Protein ubiquitination is a versatile covalent modification that controls myriad biological processes. In an analogy to kinases and phosphatases, E3 ligases and de-ubiquitinating enzymes (DUBs) provide a dynamic equilibrium for substrate modification. While it’s estimated that over 600 E3 ligase complexes exist in human cells, ≈90 DUBs control the removal of ubiquitin from substrates. A well-established, but clearly not the sole, role for protein ubiquitination is to create a signal for targeting substrates for 26S proteasomal degradation. This function is part of protein homeostasis to balance protein synthesis, to terminate protein function at defined stages of the cell cycle or cellular conditions, or to eliminate misfolded proteins upon their synthesis. Early and more recent studies indicated that up to 30% of newly synthesized proteins are rapidly ubiquitinated and degraded.1-3 Indeed, E3 ligases such as Ltn1 and Hel2 associate with ribosomes and act as quality-control factors for newly synthesized proteins.2,4 The simplest explanation for this phenomenon is that newly synthesized proteins are prone to misfolding and have to be rapidly eliminated. However, even if it was predicted, no DUBs had been reported to act on ribosomes.
In the study by Faronato et al., a DUB is now reported to control protein stability of newly synthesized proteins.5 In an unbiased DUB siRNA screen, USP15 was found as a regulator of the stability of the RE1 silencing transcription factor (REST). REST is a key transcriptional repressor that silences the expression of neuronal genes in non-neuronal cells. Targeting REST for ubiquitin-mediated proteasomal degradation is critical both for neuronal differentiation and for activation of the spindle checkpoint and mitotic exit. Phosphorylation of REST allows its recognition by the β-TrCP E3 ligase, which promotes REST ubiquitination and proteasomal degradation.6,7 Therefore, USP15 could control REST stability by opposing β-TrCP function. However, 2 key observations prompted the authors to investigate alternative mechanisms of REST stability control by USP15. As a transcriptional repressor REST is mainly found in the nucleus, O-glycosylated, with apparent molecular weight of 180–220 kDa, whereas the non-glycosylated newly synthesized form migrates as a 120 kDa protein. While USP15 controls the levels of both nuclear and cytoplasmic REST, it is mainly localized in the cytoplasm. The other key observation was that in conditions where translation was inhibited, USP15 knockdown failed to accelerate REST degradation. Therefore, the authors tested the hypothesis that USP15 controls the turnover of newly synthesized REST. Experiments addressing the re-synthesis of REST after removal of translational block showed that USP15 controls the stability of newly synthesized REST protein. Critically, ribosome-profiling experiments indicated the presence of USP15 in polysome-containing fractions. But what is the biological consequence for such regulation? As already mentioned β-TrCP-mediated degradation of REST occurs predominantly at mitosis. While USP15 does not antagonize β-TrCP-mediated REST degradation, is required for the rapid replenishment of REST upon mitotic exit for the beginning of the new cell cycle. These observations provide evidence for a co-translational role of USP15 in REST stability control through de-ubiquitination (Fig. 1). The model is further supported from the observations that inhibition of the 26S proteasome could rescue REST stability both in steady-state conditions and during translational recovery. It will be interesting to determine whether USP15 is a ribosome-associated factor or directly interacts with co-translated REST. The dynamic and reversible mode of function for protein ubiquitination predicts the presence of an E3 ligase operating along USP15. The studies by Faronato et al. also indicate that the function of the ubiquitin-proteasome pathway at the ribosome may not be restricted as a quality control mechanism to eliminate misfolded proteins, but also to provide a rapidly responding system to replenish factors with key roles in cell cycle.
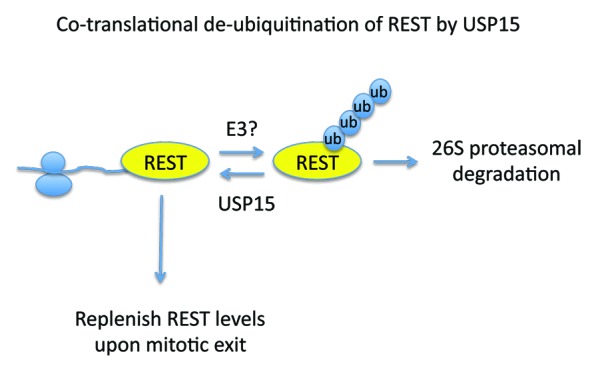
Figure 1. USP15 opposes the function of an unknown E3 ligase to stabilize newly synthesized REST protein. As REST is degraded by the β-TrCP E3 ligase at mitosis, USP15 function is critical to replenish REST levels for the new cell cycle.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25844
References
- 1.Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–4. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 2.Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013;50:379–93. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell. 2013;50:368–78. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–3. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faronato M, Patel V, Darling S, Dearden L, Clague MJ, Urbé S, et al. The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit. Cell Cycle. 2013;12:1964–77. doi: 10.4161/cc.25035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–9. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–4. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]


