required for immunoglobulin class switch recombination in mouse B cells.50 These somatic functions for 9-1-1 raise the possibility of a related role in meiotic DSB repair, since many of the key players, particularly RAD51, are essential for homologous recombination in both somatic and germ cells.
DNA Damage Responses during Meiosis
Meiosis is a specialized cell division that involves purposeful breakage and repair of meiotic chromosomes. Given the roles of 9-1-1 in DNA repair in somatic cells, it would not be surprising that the complex functions in a similar capacity during meiosis. Crucial redistribution of genetic material takes place during meiotic recombination, initiated by programmed DSBs. These are introduced by the evolutionarily conserved endonuclease SPO11 at the onset of meiosis and serve as a target for repair through the use of either homologs (inter-homolog repair) or sister chromatids (inter-sister repair) as a recombination/repair template. Repair requires that homologous sequences recognize each other, a process aided by the single-stranded overhangs produced by DSB resection. Two mammalian homologs of E. coli RecA, RAD51, and DMC1, assist the homology search by promoting strand invasion of resected DNA ends into homologous DNA sequences.51,52 The ultimate goal of this process is to generate crossovers (COs), which, together with sister chromatid cohesion, provide physical connections between maternal and paternal homologs that ensure correct segregation during the first meiotic division.
Genetic analyses in lower eukaryotes, particularly in budding yeast,13,53 worms,11 and flies,7,8 have established a role for 9-1-1 in promoting meiotic DSB repair. In yeast, rad17 (mammalian Rad1) and rad24 (a 9-1-1 clamp loader subunit) mutants exhibit delayed meiotic DSB repair that results in an altered ratio of crossover-to-noncrossover products.13 These mutants also have decreased colocalization of RAD51 and DMC1 foci as well as delayed disappearance of RAD51 but not DMC1 foci. Yeast rad17 mutants additionally exhibit elevated levels of unequal sister chromatid recombination during meiosis.53 In Drosophila hus1 mutant oocytes, DSBs are not processed efficiently, and oocyte nuclear defects are abrogated by blockage of DSB formation.7,8 Drosophila oocytes also exhibit a genetic interaction between hus1 and the DNA repair gene brca2.8
DSB repair is also coordinated and tightly coupled with dynamic changes in chromatin architecture that facilitate the homology search and stabilize interactions between homologous DNA sequences.54-56 These changes in meiotic chromosome dynamics begin with the formation of synaptonemal complexes (SCs) between pre-aligned homologs. SCs play an important role in DSB repair and CO formation.54,57 These proteinaceous structures consist of axial elements, each comprised of SYCP3 protein and sister chromatid pairs, which later become linked together by SYCP1-containing transverse filaments of the central element. Axial elements begin to form during leptonema prior to SC formation, which starts in zygonema. SCs are fully assembled in early pachynema and disassemble as cells progress through diplonema.58 In mammals, spermatocytes with defects in SC formation or DSB repair are eliminated at mid-pachynema, and coordination between SC formation and DSB repair is essential for proper meiotic progression.58-60
Throughout meiotic prophase in male mammals, large non-homologous portions of the X and Y chromosomes remain mostly unsynapsed. These regions of the sex chromosomes are remodeled into a transcriptionally silenced, phospho-histone H2AX (γH2AX)-rich chromatin domain termed the XY body or sex body.61-63 The importance of a functional XY body is demonstrated by the observation that male H2ax−/− mice, which fail to produce this structure in their pachytene spermatocytes, are sterile and exhibit meiotic disruption at pachynema.64 While a major function of the XY body is thought to be protection of the partially unsynapsed sex chromosomes from triggering the synaptic checkpoint machinery at pachynema, it is clear that aberrant events within this structure can initiate a checkpoint response.
Rad9a and Hus1 are Critical for Mammalian Fertility
Homologous chromosome synapsis and DNA repair are essential aspects of meiosis and thus organismal fertility. Consistent with the possibility that the mammalian 9-1-1 complex plays critical roles during gametogenesis, 9-1-1 subunits are highly expressed in the mouse testis,14,15,33,65-67 and both RAD1 and RAD9A localize to meiotic chromosomes.15,33 Furthermore, Hus1 gene expression is reduced in the mouse testis in the absence of SPO11,68 although it remains possible that this indicates that HUS1 function requires progression further into meiotic prophase I rather than a role specifically in meiotic recombination. In order to elucidate the germline functions of murine 9-1-1, conditional mutations were generated in Rad9a14 and Hus115 using mice that express CRE recombinase in undifferentiated spermatogonia under control of the Stra8 promoter.16 This strategy was adopted because whole-body deletion of either Rad9a44 or Hus169 results in embryonic lethality. Loss of Rad9a or Hus1 function in male germ cells resulted in disrupted fertility due to meiotic defects, indicating important roles for these factors in the germline. In both models, the numbers of mature sperm were greatly reduced, and fertility was severely compromised. Some Rad9a conditional knockout (CKO) mice lacked sperm and were infertile, while a second, sub-fertile cohort of Rad9a CKO males had sperm counts between 0.6–10% of the wild-type controls.14 Hus1 CKO adults had on average only 10% of the normal number of epididymal sperm.15 Testis size was drastically reduced in both models, and seminiferous tubules exhibited severely decreased cellularity, vacuolization, large multinucleate cells, and increased apoptosis.14,15
Despite expected defects in mitotically dividing spermatogonia concurrent with depletion of RAD9A and HUS1, no overt pre-meiotic abnormalities were observed in Rad9a CKOs (A Vasileva, DJ Wolgemuth, HB Lieberman, unpublished results).14 Some apoptotic spermatogonia were observed in Hus1 CKO mice, potentially but not conclusively indicating possible pre-meiotic defects.15 In both models, the majority of germ cells underwent apoptosis during the pachytene stage of meiosis, although there were subtle yet distinct differences in the timing. Rad9a CKO spermatocytes underwent apoptosis by early to mid-pachynema, leaving few late pachytene or diplotene cells.14 In contrast, only a subset of Hus1 CKO cells underwent apoptosis during pachynema, leaving a substantial fraction of late pachytene and diplotene cells that were cleared prior to entry into metaphase.15
The minor differences between the 2 models, which used similar gene targeting strategies, could relate to the kinetics of RAD9A or HUS1 depletion, determined by protein half-life and other factors, or could indicate functional differences between the subunits of the 9-1-1 complex. For instance, RAD9A may have additional HUS1-independent functions. In other cell types, RAD9A can function as a transcriptional activator, promoting transcription of a specific set of downstream target genes.70,71 RAD9A physically interacts with p53 binding consensus sequences in the promoter of p21, and can activate transcription of p21 in a p53-independent manner. As there is no evidence that either HUS1 or RAD1 can bind the p21 promoter in a similar fashion, this may be a unique feature of the mammalian RAD9A protein. Whether this transcriptional activity of RAD9A is needed for progression of meiosis has yet to be determined, but might explain some of the phenotypic differences between Rad9a and Hus1 CKO spermatocytes.
RAD9A and HUS1 Are Essential for a Subset of Meiotic Recombination Events
One of the most striking phenotypes in both the Hus1 and Rad9a CKO models is the presence of unrepaired meiotic DSBs. In both models, DSBs appeared to form with appropriate timing and in appropriate numbers.14,15 In Hus1 CKO leptotene spermatocytes, RAD51 foci were present in approximately wild-type numbers; however, unlike control spermatocytes in which foci disappeared during pachynema, Hus1-deficient spermatocytes harbored between 4 and 8 autosomal RAD51 foci in late pachynema that persisted into diplonema.15 Furthermore, in the Hus1 CKO study, the number of COs, as measured by numbers of pachytene MLH1 foci and bivalent chromosomes at diakinesis, was similar to that in control animals. While the majority of DSBs were successfully repaired in both models, flares of γH2AX staining on pachytene and diplotene chromosomes, persistent RAD51 foci on late pachytene and diplotene chromosomes, and flares of TOPBP1 staining on pachytene chromosomes indicated the presence of unrepaired DSBs in Hus1 CKOs. Similar patterns of γH2AX and TOPBP1 staining were seen in Rad9a CKO spermatocytes, and persistent DMC1 foci were also evident,14 suggesting a role for the entire 9-1-1 complex in meiotic DSB repair. Although these data suggest defective repair of only a small subset of meiotic DSBs in the absence of 9-1-1, this number of unrepaired DSBs likely was sufficient to cause cell death. In addition, delayed or defective DSB repair was pervasive, as all Hus1 CKO pachytene spermatocytes and 72% of diplotene spermatocytes contained persistent RAD51 foci.15 While the persistent breaks in Hus1 and Rad9a CKO spermatocytes were detected by the presence of RAD51 and DMC1 foci, respectively, it is possible that aberrant intermediates were present earlier and could contribute to other observed phenotypes, such as synapsis defects and inclusion of autosomes within the XY body domain. Altogether, this evidence supports a model in which the mammalian 9-1-1 complex plays critical roles in DSB repair.
What is unique about the small subset of meiotic DSBs that remains unrepaired in Rad9a and Hus1 CKOs? One possibility is that a DNA structure generated at these particular break sites requires 9-1-1 for recognition or processing during the course of repair. It might be that these are recombination events involving multiple chromatids, such as those seen in BLM/Sgs1 helicase and Mus81 endonuclease mutants,72-74 which would be consistent with the observation of persistent paired RAD51 foci on either side of the SC axial elements in diplotene-stage Hus1 CKO spermatocytes.15 Such a model would fit well with the recently identified role of yeast 9-1-1 in error-free DNA damage tolerance, where it functions in homologous recombination and template switching in mitotic cells, resulting in the formation of sister chromatid joint molecules that require resolution by SGS1 helicase (homolog of mammalian BLM) and TOP3 topoisomerase (homolog of mammalian TOP3A).75 It is tempting to speculate that 9-1-1 might play a similar role in inter-sister chromatid recombination during meiosis, perhaps stabilizing intermediates as they accumulate RAD51 recombinase.
Another possibility is that the persistent breaks observed in Hus1 and Rad9a CKOs are those that are repaired later in prophase I than most DSBs, and that 9-1-1 is important for repair of these late DSBs, due to differences in chromatin structure, specific DNA intermediates, or absence of particular recombination factors. Such a possibility would be consistent with the fact that RAD9A foci normally persist on the sex chromosome cores during pachynema, colocalizing with RAD51 foci at these sites.15 Although RAD9A can interact with MLH1,42 it seems unlikely that the persistent DSBs in Rad9a and Hus1 CKOs result from failed crossing over, as approximately normal numbers of pachytene MLH1 foci and bivalent diakinesis chromosomes were observed in the absence of HUS1. We also cannot exclude the possibility that HUS1 is involved in partner choice during the homology search, and that perhaps some of the DSB sites indicated by MLH1 foci in the Hus1 CKO are resolved by inter-sister chromatid rather than inter-homolog chromosome recombination events. This might explain the large percentage of Hus1 CKO spermatocytes that undergo apoptosis near meiotic metaphase, but appears inconsistent with the observation of normal numbers of bivalents at diakinesis.
ATR Pathway Functions during Meiosis: Relevance to the 9-1-1 Complex
ATR and its activator, TOPBP1, are present on meiotic chromosomes and have been implicated in genome surveillance during mammalian meiosis.34,35,76,77 In particular, both factors localize to sites undergoing meiotic silencing of unsynapsed chromatin (MSUC), which occurs on the autosomes, and meiotic sex chromosome inactivation (MSCI), which occurs in the XY body,76,78 suggesting that ATR kinase activation occurs in response to asynapsis. Currently it is thought that ATR phosphorylates histone H2AX at sites of MSCI and MSUC, including the XY body domain, while the ATM kinase phosphorylates H2AX at DSBs.78,79 However, this has been difficult to assess, since inactivation of Atr and other ATR pathway components results in embryonic lethality in mice.44,69,80-82 Interestingly, ATM-deficient spermatocytes accumulate massive amounts of ATR on meiotic chromosomes,35 supporting a role for ATR in monitoring meiotic chromosome abnormalities. The germ cell-specific conditional knockout strategies described here for Rad9a14 and Hus115 provide new evidence for ATR pathway components playing a role in the mammalian germline, though not necessarily in an ATR-dependent manner, as elaborated below.
Mouse RAD9A and HUS1 Are Largely Dispensable for Homolog Synapsis
In both Hus1 and Rad9a CKO spermatocytes, synapsis and SC defects were observed at low frequency. Specifically, 14% of pachytene-like Hus1 CKO spermatocytes had improperly synapsed autosomes,15 while asynapsis or incomplete synapsis was observed in ~10–15% of pachytene-like Rad9a CKO spermatocytes (A Vasileva, DJ Wolgemuth, and HB Lieberman, unpublished results). In addition, ~36% of diplotene-stage Hus1 CKO spermatocytes15 and ~22–25% of pachytene-like Rad9a CKO spermatocytes14 harbored ruptured SCs. Could these synapsis defects be due to an inherent defect in SC structure in the absence of 9-1-1? Alternatively, could ruptured SCs be due to unrepaired DSBs that lead to SC discontinuities? In yeast, interaction of 2 9-1-1 subunits, MEC3 (HUS1) and DDC1 (RAD9A), with the SC lateral element structural protein RED1 is required for SC formation.4 Drosophila hus1 mutant oocytes also exhibit abnormal SC disassembly.8 Thus, it would not be unexpected for the mammalian 9-1-1 complex to have roles related to SC formation, maintenance, or disassembly. However, the relatively infrequent occurrence of improperly synapsed chromosomes in Rad9a and Hus1 CKO spermatocytes suggests that 9-1-1 function is not essential for synapsis per se.
Some phenotypes in meiotic Hus1 and Rad9a CKOs may indicate more subtle defects in the SC or in chromosome structure. For example, phosphorylated γH2AX persists in flares perpendicular to the SC,14,15 which are similar to previously described L-foci,83 rather than in simple punctate foci along the chromosome cores. This suggests that in the absence of 9-1-1, DSBs may either become uncoupled from the SC, or be associated with extensive resection or altered chromatin configurations. An aberrant chromatin configuration at some meiotic DSBs in 9-1-1 CKOs might not be surprising, as it has been reported that both RAD9A and HUS1 can form a complex with the HDAC1 histone deacetylase.84 The large, flared γH2AX foci (and similar TOPBP1 foci) could also be due to formation of multi-chromatid recombination intermediates at some DSBs, structures that might also lead to the presence of paired RAD51 foci on each side of the SC as noted above.15 Additionally, we cannot rule out the possibility that the aberrant γH2AX and TOPBP1 localization is due to altered DNA damage response signaling at some DSBs in the absence of an intact 9-1-1 complex. In C. elegans, disruption of the sister chromatid cohesion gene scc-2 leads to failure to load HUS-1 at sites of early meiotic DNA damage, as well as failure of cohesin loading and defective DNA repair,85 raising the possibility that 9-1-1 might interact with meiotic cohesin proteins and coordinate DSB repair.
New Insights into ATR Kinase Activation: Disruption of 9-1-1 Does Not Preclude MSCI or XY Body Formation
As part of the 9-1-1 complex, RAD9A and HUS1 are predicted to function in ATR kinase activation through interaction with TOPBP1 at DNA damage sites. It was therefore somewhat surprising that TOPBP1 localization to the XY body appeared unperturbed (Hus1 CKO)15 or only slightly affected (Rad9a CKO)14 in primary spermatocytes in the absence of the 9-1-1 complex. H2AX phosphorylation at presumptive autosomal DSBs also proceeded without RAD9A or HUS1 as noted above. Similarly, gene silencing via MSCI appeared intact as evidenced by the normal exclusion of RNA polymerase II from the XY body domain in Hus1 CKO spermatocytes.15 That TOPBP1 may activate ATR in a RAD9A- and HUS1-independent manner is novel, and distinct even from alternative modes of ATR activation seen during mitotic DNA damage checkpoint activation (discussed above).
The XY body formed in all Rad9a and Hus1 CKO cells, though both studies reported XY body defects, such as extended γH2AX domains and inclusion of whole or partial autosomes. While such defects can have severe consequences, particularly with respect to inappropriate gene silencing, these abnormalities are likely not due to primary synapsis defects in the absence of RAD9A or HUS1, but rather to secondary effects related to unrepaired or perhaps aberrantly processed DSBs.
Does 9-1-1 Play a Role in Meiotic Checkpoint Signaling?
In budding yeast5,9,12 and other model organisms,7,11,86,87 meiotic checkpoints have been well described and contain at least 2 major (yet intertwined) branches—synapsis monitoring and DSB repair monitoring—both of which primarily occur during the pachytene stage and thus have been termed branches of the pachytene checkpoint. It is not clear whether or how meiotic checkpoints operate in mammals, though it seems likely that there are multiple mechanisms that monitor and regulate progression through meiosis. Additionally, many of the same proteins involved in somatic cell checkpoints, including ATM, H2AX, ATR, TOPBP1, RAD9A, RAD1, and HUS1, are expressed during meiosis, and most are known to localize along meiotic chromosomes.5,14,15,33-35,76,83
A variety of meiotic defects have been shown to lead to cell cycle arrest and apoptosis between zygonema and mid-pachynema in stage IV of the seminiferous epithelium.77 These include cases of complete absence of DSBs (such as in Spo11 mutants),88 impaired DSB repair (such as in Dmc1 and Msh5 mutants),89-91 autosomal asynapsis,92,93 or impaired MSCI.94 This period has been loosely termed the pachytene checkpoint in mammals, despite being less well defined than in lower eukaryotes.5 As previously mentioned, meiotic gene silencing via MSUC and MSCI involves factors that have checkpoint activity in somatic cells, and it is reasonable to predict that these may be components of a mammalian synaptic checkpoint.
A possible role for the 9-1-1 complex in a putative pachytene checkpoint is supported by the significant fraction of Hus1 CKO spermatocytes that progressed beyond pachynema into the diplotene stage despite containing abnormal chromosomes, including those with ruptured SCs, persistent γH2AX and RAD51 foci, and extended XY body domains marked by γH2AX.15 If meiotic checkpoints exist and are intact, such abnormal cells should be cleared during pachynema, suggesting that a subset of Hus1 CKO spermatocytes do not have the capacity to undergo pachytene-stage apoptosis. Because most Rad9a CKO cells did not progress as far into meiosis, it is difficult to assess whether such a proposed checkpoint role would involve the entire 9-1-1 complex, though it is possible that Rad9a CKO cells simply have more DNA damage or additional defects compared with Hus1 CKO cells, and thus die earlier.
Does this mean that HUS1 is required for a pachytene checkpoint? Interestingly, we observed a loss of phosphorylated CHK2 in Hus1 CKO whole-testis lysates,15 suggesting that HUS1, and potentially 9-1-1, may be required for CHK2 phosphorylation, perhaps in response to DSBs. In mouse somatic cells, HUS195 and RAD9A96,97 each show strong genetic interactions with the ATM kinase, which is activated in response to DSBs and phosphorylates CHK2. While functional interactions during meiotic DSB repair would not be surprising, we expect that 9-1-1 checkpoint functions primarily involve ATR activation. We observed that TOPBP1 localizes to meiotic chromosomes in Hus1 and Rad9a CKOs, indicating that TOPBP1 recruitment to DSBs is not mediated via 9-1-1, and in light of recent studies in Xenopus extracts and cultured mammalian cells,25-27 it may be that TOPBP1 recruitment to meiotic DSBs is instead facilitated by the MRN complex.
If synapsis and DSB repair checkpoints do exist during mammalian meiosis, there are clearly aspects of signaling that appear to be HUS1- and RAD9A-independent. For example, in both models, H2AX phosphorylation appeared normal,14,15 though this is not surprising, since H2AX phosphorylation can be ATM-dependent,79 or as seen during responses to replication stress, ATR-dependent but 9-1-1-independent.98,99 Additionally, increased total CHK1 and P-CHK1 were observed in the absence of HUS1,15 suggesting that signaling to CHK1 during meiosis does not require HUS1.
In yeast, interaction of 9-1-1 with the SC protein RED1 couples synapsis to meiotic checkpoint activation.4 As there is little evidence that mammalian 9-1-1 participates in checkpoint monitoring of synapsis, it is unclear whether direct interaction between 9-1-1 and SC proteins would promote a checkpoint response during spermatogenesis. Rather, the evidence to date suggests that if there is a true meiotic checkpoint function of mammalian RAD9A-RAD1-HUS1, it would primarily respond to unrepaired DSBs. As discussed below, a distinct RAD1-containing complex could perform synapsis checkpoint functions. While direct interaction of RAD1 with SC structural proteins has not been reported, the meiotic localization of RAD1, like BRCA1, TOPBP1, and ATR, but not RAD9A, is highly similar to that of the HORMA-domain-containing SC protein, HORMAD1,15,33,34,100 and in fact, HORMAD1 is required for loading of γH2AX, BRCA1, and ATR in the XY body domain.100
RAD1 Can Function Independently of RAD9A and HUS1 during Meiosis
Consistent with a previous report,33 we observed that RAD1 localized to many foci along chromosome cores during zygonema and pachynema, and was particularly abundant along the X and Y axial elements (Fig. 1).15 The localization of RAD1 is reminiscent to that of TOPBP1 and ATR,34,76 with strong staining on the unsynapsed axial elements of the X and Y and on asynapsed autosomes. Together, this might indicate that RAD1 functions along with ATR to monitor ongoing or aberrant chromosomal synapsis and could also implicate the RAD1 protein in the MSUC and MSCI meiotic silencing processes. Surprisingly, RAD9A and RAD1 showed distinct, though overlapping, patterns along pachytene chromosomes. Most RAD9A foci coincided with RAD1, but the majority of RAD1 foci lacked RAD9A (Fig. 1). Additionally, RAD1 localization was not disrupted in Hus1 CKO spermatocytes, whereas RAD9A foci were completely lost in the absence of HUS1.15 In the reciprocal experiment in C. elegans, MRT-2 (RAD1) was found to be required for the nuclear localization of HUS-1 in germ cells.86 Together, these results strongly suggest that RAD1 participates in canonical 9-1-1 complexes but also may have RAD9A- and HUS1-independent meiotic functions, potentially overlapping with those of TOPBP1 and ATR.
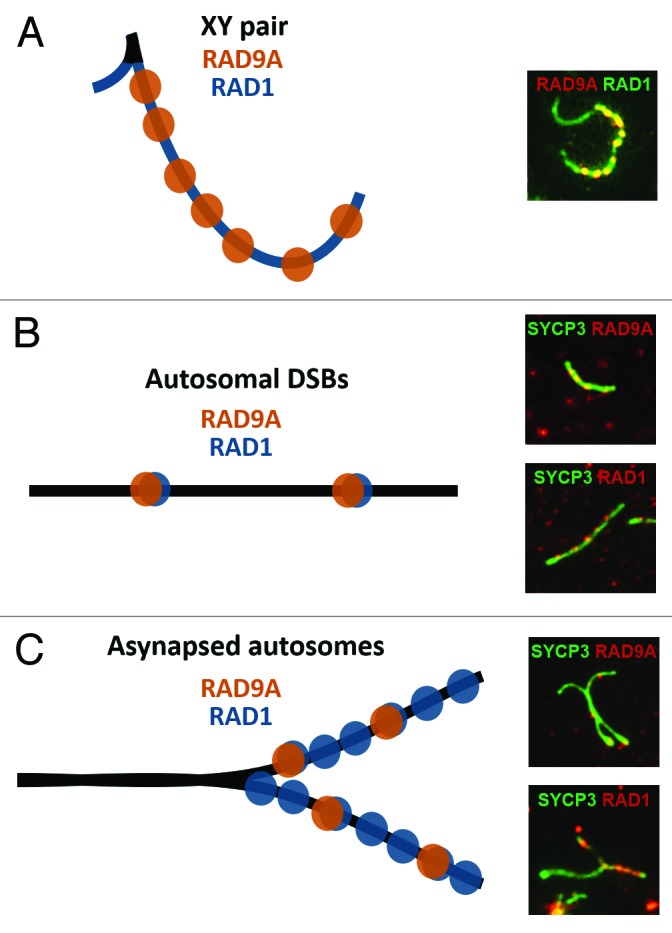
Figure 1. RAD9A and RAD1 proteins localize in distinct yet overlapping patterns on mammalian meiotic chromosomes. The localization patterns of RAD9A and RAD1 are shown in schematic form (left) and in representative immunofluorescence images (right).(A)During late pachynema, unsynapsed regions of the X and Y axial elements are continuously coated with RAD1, while the X chromosome axial element additionally harbors discrete RAD9A foci, presumably at sites of DSBs.(B)On early pachytene-stage autosomes, colocalization of RAD9A and RAD1 is observed in a focal pattern, likely marking DSB sites.(C)Asynapsed regions of autosomes show extensive RAD1 staining and discrete RAD9A foci. Similar to what occurs on the unsynapsed X and Y (A), RAD9A is present at sites that also contain RAD1, whereas RAD1 displays a broader staining pattern that includes regions without detectable RAD9A. See reference 15 for additional examples of these staining patterns.
To B or Not to B: A Hypothetical Model for Alternative 9-1-1 Complexes Functioning in Meiosis
The 9-1-1 complex in lower eukaryotes participates in the checkpoint-dependent monitoring of both synapsis and recombination during prophase I of meiosis.5,9,86,87,101,102 Results from both the Rad9a and Hus1 CKO mouse studies strongly support roles for the canonical RAD9A-RAD1-HUS1 complex in events of meiotic recombination but provide little, if any, evidence of direct roles for RAD9A or HUS1 in synapsis or in the checkpoint monitoring of synapsis, which ordinarily leads to meiotic silencing via MSUC or MSCI. Since RAD9A and HUS1 in mammals have paralogs, RAD9B and HUS1B, respectively, which coincidentally are highly expressed in germ cells,67,103,104 it is tempting to speculate that the meiotic functions of the ancestral 9-1-1 complex, such as that in budding yeast, have diverged in mammals and have been split between the canonical RAD9A-RAD1-HUS1 complex and an alternative RAD1-containing complex, potentially RAD9B-RAD1-HUS1B.
The human HUS1B protein is 48% identical and 69% similar to HUS1,103 while human RAD9B is approximately 35% identical and 55% similar to RAD9A.67,104 These paralogs are expected to assemble in a similar manner to canonical 9-1-1 subunits, as depicted in Figure 2A, leaving open the possibility that RAD9B and HUS1B could also form heterotrimeric complexes with other 9-1-1 subunits,67,103,104 resulting in RAD9B-RAD1-HUS1B or RAD9B-RAD1-HUS1 complexes. HUS1B reportedly cannot interact with RAD9A,103 arguing against the possibility of RAD9A-RAD1-HUS1B trimer formation.
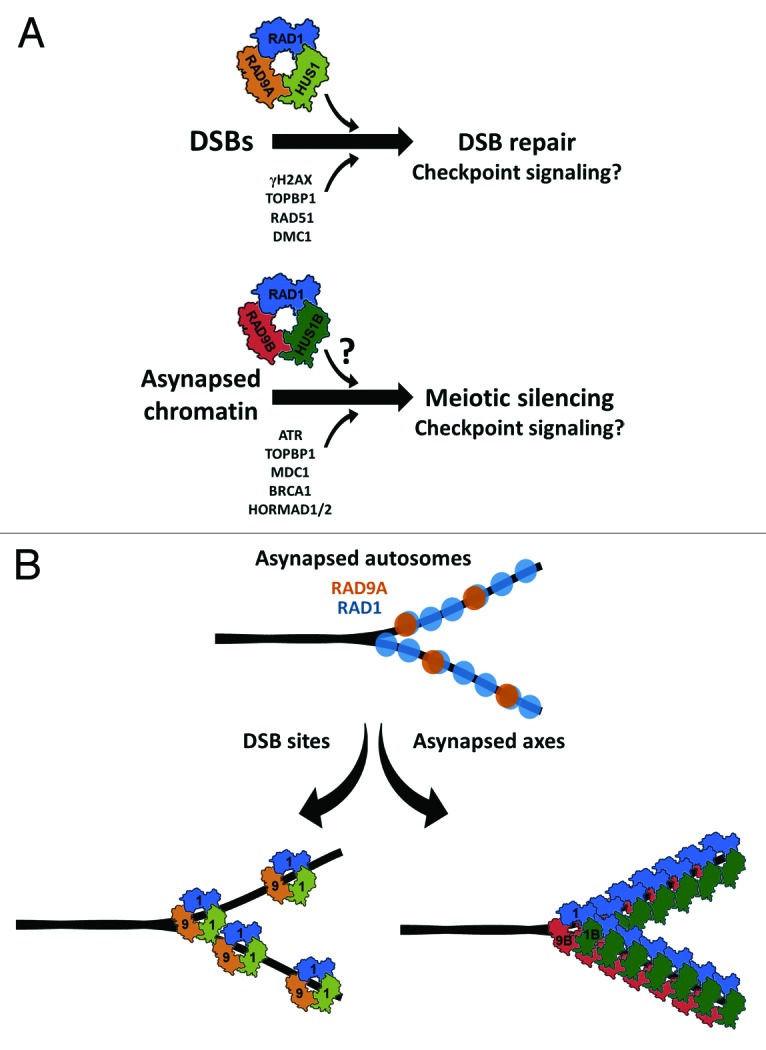
Figure 2. Proposed functions for canonical and alternative 9-1-1 complexes during mammalian meiosis.(A)We propose that the canonical RAD9A-RAD1-HUS1 complex is loaded on at least a subset of DSB sites and facilitates DSB repair and potentially checkpoint signaling. Alternative trimeric 9-1-1 complexes, containing RAD1 and a combination of RAD9B, RAD9A, HUS1B, or HUS1, may form in mouse germ cells and localize to asynapsed chromatin to facilitate meiotic silencing through MSUC and MSCI. The composition of this complex has yet to be determined, but here we propose a model involving RAD9B-RAD1-HUS1B. Depiction is based on the crystal structure of human 9-1-1.108-110(B)Based on the observed localization of RAD9A and RAD1 along asynapsed autosomes, we hypothesize that discrete RAD9A foci represent conventional 9-1-1 complexes at DSB sites, while more extensive RAD1 staining at regions of asynapsis reflects the presence of alternative clamps that include RAD1 in conjunction with paralogs HUS1B and/or RAD9B, without HUS1 or RAD9A.
Relatively little is known about the functions of RAD9B and HUS1B. In embryonic stem cells, RAD9B aids in mediating resistance to DNA-damaging agents,105 and its overexpression in human U2OS cells results in cell cycle delay in G1 phase.106 RAD9B is also expressed in tumor cells, where it co-precipitates with HUS1.104 Additionally, Rad9b is an essential gene,105 like Rad9a,44 Rad1,81 and Hus1,69 whereas Hus1b is not (H Hang, KM Hopkins, and HB Lieberman, unpublished results). Rad9b expression is increased in Rad9a CKO mouse keratinocytes, suggesting that some functional overlap between the paralogs may allow for compensatory responses in some circumstances.107 HUS1B overexpression in cultured cells induces clonogenic cell death, whereas HUS1 overexpression does not.103 Altogether, these results suggest that RAD9B and HUS1B have distinct functions relative to the conventional 9-1-1 subunits (Fig. 2). Given the scaffolding functions assigned to the family of PCNA-like clamps, it seems likely that alternative, 9-1-1-related clamps may have diverged to recognize distinct DNA structures or to associate with a different set of effector proteins.
RAD1, TOPBP1, and ATR all localize to sites of unsynapsed chromatin,15,33,34 and in the case of RAD115 and TOPBP1,14,15 this localization is independent of HUS115 and RAD9A.14 RAD1 foci are also much more abundant than RAD9A foci15 or even DMC1 foci,33 and only a subset of RAD1 foci also contain RAD9A.15 Together, these results strongly support a model in which RAD1 participates in both the canonical 9-1-1 complex, functioning in DSB repair and possibly in checkpoint signaling, as well as in a distinct RAD9A- and HUS1-independent complex that localizes to asynaptic sites containing TOPBP1 and ATR (Fig. 2A). We propose that an alternative RAD9B-RAD1-HUS1B (or RAD9B-RAD1-HUS1) clamp might be loaded at asynaptic regions to activate ATR pathway checkpoint signaling and promote silencing of unsynapsed chromatin. While RAD9B is reported not to interact with TOPBP1 in human U2OS cells,106 serine 387 of RAD9A, which is important for interaction with TOPBP1,20 is conserved in RAD9B (PX Lim and RS Weiss, unpublished results). Based on the observed localization patterns for 9-1-1 subunits in mouse spermatocytes, we speculate that both canonical and alternative 9-1-1 complexes would localize to meiotic chromosomes as shown in Figure 2B, with 9-1-1 at DSB sites and an alternative 9-1-1 complex at asynaptic sites.
Conclusions and Unanswered Questions
Our findings independently demonstrate that RAD9A and HUS1 are essential for male fertility by ensuring effective repair of DSBs during meiosis. Many unanswered questions remain regarding the function of the 9-1-1 complex and potential alternative RAD1-containing complexes in this process. What is the precise composition of the putative alternative 9-1-1 complex(es)? Do the high protein levels of RAD9B, HUS1B, and RAD1 in the mammalian testis reflect functions specific to meiosis? Additionally, what is the DNA substrate recognized by the alternative complex(es)? Canonical 9-1-1 recognizes single-strand/double-strand DNA junctions containing 5′ recessed ends, which would be present at resected meiotic DSBs, but what would be the signal for loading or recruiting an alternative complex? Do the 9-1-1 complexes share the same clamp loader? Is there a meiosis-specific clamp loader? Finally, what are the key effectors, both for the conventional 9-1-1 complex at DSB sites and the alternative 9-1-1 complex at asynaptic sites? Clearly much remains to be elucidated regarding the biology of these fascinating and functionally distinct DNA damage response proteins in the mammalian germline.
Acknowledgments
The authors thank Pei Xin Lim for generating schematic images of the checkpoint clamps. This work was supported by NIH grants R01 CA108773 (to RSW) and GM079107 (to HBL and DJW), and AML was supported by NIH training grant T32 HD052471.
Glossary
Abbreviations:
- 9-1-1
RAD9A-RAD1-HUS1
- DSB
double-strand break
- SC
synaptonemal complex
- CO
crossover
- CKO
conditional knockout
- γH2AX
phosphorylated histone H2AX
- MSCI
meiotic sex chromosome inactivation
- MSUC
meiotic silencing of unsynapsed chromatin
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26061
References
- 1.Broustas CG, Lieberman HB. Contributions of Rad9 to tumorigenesis. J Cell Biochem. 2012;113:742–51. doi: 10.1002/jcb.23424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helt CE, Wang W, Keng PC, Bambara RA. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–32. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- 3.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, Gupta A, Wellinger RJ, Zhang J, Powell SN, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol. 2006;26:1850–64. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichinger CS, Jentsch S. Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc Natl Acad Sci U S A. 2010;107:11370–5. doi: 10.1073/pnas.1004248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacQueen AJ, Hochwagen A. Checkpoint mechanisms: the puppet masters of meiotic prophase. Trends Cell Biol. 2011;21:393–400. doi: 10.1016/j.tcb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Hong E-JE, Roeder GS. A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev. 2002;16:363–76. doi: 10.1101/gad.938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdu U, Klovstad M, Butin-Israeli V, Bakhrat A, Schüpbach T. An essential role for Drosophila hus1 in somatic and meiotic DNA damage responses. J Cell Sci. 2007;120:1042–9. doi: 10.1242/jcs.03414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peretz G, Arie LG, Bakhrat A, Abdu U. The Drosophila hus1 gene is required for homologous recombination repair during meiosis. Mech Dev. 2009;126:677–86. doi: 10.1016/j.mod.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 2009;8:1127–38. doi: 10.1016/j.dnarep.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–3. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 11.Jaramillo-Lambert A, Harigaya Y, Vitt J, Villeneuve A, Engebrecht J. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr Biol. 2010;20:2078–89. doi: 10.1016/j.cub.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H-Y, Burgess SM. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr Biol. 2006;16:2473–9. doi: 10.1016/j.cub.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinohara M, Sakai K, Ogawa T, Shinohara A. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics. 2003;164:855–65. doi: 10.1093/genetics/164.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasileva A, Hopkins KM, Wang X, Weissbach MM, Friedman RA, Wolgemuth DJ, Lieberman HB. The DNA damage checkpoint protein RAD9A is essential for male meiosis in the mouse. J Cell Sci. 2013 doi: 10.1242/jcs.126763. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyndaker AM, Lim PX, Mleczko JM, Diggins CE, Holloway JK, Holmes RJ, Kan R, Schlafer DH, Freire R, Cohen PE, et al. Conditional inactivation of the DNA damage response gene Hus1 in mouse testis reveals separable roles for components of the RAD9-RAD1-HUS1 complex in meiotic chromosome maintenance. PLoS Genet. 2013;9:e1003320. doi: 10.1371/journal.pgen.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–42. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navadgi-Patil VM, Burgers PM. A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst) 2009;8:996–1003. doi: 10.1016/j.dnarep.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu YJ, Leffak M. ATRIP from TopBP1 to ATR--in vitro activation of a DNA damage checkpoint. Proc Natl Acad Sci U S A. 2010;107:13561–2. doi: 10.1073/pnas.1008909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majka J, Burgers PM. Clamping the Mec1/ATR checkpoint kinase into action. Cell Cycle. 2007;6:1157–60. doi: 10.4161/cc.6.10.4221. [DOI] [PubMed] [Google Scholar]
- 20.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K-i, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–7. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–44. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 22.Greer DA, Besley BDA, Kennedy KB, Davey S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II β binding protein 1 focus formation. Cancer Res. 2003;63:4829–35. [PubMed] [Google Scholar]
- 23.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–64. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotta-Ramusino C, McDonald ER, 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–7. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duursma AM, Driscoll R, Elias JE, Cimprich KA. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell. 2013;50:116–22. doi: 10.1016/j.molcel.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiotani B, Nguyen HD, Håkansson P, Maréchal A, Tse A, Tahara H, Zou L. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep. 2013;3:1651–62. doi: 10.1016/j.celrep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Dunphy WG. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell. 2013;24:1343–53. doi: 10.1091/mbc.E13-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan S, Michael WM. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle. 2009;8:2877–84. doi: 10.4161/cc.8.18.9485. [DOI] [PubMed] [Google Scholar]
- 29.Yan S, Michael WM. TopBP1 and DNA polymerase-α directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou L. Four pillars of the S-phase checkpoint. Genes Dev. 2013;27:227–33. doi: 10.1101/gad.213306.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Burgers PM. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013;27:313–21. doi: 10.1101/gad.204750.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freire R, Murguía JR, Tarsounas M, Lowndes NF, Moens PB, Jackson SP. Human and mouse homologs of Schizosaccharomyces pombe rad1(+) and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev. 1998;12:2560–73. doi: 10.1101/gad.12.16.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perera D, Perez-Hidalgo L, Moens PB, Reini K, Lakin N, Syväoja JE, San-Segundo PA, Freire R. TopBP1 and ATR colocalization at meiotic chromosomes: role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol Biol Cell. 2004;15:1568–79. doi: 10.1091/mbc.E03-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moens PB, Tarsounas M, Morita T, Habu T, Rottinghaus ST, Freire R, Jackson SP, Barlow C, Wynshaw-Boris A. The association of ATR protein with mouse meiotic chromosome cores. Chromosoma. 1999;108:95–102. doi: 10.1007/s004120050356. [DOI] [PubMed] [Google Scholar]
- 36.Friedrich-Heineken E, Toueille M, Tännler B, Bürki C, Ferrari E, Hottiger MO, Hübscher U. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J Mol Biol. 2005;353:980–9. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Park MJ, Park J-H, Hahm S-H, Ko SI, Lee YR, Chung JH, Sohn SY, Cho Y, Kang LW, Han YS. Repair activities of human 8-oxoguanine DNA glycosylase are stimulated by the interaction with human checkpoint sensor Rad9-Rad1-Hus1 complex. DNA Repair (Amst) 2009;8:1190–200. doi: 10.1016/j.dnarep.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Shi G, Chang D-Y, Cheng C-C, Guan X, Venclovas C, Lu A-L. Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hübscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase β and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–24. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smirnova E, Toueille M, Markkanen E, Hübscher U. The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem J. 2005;389:13–7. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai H, Madabushi A, Guan X, Lu AL. Interaction between human mismatch repair recognition proteins and checkpoint sensor Rad9-Rad1-Hus1. DNA Repair (Amst) 2010;9:478–87. doi: 10.1016/j.dnarep.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He W, Zhao Y, Zhang C, An L, Hu Z, Liu Y, Han L, Bi L, Xie Z, Xue P, et al. Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucleic Acids Res. 2008;36:6406–17. doi: 10.1093/nar/gkn686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T, Wang Z, Zhao Y, He W, An L, Liu S, Liu Y, Wang H, Hang H. Checkpoint protein Rad9 plays an important role in nucleotide excision repair. DNA Repair (Amst) 2013;12:284–92. doi: 10.1016/j.dnarep.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, Joyner AL, Lieberman HB. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–48. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kidane D, Jonason AS, Gorton TS, Mihaylov I, Pan J, Keeney S, de Rooij DG, Ashley T, Keh A, Liu Y, et al. DNA polymerase β is critical for mouse meiotic synapsis. EMBO J. 2010;29:410–23. doi: 10.1038/emboj.2009.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plug AW, Clairmont CA, Sapi E, Ashley T, Sweasy JB. Evidence for a role for DNA polymerase β in mammalian meiosis. Proc Natl Acad Sci U S A. 1997;94:1327–31. doi: 10.1073/pnas.94.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Hu B, Weiss RS, Wang Y. The effect of Hus1 on ionizing radiation sensitivity is associated with homologous recombination repair but is independent of nonhomologous end-joining. Oncogene. 2006;25:1980–3. doi: 10.1038/sj.onc.1209212. [DOI] [PubMed] [Google Scholar]
- 48.Brandt PD, Helt CE, Keng PC, Bambara RA. The Rad9 protein enhances survival and promotes DNA repair following exposure to ionizing radiation. Biochem Biophys Res Commun. 2006;347:232–7. doi: 10.1016/j.bbrc.2006.06.064. [DOI] [PubMed] [Google Scholar]
- 49.Saberi A, Nakahara M, Sale JE, Kikuchi K, Arakawa H, Buerstedde J-M, Yamamoto K, Takeda S, Sonoda E. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol Cell Biol. 2008;28:6113–22. doi: 10.1128/MCB.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.An L, Wang Y, Liu Y, Yang X, Liu C, Hu Z, He W, Song W, Hang H. Rad9 is required for B cell proliferation and immunoglobulin class switch recombination. J Biol Chem. 2010;285:35267–73. doi: 10.1074/jbc.M110.161208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci. 2007;120:1017–27. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- 52.Tarsounas M, Morita T, Pearlman RE, Moens PB. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol. 1999;147:207–20. doi: 10.1083/jcb.147.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–41. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–9. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 55.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–94. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 56.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–58. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 57.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–77. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 58.Hamer G, Wang H, Bolcun-Filas E, Cooke HJ, Benavente R, Höög C. Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J Cell Sci. 2008;121:2445–51. doi: 10.1242/jcs.033233. [DOI] [PubMed] [Google Scholar]
- 59.Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, Benavente R, Cooke HJ. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5:e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamer G, Novak I, Kouznetsova A, Höög C. Disruption of pairing and synapsis of chromosomes causes stage-specific apoptosis of male meiotic cells. Theriogenology. 2008;69:333–9. doi: 10.1016/j.theriogenology.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 61.Turner JMA, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng C-X, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–7. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 62.Turner JMA, Mahadevaiah SK, Ellis PJI, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell. 2006;10:521–9. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Handel MA. The XY body: a specialized meiotic chromatin domain. Exp Cell Res. 2004;296:57–63. doi: 10.1016/j.yexcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss RS, Kostrub CF, Enoch T, Leder P. Mouse Hus1, a homolog of the Schizosaccharomyces pombe hus1+ cell cycle checkpoint gene. Genomics. 1999;59:32–9. doi: 10.1006/geno.1999.5865. [DOI] [PubMed] [Google Scholar]
- 66.Hang H, Rauth SJ, Hopkins KM, Davey SK, Lieberman HB. Molecular cloning and tissue-specific expression of Mrad9, a murine orthologue of the Schizosaccharomyces pombe rad9+ checkpoint control gene. J Cell Physiol. 1998;177:241–7. doi: 10.1002/(SICI)1097-4652(199811)177:2<241::AID-JCP6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 67.Hopkins KM, Wang X, Berlin A, Hang H, Thaker HM, Lieberman HB. Expression of mammalian paralogues of HRAD9 and Mrad9 checkpoint control genes in normal and cancerous testicular tissue. Cancer Res. 2003;63:5291–8. [PubMed] [Google Scholar]
- 68.Smirnova NA, Romanienko PJ, Khil PP, Camerini-Otero RD. Gene expression profiles of Spo11-/- mouse testes with spermatocytes arrested in meiotic prophase I. Reproduction. 2006;132:67–77. doi: 10.1530/rep.1.00997. [DOI] [PubMed] [Google Scholar]
- 69.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–98. [PMC free article] [PubMed] [Google Scholar]
- 70.Lieberman HB, Yin Y. A novel function for human Rad9 protein as a transcriptional activator of gene expression. Cell Cycle. 2004;3:1008–10. doi: 10.4161/cc.3.8.1069. [DOI] [PubMed] [Google Scholar]
- 71.Yin Y, Zhu A, Jin YJ, Liu Y-X, Zhang X, Hopkins KM, Lieberman HB. Human RAD9 checkpoint control/proapoptotic protein can activate transcription of p21. Proc Natl Acad Sci U S A. 2004;101:8864–9. doi: 10.1073/pnas.0403130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oh SD, Lao JP, Hwang PY-H, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–72. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31:313–23. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 2010;188:779–89. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karras GI, Fumasoni M, Sienski G, Vanoli F, Branzei D, Jentsch S. Noncanonical role of the 9-1-1 clamp in the error-free DNA damage tolerance pathway. Mol Cell. 2013;49:536–46. doi: 10.1016/j.molcel.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 76.Keegan KS, Holtzman DA, Plug AW, Christenson ER, Brainerd EE, Flaggs G, Bentley NJ, Taylor EM, Meyn MS, Moss SB, et al. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes Dev. 1996;10:2423–37. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 77.Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, Burgoyne PS, Jasin M, Keeney S. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol. 2005;25:7203–15. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turner JMA, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GVR, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–42. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 79.Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/- spermatocytes. J Cell Sci. 2005;118:3233–45. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 80.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 81.Han L, Hu Z, Liu Y, Wang X, Hopkins KM, Lieberman HB, Hang H. Mouse Rad1 deletion enhances susceptibility for skin tumor development. Mol Cancer. 2010;9:67. doi: 10.1186/1476-4598-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–82. doi: 10.1016/S0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 83.Chicheportiche A, Bernardino-Sgherri J, de Massy B, Dutrillaux B. Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J Cell Sci. 2007;120:1733–42. doi: 10.1242/jcs.004945. [DOI] [PubMed] [Google Scholar]
- 84.Cai RL, Yan-Neale Y, Cueto MA, Xu H, Cohen D. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J Biol Chem. 2000;275:27909–16. doi: 10.1074/jbc.M000168200. [DOI] [PubMed] [Google Scholar]
- 85.Lightfoot J, Testori S, Barroso C, Martinez-Perez E. Loading of meiotic cohesin by SCC-2 is required for early processing of DSBs and for the DNA damage checkpoint. Curr Biol. 2011;21:1421–30. doi: 10.1016/j.cub.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 86.Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, Vidal M, Hengartner MO. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12:1908–18. doi: 10.1016/S0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 87.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–43. doi: 10.1016/S1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 88.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–87. doi: 10.1016/S1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 89.Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–18. doi: 10.1016/S1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 90.Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/S1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 91.de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P, te Riele H. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 1999;13:523–31. doi: 10.1101/gad.13.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kogo H, Tsutsumi M, Ohye T, Inagaki H, Abe T, Kurahashi H. HORMAD1-dependent checkpoint/surveillance mechanism eliminates asynaptic oocytes. Genes Cells. 2012;17:439–54. doi: 10.1111/j.1365-2443.2012.01600.x. [DOI] [PubMed] [Google Scholar]
- 93.Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, Anastassiadis K, Stewart AF, Reményi A, Turner JM, et al. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev. 2012;26:958–73. doi: 10.1101/gad.187559.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JM. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol. 2010;20:2117–23. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 95.Balmus G, Zhu M, Mukherjee S, Lyndaker AM, Hume KR, Lee J, Riccio ML, Reeves AP, Sutter NB, Noden DM, et al. Disease severity in a mouse model of ataxia telangiectasia is modulated by the DNA damage checkpoint gene Hus1. Hum Mol Genet. 2012;21:3408–20. doi: 10.1093/hmg/dds173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kleiman NJ, David J, Elliston CD, Hopkins KM, Smilenov LB, Brenner DJ, Worgul BV, Hall EJ, Lieberman HB. Mrad9 and atm haploinsufficiency enhance spontaneous and X-ray-induced cataractogenesis in mice. Radiat Res. 2007;168:567–73. doi: 10.1667/rr1122.1. [DOI] [PubMed] [Google Scholar]
- 97.Smilenov LB, Lieberman HB, Mitchell SA, Baker RA, Hopkins KM, Hall EJ. Combined haploinsufficiency for ATM and RAD9 as a factor in cell transformation, apoptosis, and DNA lesion repair dynamics. Cancer Res. 2005;65:933–8. [PubMed] [Google Scholar]
- 98.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–62. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 99.Kinner A, Wu W, Staudt C, Iliakis G. γ-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–94. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shin Y-H, Choi Y, Erdin SU, Yatsenko SA, Kloc M, Yang F, Wang PJ, Meistrich ML, Rajkovic A. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 2010;6:e1001190. doi: 10.1371/journal.pgen.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murakami H, Nurse P. Meiotic DNA replication checkpoint control in fission yeast. Genes Dev. 1999;13:2581–93. doi: 10.1101/gad.13.19.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shimada M, Nabeshima K, Tougan T, Nojima H. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 2002;21:2807–18. doi: 10.1093/emboj/21.11.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hang H, Zhang Y, Dunbrack RL, Jr., Wang C, Lieberman HB. Identification and characterization of a paralog of human cell cycle checkpoint gene HUS1. Genomics. 2002;79:487–92. doi: 10.1006/geno.2002.6737. [DOI] [PubMed] [Google Scholar]
- 104.Dufault VM, Oestreich AJ, Vroman BT, Karnitz LM. Identification and characterization of RAD9B, a paralog of the RAD9 checkpoint gene. Genomics. 2003;82:644–51. doi: 10.1016/S0888-7543(03)00200-3. [DOI] [PubMed] [Google Scholar]
- 105.Leloup C, Hopkins KM, Wang X, Zhu A, Wolgemuth DJ, Lieberman HB. Mouse Rad9b is essential for embryonic development and promotes resistance to DNA damage. Dev Dyn. 2010;239:2837–50. doi: 10.1002/dvdy.22415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pérez-Castro AJ, Freire R. Rad9B responds to nucleolar stress through ATR and JNK signalling, and delays the G1-S transition. J Cell Sci. 2012;125:1152–64. doi: 10.1242/jcs.091124. [DOI] [PubMed] [Google Scholar]
- 107.Hu Z, Liu Y, Zhang C, Zhao Y, He W, Han L, Yang L, Hopkins KM, Yang X, Lieberman HB, et al. Targeted deletion of Rad9 in mouse skin keratinocytes enhances genotoxin-induced tumor development. Cancer Res. 2008;68:5552–61. doi: 10.1158/0008-5472.CAN-07-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Doré AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol Cell. 2009;34:735–45. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 109.Sohn SY, Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 110.Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J Biol Chem. 2009;284:20457–61. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]


