Abstract
Purpose
The -159C/T polymorphism in the cluster of differentiation (CD)14 gene has been extensively studied for an association with cancer; however, results from replication studies have been inconclusive. The aim of this study was to perform a comprehensive assessment of the possible association between the -159C/T polymorphism in the CD14 gene and cancer risk, by meta-analysis.
Methods
We searched in PubMed, Embase, and other databases, covering all case-control studies on the possible association between CD14 -159C/T gene polymorphism and cancer risk. Data were extracted and statistical analyses were performed using RevMan 5.0 and STATA 12.0 software.
Results
A total of 12 case-control studies met our inclusion criteria, including 2,498 cases and 2,696 controls. The combined analysis indicated that the CD14 -159C/T gene polymorphism didn’t confer risk for cancer – the recessive model (TT versus (vs) CT + CC), showed odds ratio (OR) =1.01, 95% confidence interval (CI) =0.82–1.23 (P=0.94), while the dominant model (TT + TC vs CC) showed OR =0.81, 95% CI =0.66–1.00 (P=0.05). A subgroup analysis by ethnicity showed that the cancer risk associated with CD14 -159C/T gene polymorphism was significantly decreased among Caucasians for the TC + TT vs CC comparison (OR =0.83, 95% CI =0.70–0.98 [P=0.03]). The subgroup analysis by cancer type suggested that the CD14 -159C/T gene polymorphism was not associated with gastric cancer risk.
Conclusion
The evidence from the present meta-analysis did not support the CD14 -159C/T gene polymorphism as a genetic risk factor for cancer. Further studies on different cancer types and ethnicities are needed to validate our findings.
Keywords: CD14, cancer, gene, polymorphism, meta-analysis
Introduction
Cancer remains a major public health problem around the world. It is the leading cause of death in economically developed countries and the second leading cause of death in developing countries.1 Although the etiology of cancer is complicated and has not been fully elucidated, recent studies have suggested an association between chronic inflammation and the increased risk of developing several types of cancers, and a hypothesized sequence of pathogenesis beginning with inflammation and progressing to atrophy, metaplasia, dysplasia, carcinoma in situ, and finally, to cancer.2 A growing number of studies have suggested that genetic variations in the inflammatory factor genes, which play an important role in the regulation of inflammation, are correlated with increased risk in several malignant tumors and may play critical roles in the pathogenesis of cancer.3 Repeated studies have investigated the association of genetic variants of the inflammatory factor genes with cancer susceptibility,3,4 and among them, the -159 cytosine/thymine (C/T) polymorphism in the cluster of differentiation 14 (CD14) gene has been highlighted.
CD14 is encoded by a single-copy gene that is located at bands 5q23q31, with about 3,900 bp harbored in two exons.5 CD14 is a pattern-recognition receptor that plays a central role in immune regulation; CD14 acts by transferring lipopolysaccharide to the Toll-like receptor 4, which results in the activation of inflammatory genes, production of proinfammatory cytokines, the generation of oxygen free radicals, and the beginning of the inflammation process.6,7 Thus, the regulation of CD14 expression is important in determining whether growth-arrested cells are induced to undergo differentiation or apoptosis, which are the critical process of carcinogenesis.7,8 Studies have proposed CD14 gene polymorphism to increase CD14 production, leading to increased expression levels of CD14,9,10 thus, a growing number of studies have studied the relationship between CD14 gene polymorphism and cancer susceptibility.
Recent studies suggest that the gene variant within the CD14 -159C/T (rs2569190) polymorphism, also called CD14 -260C/T, is critical for CD14 expression and has been suggested as a genetic factor for cancer susceptibility.6–8 Several studies6–8 have investigated whether the -159C/T polymorphism in the CD14 gene is associated with cancer risk, and the results were inconclusive. Since pooled estimates based on meta-analysis have proven useful in determining the overall risk of certain cancer polymorphisms when results of individual studies were inconsistent,11 we decided to perform a meta-analysis to investigate the potential association between the CD14 -159C/T gene polymorphism and cancer risk.
Methods
Literature search
Two authors (W Zhou and L Jia) independently performed systematic searches of the PubMed, Embase, China National Knowledge Infrastructure (CNKI), Weipu, and Wanfang databases to identify studies examining the possible association between the -159C/T polymorphism in the CD14 gene and cancer risk. The search terms were as follows: “cancer or carcinoma or neoplasm” in combination with “CD14 or cluster of differentiation 14” in combination with “polymorphism or variant or mutation.” The search was conducted for all terms updated until July 10, 2013. The reference lists of the identified original studies and review articles were also manually searched to find additional relevant publications.
Study selection
A study was included in this meta-analysis if it satisfied the following inclusion criteria: (1) it evaluated the potential association between the CD14 -159C/T gene polymorphism and cancer risk; (2) it was a case-control study; (3) genotype distributions in the cases and controls were available for estimating an odds ratio (OR) with 95% confidence interval (CI); (4) the distribution of genotypes in the control group was consistent with Hardy–Weinberg equilibrium (HWE);12 and (5) it was published in the English language. In studies of several types of cancer, each type of cancer was treated as a single study in this meta-analysis. Reviews, conference abstracts, and studies in which the genotype frequencies were not reported were excluded. When studies involved the same or overlapping data sets, only the study with the largest number of participants was included.
Data extraction
Two reviewers (W Zhou and L Jia) independently extracted data from the final set of included studies. The following data were extracted: the name of the first author, year of publication, country of origin, ethnicity, sample size, cancer type, genotyping method, and the genotype frequencies in the cancer cases and controls. Disagreements between authors were resolved by consensus.
Statistical analysis
Crude ORs and 95% CIs were used to determine the strength of the association between the CD14 gene -159C/T polymorphism and the risk of cancer. The significance of the pooled OR was determined using the Z-test, and P<0.05 was considered statistically significant. First, we evaluated the recessive model (TT versus (vs) TC + CC) and the dominant model (TT + TC vs CC), followed by the additive model (TT vs CC). The allelic model (T vs C) was also used to estimate the potential association. To evaluate whether the association showed any ethnicity– or cancer type–specific effects, we analyzed the data for subgroups defined by ethnicity and cancer type.
Heterogeneity was evaluated using a χ2-based Q statistic and I2 statistic, with P<0.10 and I2>50% as considered statistically significant. When P≥0.10 and I2<50%, the pooled OR of each study was calculated using a fixed effects model; otherwise, a random effects model was used.13
Begg’s funnel plots and Egger’s test were used to detect the potential publication bias among the included studies.13 All statistical tests were performed using RevMan 5.0 (Nordic Cochrane Centre, Copenhagen, Denmark) and STATA 12.0 (StataCorp LP, College Station, TX, USA) software.
Results
Study inclusion and characteristics
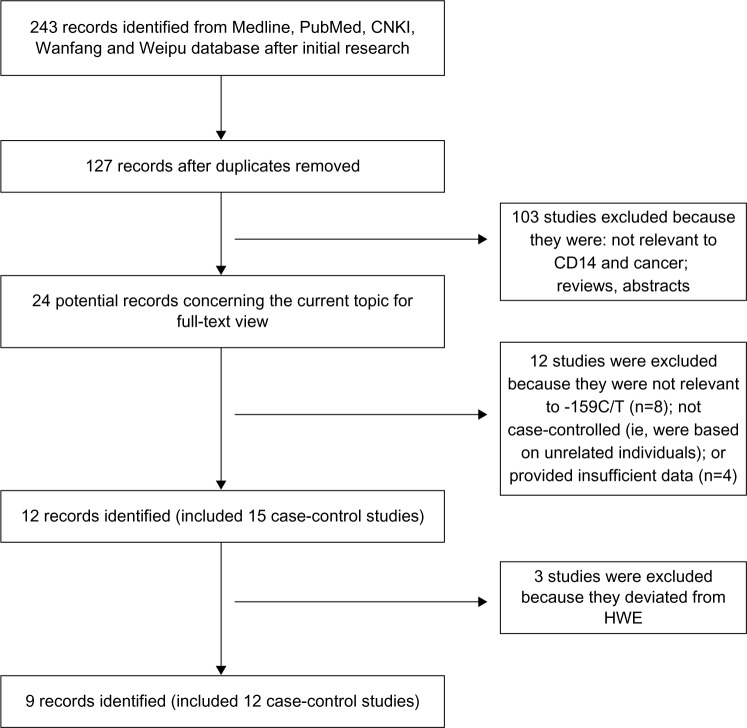
A total of 243 articles were identified after an initial search from the PubMed, Embase, CNKI, Weipu, and Wanfang databases. After removing duplications, 127 publications were identified. Then, 103 articles were excluded that were abstracts or reviews, or that were irrelevant to CD14 polymorphisms and cancer risk. After reading the full texts of the remaining 24 articles, 12 of these were excluded: eight studies that were not concerned with the -159C/T polymorphism, two prognostic studies, and two studies for which there was insufficient data. After the HWE test, three more articles were excluded. Finally, a total of nine articles reporting on 12 case-control studies that evaluated the possible association between the CD14 -159C/T gene polymorphism and cancer risk were included in the meta-analysis (Figure 1).14–22
Figure 1.
The fow diagram of included and excluded studies.
Abbreviations: -159C/T, CD14 gene polymorphism; CD14, cluster of differentiation 14; CNKI, China National Knowledge infrastructure (database); HWE, Hardy–Weinberg equilibrium.
A total of nine publications met our inclusion criteria and involved 5,194 subjects: 2,498 cases and 2,696 controls. There were seven case-control studies of Asians and five of Caucasians. The cancer types included gastric cancer (n=6), colorectal cancer (n=2), acute lymphoblastic leukemia (n=1), lymphoma (n=1), esophageal cancer (n=1), and mucosal-associated lymphoid tissue lymphoma (n=1). The characteristics of each study, including author information, ethnicity, sample size, cancer type, genotyping method country of origin, and other data in this meta-analysis are presented in Table 1. The genotype frequencies and HWE examination results are listed in Table 2.
Table 1.
Characteristics of studies included in the present meta-analysis
| Author | Year | Country | Ethnicity | Cancer | Cases/controls | Genotyping method |
|---|---|---|---|---|---|---|
| Andrie et al14 | 2009 | Greece | Caucasian | Lymphoma | 83/83 | PCR |
| Castaño-Rodríguez et al15 | 2013 | Singapore Malaysia |
Asian | Gastric cancer | 70/214 | PCR |
| Guo et al16 | 2006 | China | Asian | Colorectal cancer | 110/160 | PCR-RFLP |
| Hold et al17 | 2009 | Poland | Caucasian | Gastric cancer | 327/389 | TaqMan® (Life Technologies Corp, Carlsbad, CA, USA) |
| Hold et al17,* | 2009 | USA | Caucasian | Gastric cancer | 306/211 | TaqMan |
| Hold et al17,* | 2009 | USA | Caucasian | Esophageal cancer | 158/211 | TaqMan |
| Landi et al18 | 2006 | Spain | Caucasian | Colorectal cancer | 377/326 | TaqMan |
| Tahara et al19 | 2007 | Japan | Asian | Gastric cancer | 149/94 | PCR-RFLP |
| Wu et al20 | 2006 | China | Asian | MALT | 70/210 | PCR |
| Wu et al20,* | 2006 | China | Asian | Gastric cancer | 204/210 | PCR |
| Yu et al21 | 2011 | China | Asian | Acute lymphoblastic leukemia | 174/539 | PCR-RFLP |
| Zhao et al22 | 2007 | China | Asian | Gastric cancer | 470/470 | PCR-RFLP |
Note:
Different cancers were studied in one publication and each was treated to a separate analysis.
Abbreviations: MALT, mucosal-associated lymphoid tissue lymphoma; PCR, polymerase chain reaction; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Table 2.
Distributions of CD14 -159c/T polymorphism genotypes and alleles among cancer patients and controls
| Author | Case
|
Control
|
Case
|
Control
|
HWE
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | TT | CT | CC | T | C | T | C | P-value | |
| Andrie et al14 | 13 | 39 | 31 | 23 | 42 | 18 | 65 | 101 | 88 | 78 | 0.89 |
| Castaño-Rodríguez et al15 | 14 | 38 | 18 | 72 | 108 | 34 | 66 | 74 | 252 | 176 | 0.54 |
| Guo et al16 | 41 | 34 | 35 | 58 | 77 | 25 | 116 | 104 | 193 | 127 | 0.95 |
| Hold et al17 | 83 | 134 | 110 | 82 | 176 | 131 | 300 | 354 | 340 | 438 | 0.11 |
| Hold et al17,* | 68 | 147 | 91 | 51 | 108 | 52 | 283 | 329 | 210 | 212 | 0.73 |
| Hold et al17,* | 34 | 74 | 50 | 51 | 108 | 52 | 142 | 174 | 210 | 212 | 0.73 |
| Landi et al18 | 68 | 151 | 62 | 63 | 137 | 65 | 287 | 275 | 263 | 267 | 0.58 |
| Tahara et al19 | 32 | 80 | 37 | 27 | 53 | 14 | 144 | 154 | 107 | 81 | 0.15 |
| Wu et al20 | 24 | 29 | 17 | 54 | 102 | 54 | 77 | 63 | 210 | 210 | 0.68 |
| Wu et al20,* | 50 | 102 | 52 | 54 | 102 | 54 | 202 | 206 | 210 | 210 | 0.68 |
| Yu et al21 | 90 | 55 | 29 | 200 | 259 | 80 | 235 | 113 | 659 | 419 | 0.80 |
| Zhao et al22 | 212 | 225 | 33 | 187 | 227 | 56 | 649 | 291 | 601 | 339 | 0.30 |
Note:
Different cancers were studied in one publication and each was treated to a separate analysis.
Abbreviations: C, cytosine; CD, cluster of differentiation; T, thymine; HWE, Hardy–Weinberg equilibrium.
Quantitative data synthesis
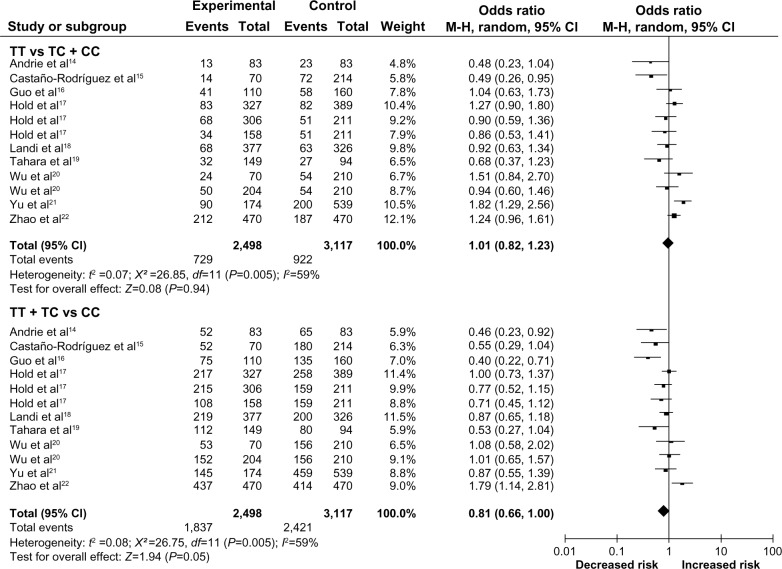
A total of 2,498 cancer cases and 2,696 controls in 12 case-control studies were included. First, we analyzed the heterogeneity of the recessive model (TT vs TC + CC) in order to identify the best risk model. The value of χ2 was 26.85, with eleven degrees of freedom and P=0.005, suggesting significant heterogeneity among the included studies. Thus, we chose the random effects model to analyze the data. For the recessive model, the pooled OR was 1.01 (95% CI =0.82–1.23), and the Z–value, for overall effect, was 0.08 (P=0.94) (Figure 2). For the dominant model (TT + TC vs CC), the OR was 0.81 (95% CI =0.66–1.00) and the test for overall effect Z-value was 1.94 (P=0.05) (Figure 2). These results suggested that the TT homozygote and TC heterozygote carriers did not have an increased risk of cancer compared with those individuals with the CC homozygote. For the additive model (TT vs CC), the OR was 0.94 (95% CI =0.81–1.08), and the test for overall effect Z-value was 0.90 (P=0.37). These results showed no association between the -159C/T polymorphisms of the CD14 gene and cancer risk. A summary of results from other comparisons is listed in Table 3.
Figure 2.
Meta-analysis with a random-effects model, for the association between cancer risk and the CD14 -159C/T polymorphism (TT vs TC + TT) and (TT + TC vs CC).
Abbreviations: -159C/T, CD14 gene polymorphism; CD14, cluster of differentiation 14; C, cytosine; CI, confidence interval; df, degrees of freedom; M-H, Mantel-Haenszel; T, thymine; vs, versus.
Table 3.
Summary of results from different comparative genetic models
| N* | Case/control | TT vs TC + CC
|
TT + TC vs CC
|
T vs C
|
TT vs CC
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P** | OR (95% CI) | P** | OR (95% CI) | P** | OR (95% CI) | P** | |||
| Total | 12 | 2,498/2,696 | 1.01 (0.82, 1.23) | 0.94 | 0.81 (0.66, 1.00) | 0.05 | 0.94 (0.81, 1.08) | 0.37 | 0.86 (0.64, 1.14) | 0.29 |
| By ethnicity | ||||||||||
| Asian | 7 | 1,247/1,687 | 1.07 (0.80, 1.44) | 0.65 | 0.82 (0.55, 1.21) | 0.31 | 0.96 (0.77, 1.20) | 0.73 | 0.87 (0.55, 1.38) | 0.56 |
| Caucasian | 5 | 1,251/1,009 | 0.96 (0.79, 1.16) | 0.66 | 0.83 (0.70, 0.98) | 0.03 | 0.91 (0.75, 1.09) | 0.29 | 0.84 (0.58, 1.20) | 0.33 |
| By cancer type | ||||||||||
| Gastric cancer | 6 | 1,526/1,588 | 0.96 (0.74, 1.23) | 0.72 | 0.91 (0.66, 1.25) | 0.55 | 0.93 (0.76, 1.13) | 0.48 | 0.87 (0.56, 1.36) | 0.55 |
Notes:
number of included studies
P-value for the Z-test.
Abbreviations: C, cytosine; CD, cluster of differentiation; CI, confidence interval; T, thymine; HWE, Hardy-Weinberg equilibrium; Or, odds ratio; vs, versus.
Subgroup analysis
In the subgroup analysis by ethnicity, the OR was 1.07 (95% CI =0.80–1.44) (P=0.65) among Asians and was 0.96 (95% CI =0.79–1.16) (P=0.66) among Caucasians, based on the recessive model (TT vs TC + CC), thus both showed on association between the -159C/T polymorphisms of the CD14 gene and cancer risk. When analyzed by the dominant model (TT + TC vs CC), the OR was 0.82 (95% CI =0.55–1.21) (P=0.31) among Asians and was 0.83 (95% CI =0.70–0.98) (P=0.03) among Caucasians. These results suggested that TT and TC carriers have a 17% decreased risk of cancer compared with those individuals with the CC homozygote in Caucasians. The other models didn’t show any association between this polymorphism and cancer risk in Caucasians.
In the subgroup analysis by cancer type, gastric cancer was investigated in six studies,15,17,19,20,22 involving 1,526 gastric cancer cases and 1,588 controls. The pooled OR was 0.96 (95% CI =0.74–1.23) (P=0.72) for the recessive TT vs TC + CC model (Figure 3), and was 0.91 (95% CI =0.66–1.25) (P=0.55) for the TT + TC vs CC model and therefore, showed no association. Due the limited number of included studies, we didn’t perform a subgroup analysis of the other cancer types.
Figure 3.
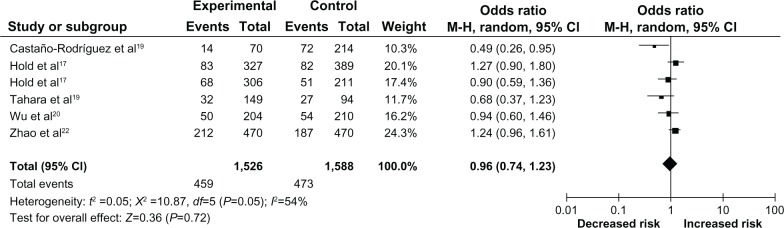
Meta-analysis with a random-effects model for the association between gastric cancer risk and the CD14 -159C/T polymorphism (TT vs TC + CC).
Abbreviations: CD14, cluster of differentiation 14; C, cytosine; CI, confidence interval; df, degrees of freedom; M-H, Mantel-Haenszel; T, thymine; vs, versus.
Publication bias
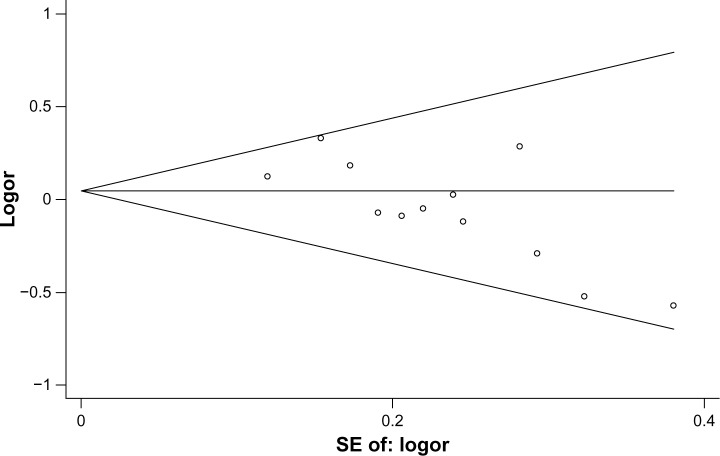
Publication bias was analyzed by using the Begg’s funnel plots and Egger’s test. The shape of the funnel plots seemed asymmetrical in the TT vs TC + CC comparator genetic model, suggesting the presence of a publication bias (Figure 4). The Egger’s test was performed to provide statistical evidence of funnel plot asymmetry. The result indicated a significant publication bias (P=0.008).
Figure 4.
Begg’s funnel plot for publication bias in selection of studies on the CD14 -159C/T polymorphism (TT vs TC + CC).
Note: Pseudo 95% confidence limits.
Abbreviations: CD14, cluster of differentiation 14; C, cytosine; T, thymine; SE, standard error; v, versus.
Discussion
Chronic inflammation is associated with the development and progression of several types of cancer.2 CD14 is an important inflammation mediator, and repeated studies have confirmed that the genetic variations of CD14 are associated with classical inflammatory diseases, such as asthma and inflammatory bowel disease.23,24 Studies have reported that CD14 may be involved in cancer development and progression, and a growing number of studies have investigated the potential relationship between the CD14 -159C/T gene polymorphism and cancer risk. However, the results from different published studies have been inconsistent. Thus, we performed this meta-analysis to comprehensively analyze their associations.
In present study, the effect of dominant/recessive models and the effect of allele frequency were all assessed to investigate the precise role of the CD14 -159C/T gene polymorphism in cancer. What’s more, the consistency of the genetic effects across populations of different ethnicities and cancer types was also investigated. The findings from present study indicated that -159C/T polymorphism in the CD14 gene showed no association with cancer risk. In addition, the dominant allele model showed a potential decreased risk of cancer for this polymorphism (OR =0.81, 95% CI =0.66–1.00). Since the genetic–protein interaction is complicated, it is not surprising that there was no association found between the CD14 -159C/T gene polymorphism and cancer risk. Growing studies suggest a complex interaction between gene polymorphism and cancer. The contributions of genetic factors to cancer are quite complicated, except for the current known gene polymorphisms, and uncommon polymorphisms may also contribute to cancer.25 In addition, the effects of gene-gene and gene-environment interactions remain questions.26 For a deeper understanding of the precise role of the CD14 -159C/T gene polymorphism in cancer risk, more studies are needed.
Our study included several types of cancer, and we did a subgroup analysis on gastric cancer to investigate whether the relationship between CD14 -159C/T gene polymorphism and cancer risk is tumor origin-specific. A total of six studies on gastric cancer were included. However, the results failed to show any association (OR =0.91, 95% CI =0.66–1.25) (P=0.55) for dominant model. Since Helicobacter pylori (H. pylori) infection plays an important role in the pathogenic process of gastric cancer, Zhao et al investigated this. The researchers reported that the CD14 -159C/T gene polymorphism is associated with a greater risk of H. pylori–related gastric cancer and postulated that this polymorphism might play a role in the outcome of H. pylori infection, further suggesting that the effect of CD14 -159C/T gene polymorphism on gastric cancer is quite complex and that other pathogenic factors should be taken into consideration.22 In addition, we also have noticed that the association between the CD14 -159C/T gene polymorphism and cancer risk may also be cancer subtype-specific – in a study by Andrie et al, a significantly increased risk for Hodgkin lymphoma was found with presence of the CD14 -159C/T polymorphism, while not for non-Hodgkin lymphoma.14 These results indicated that the effect of CD14 -159C/T gene polymorphism on cancer risk may be tumor origin-specific and subtype-specific.
Different ethnicities have different genetic backgrounds, which may influence the association between polymorphism and cancer susceptibility in those groups. In this meta-analysis, data were also stratified by ethnicity. For Asians, no potential association between the CD14 -159C/T gene polymorphism and cancer risk was identified in any of the models, and for Caucasians, the results suggested that TT and TC carriers have a 17% decreased risk of cancer compared with those individuals with the CC homozygote (P=0.03). However, it should be pointed out that there may be differences in the genes involved, in different populations; different polymorphisms within a specific locus; and different loci within the same gene and that environmental factors may even play a role in the expression of these changes.27 The identification of susceptibility genes in cancer patients of different ethnicities provides an opportunity to explore new mechanisms of disease that are specific in different populations.28
The objective of our meta-analysis was to integrate the results from comparable studies on the potential association between CD14 -159C/T gene polymorphism and cancer risk, in order to increase their sample size and statistical power and to draw more valid conclusions. However, the present meta-analysis also has several limitations that should be addressed. First, strict inclusion and exclusion criteria were used in order to reduce selection bias, and the resulting limited number of studies and subjects may not have enough power to explore the association between CD14 -159C/T gene polymorphism and cancer susceptibility. Second, the included studies were based on Asian and Caucasian population, and additional studies are warranted to evaluate the effect of this functional polymorphism on cancer risk among different populations, eg, in Latinos. Third, only published studies in the selected databases were included in this meta-analysis, and our meta-analysis identified significant publication bias among included studies. It is possible that some studies that were not included in these databases or some unpublished studies with null results were not identified, and this may have biased our results.
Conclusion
Taken together, the findings from the current meta-analysis don’t support the CD14 -159C/T gene polymorphism as a risk factor for cancer. To further investigate gene–gene and gene–environment interactions between this polymorphism and cancer risk, additional case-control studies of different cancer types and ethnicities should be performed.
Acknowledgment
This work was supported by a grant (number: 81300032) from the National Natural Science Foundation of China. We are indebted to the authors of the primary studies; without their contributions, this work would have been impossible.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 3.Aiello-Laws L. Genetic cancer risk assessment. Semin Oncol Nurs. 2011;27(1):13–20. doi: 10.1016/j.soncn.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Tang L, Zhao Y, Nie W, Wang Z, Guan X. 3′ untranslated region 1630 C > T polymorphism of prohibitin increases risk of breast cancer. Onco Targets Ther. 2013;6:177–182. doi: 10.2147/OTT.S40997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Shen Y, Chen L, et al. The -159C/T Polymorphism in the CD14 gene and tuberculosis risk: a meta-analysis. Int J Med Sci. 2013;10(11):1524–1529. doi: 10.7150/ijms.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason TE, Ricks-Santi L, Chen W, et al. Association of CD14 variant with prostate cancer in African American men. Prostate. 2010;70(3):262–269. doi: 10.1002/pros.21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Türe-Ozdemir F, Gazouli M, Tzivras M, et al. Association of polymorphisms of NOD2, TLR4 and CD14 genes with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. Anticancer Res. 2008;28(6A):3697–3700. [PubMed] [Google Scholar]
- 8.Chen R, Luo FK, Wang YL, Tang JL, Liu YS. LBP and CD14 polymorphisms correlate with increased colorectal carcinoma risk in Han Chinese. World J Gastroenterol. 2011;17(18):2326–2331. doi: 10.3748/wjg.v17.i18.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A polymorphism* in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20(5):976–983. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- 10.Karhukorpi J, Yan Y, Niemela S, et al. Effect of CD14 promoter polymorphism and H. pylori infection and its clinical outcomes on circulating CD14. Clin Exp Immunol. 2002;128(2):326–332. doi: 10.1046/j.1365-2249.2002.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. PLoS Med. 2009;6(3):e1000028. doi: 10.1371/journal.pmed.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13(7):840–848. doi: 10.1038/sj.ejhg.5201410. [DOI] [PubMed] [Google Scholar]
- 13.Shen Y, Guo S, Yang T, et al. The -173G/C Polymorphism of the MIF gene and inflammatory bowel disease risk: a meta-analysis. Int J Mol Sci. 2013;14(6):11392–11401. doi: 10.3390/ijms140611392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrie E, Michos A, Kalampoki V, et al. Genetic variants in immuno-regulatory genes and risk for childhood lymphomas. Eur J Haematol. 2009;83(4):334–342. doi: 10.1111/j.1600-0609.2009.01288.x. [DOI] [PubMed] [Google Scholar]
- 15.Castaño-Rodríguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM. The role of TLR2, TLR4 and CD14 genetic polymorphisms in gastric carcinogenesis: a case-control study and meta-analysis. PLoS One. 2013;8(4):e60327. doi: 10.1371/journal.pone.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Q, Zhu J, Xia B. Polymorphism of CD14 gene but not the mutation of TLR4 gene is associated with colorectal cancer in Chinese patients. J Gastroenterol Hepatol. 2006;21(1 Pt 1):92–97. doi: 10.1111/j.1440-1746.2005.04156.x. [DOI] [PubMed] [Google Scholar]
- 17.Hold GL, Rabkin CS, Gammon MD, et al. CD14-159C/T and TLR9-1237T/C polymorphisms are not associated with gastric cancer risk in Caucasian populations. Eur J Cancer Prev. 2009;18(2):117–119. doi: 10.1097/CEJ.0b013e3283101292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landi S, Gemignani F, Bottari F, et al. Polymorphisms within infammatory genes and colorectal cancer. J Negat Results Biomed. 2006;5:15. doi: 10.1186/1477-5751-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahara T, Arisawa T, Shibata T, Hirata I, Nakano H. Association of polymorphism of TLR4 and CD14 genes with gastroduodenal diseases in Japan. Inflammopharmacology. 2007;15(3):124–128. doi: 10.1007/s10787-006-1567-8. [DOI] [PubMed] [Google Scholar]
- 20.Wu MS, Cheng TY, Shun CT, Lin MT, Chen LC, Lin JT. Functional polymorphisms of CD14 and toll-like receptor 4 in Taiwanese Chinese with Helicobacter pylori-related gastric malignancies. Hepatogastroenterolog y. 2006;53(71):807–810. [PubMed] [Google Scholar]
- 21.Yu X, Zhang C, Sun A, et al. Genetic variations in CD14 promoter and acute lymphoblastic leukemia susceptibility in a Chinese population. DNA Cell Biol. 2011;30(10):777–782. doi: 10.1089/dna.2011.1223. [DOI] [PubMed] [Google Scholar]
- 22.Zhao D, Sun T, Zhang X, et al. Role of CD14 promoter polymorphisms in Helicobacter pylori infection – related gastric carcinoma. Clin Cancer Res. 2007;13(8):2362–2368. doi: 10.1158/1078-0432.CCR-06-2612. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, Bracken MB. Association of CD14 -260 (-159) C > T and asthma: a systematic review and meta-analysis. BMC Med Genet. 2011;12:93. doi: 10.1186/1471-2350-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Hu J, Fan R, Zhou J, Zhong J. Association between CD14 gene C-260T polymorphism and inflammatory bowel disease: a meta-analysis. PLoS One. 2012;7(9):e45144. doi: 10.1371/journal.pone.0045144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodmer W, Tomlinson I. Rare genetic variants and the risk of cancer. Curr Opin Genet Dev. 2010;20(3):262–267. doi: 10.1016/j.gde.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 26.Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299(20):2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Losada J, Castellanos-Martín A, Mao JH. Cancer evolution and individual susceptibility. Integr Biol (Camb) 2011;3(4):316–328. doi: 10.1039/c0ib00094a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haiman CA, Stram DO. Exploring genetic susceptibility to cancer in diverse populations. Curr Opin Genet Dev. 2010;20(3):330–335. doi: 10.1016/j.gde.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]