Abstract
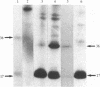
We have found that, when isolated rat liver mitochondria are incubated with [gamma-32P]ATP, there is phosphorylation of 36- and 17-kDa proteins. These proteins together with their protein kinase(s) are released as a complex by incubation of the isolated rat liver mitochondria at 20 degrees C for 30 min with 10 mM glucose 6-phosphate, 0.5 mM inositol phosphate, or 0.01 mM inositol triphosphate. Phosphorylation of the 36- and 17-kDa proteins in this soluble protein fraction is modulated by p21 proteins encoded by ras oncogenes and synthesized in Escherichia coli via recombinant DNA methods. A normal p21 ras protein stimulates phosphorylation of the 36-kDa protein and inhibits phosphorylation of the 17-kDa protein, whereas two transforming p21 ras proteins inhibit phosphorylation of both the 36- and 17-kDa proteins. Although GDP and 5'-guanylyl imidodiphosphate also influence the phosphorylation of these proteins, we present evidence that the effects of p21 ras protein are not simply due to their bound GDP. This novel system may be useful for further studies on the biochemical functions of the p21 ras proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birchmeier C., Broek D., Wigler M. ras proteins can induce meiosis in Xenopus oocytes. Cell. 1985 Dec;43(3 Pt 2):615–621. doi: 10.1016/0092-8674(85)90233-8. [DOI] [PubMed] [Google Scholar]
- Bourne H. R. Transducing proteins. Yeast RAS and Tweedledee's logic. Nature. 1985 Sep 5;317(6032):16–17. doi: 10.1038/317016b0. [DOI] [PubMed] [Google Scholar]
- Der C. J., Finkel T., Cooper G. M. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986 Jan 17;44(1):167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Furth M. E., Davis L. J., Fleurdelys B., Scolnick E. M. Monoclonal antibodies to the p21 products of the transforming gene of Harvey murine sarcoma virus and of the cellular ras gene family. J Virol. 1982 Jul;43(1):294–304. doi: 10.1128/jvi.43.1.294-304.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Ellis R. W., Scolnick E. M. Autophosphorylation of v-Ha-ras p21 is modulated by amino acid residue 12. Proc Natl Acad Sci U S A. 1984 May;81(9):2674–2678. doi: 10.1073/pnas.81.9.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Sigal I. S., Poe M., Scolnick E. M. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman A. G. G proteins and dual control of adenylate cyclase. Cell. 1984 Mar;36(3):577–579. doi: 10.1016/0092-8674(84)90336-2. [DOI] [PubMed] [Google Scholar]
- Greenawalt J. W. The isolation of outer and inner mitochondrial membranes. Methods Enzymol. 1974;31:310–323. doi: 10.1016/0076-6879(74)31033-6. [DOI] [PubMed] [Google Scholar]
- Hurley J. B., Simon M. I., Teplow D. B., Robishaw J. D., Gilman A. G. Homologies between signal transducing G proteins and ras gene products. Science. 1984 Nov 16;226(4676):860–862. doi: 10.1126/science.6436980. [DOI] [PubMed] [Google Scholar]
- Lochrie M. A., Hurley J. B., Simon M. I. Sequence of the alpha subunit of photoreceptor G protein: homologies between transducin, ras, and elongation factors. Science. 1985 Apr 5;228(4695):96–99. doi: 10.1126/science.3856323. [DOI] [PubMed] [Google Scholar]
- Madaule P., Axel R. A novel ras-related gene family. Cell. 1985 May;41(1):31–40. doi: 10.1016/0092-8674(85)90058-3. [DOI] [PubMed] [Google Scholar]
- McGrath J. P., Capon D. J., Goeddel D. V., Levinson A. D. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 1984 Aug 23;310(5979):644–649. doi: 10.1038/310644a0. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schejter E., Hoffmann F. M., Shilo B. Z. The Drosophila ras oncogenes: structure and nucleotide sequence. Cell. 1984 Jul;37(3):1027–1033. doi: 10.1016/0092-8674(84)90437-9. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984 Apr 19;308(5961):693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- Papageorge A. G., Defeo-Jones D., Robinson P., Temeles G., Scolnick E. M. Saccharomyces cerevisiae synthesizes proteins related to the p21 gene product of ras genes found in mammals. Mol Cell Biol. 1984 Jan;4(1):23–29. doi: 10.1128/mcb.4.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe M., Scolnick E. M., Stein R. B. Viral Harvey ras p21 expressed in Escherichia coli purifies as a binary one-to-one complex with GDP. J Biol Chem. 1985 Apr 10;260(7):3906–3909. [PubMed] [Google Scholar]
- Reymond C. D., Gomer R. H., Mehdy M. C., Firtel R. A. Developmental regulation of a Dictyostelium gene encoding a protein homologous to mammalian ras protein. Cell. 1984 Nov;39(1):141–148. doi: 10.1016/0092-8674(84)90199-5. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Papageorge A. G., Stokes P. E., Weeks M. O., Scolnick E. M. Guanine nucleotide-binding and autophosphorylating activities associated with the p21src protein of Harvey murine sarcoma virus. Nature. 1980 Oct 23;287(5784):686–691. doi: 10.1038/287686a0. [DOI] [PubMed] [Google Scholar]
- Stein R. B., Robinson P. S., Scolnick E. M. Photoaffinity labeling with GTP of viral p21 ras protein expressed in Escherichia coli. J Virol. 1984 May;50(2):343–351. doi: 10.1128/jvi.50.2.343-351.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet R. W., Yokoyama S., Kamata T., Feramisco J. R., Rosenberg M., Gross M. The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature. 1984 Sep 20;311(5983):273–275. doi: 10.1038/311273a0. [DOI] [PubMed] [Google Scholar]
- Tabin C. J., Bradley S. M., Bargmann C. I., Weinberg R. A., Papageorge A. G., Scolnick E. M., Dhar R., Lowy D. R., Chang E. H. Mechanism of activation of a human oncogene. Nature. 1982 Nov 11;300(5888):143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- Tanabe T., Nukada T., Nishikawa Y., Sugimoto K., Suzuki H., Takahashi H., Noda M., Haga T., Ichiyama A., Kangawa K. Primary structure of the alpha-subunit of transducin and its relationship to ras proteins. Nature. 1985 May 16;315(6016):242–245. doi: 10.1038/315242a0. [DOI] [PubMed] [Google Scholar]
- Taparowsky E., Shimizu K., Goldfarb M., Wigler M. Structure and activation of the human N-ras gene. Cell. 1983 Sep;34(2):581–586. doi: 10.1016/0092-8674(83)90390-2. [DOI] [PubMed] [Google Scholar]
- Thomas A. P., Alexander J., Williamson J. R. Relationship between inositol polyphosphate production and the increase of cytosolic free Ca2+ induced by vasopressin in isolated hepatocytes. J Biol Chem. 1984 May 10;259(9):5574–5584. [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Wilson J. E. Brain hexokinase, the prototype ambiquitous enzyme. Curr Top Cell Regul. 1980;16:1–54. doi: 10.1016/b978-0-12-152816-4.50005-4. [DOI] [PubMed] [Google Scholar]
- Yuasa Y., Srivastava S. K., Dunn C. Y., Rhim J. S., Reddy E. P., Aaronson S. A. Acquisition of transforming properties by alternative point mutations within c-bas/has human proto-oncogene. Nature. 1983 Jun 30;303(5920):775–779. doi: 10.1038/303775a0. [DOI] [PubMed] [Google Scholar]