Abstract
Identification of reference genes with stable levels of gene expression is an important prerequisite for obtaining reliable results in analysis of gene expression data using quantitative real time PCR (RT-qPCR). Since the underlying assumption of reference genes is that expressed at the exact same level in all sample types, in this study, we evaluated the expression stability of nine most commonly used endogenous controls (GAPDH, ACTB, 18S rRNA, RPS18, HSP-90, ALAS, HMBS, ACAC, and B2M) in four different tissues of the domestic goat, Capra hircus, including liver, visceral, subcutaneous fat and longissimus muscles, across different experimental treatments (a standard diet prepared using the NRC computer software as control and the same diet plus one mg chromium/day). We used six different software programs for ranking of reference genes and found that individual rankings of the genes differed among them. Additionally, there was a significant difference in ranking patterns of the studied genes among different tissues. A rank aggregation method was applied to combine the ranking lists of the six programs to a consensus ranking. Our results revealed that HSP-90 was nearly always among the two most stable genes in all studied tissues. Therefore, it is recommended for accurate normalization of RT-qPCR data in goats, while GAPDH, ACTB, and RPS18 showed the most varied expressions and should be avoided as reference genes.
Introduction
Over 830 million of the domestic goats, Capra hircus, belonging to more than 1000 breeds are reared throughout the world because of their importance as sources of meat, milk, fiber and pelts (http://www.fao.org/corp/statistics/en). In addition to their value as domestic animals, goats have been extensively used as animal models for biomedical studies, to investigate the genetic basis of complex traits as well as in the transgene production of peptide medicines[1,2].The recent report of whole genome sequence of the domestic goat [1] has led to a striking increase in its use as a model species in a wide range of genetic studies including gene expression analyses.
Quantitative real-time PCR (RT-qPCR) is the most widely used method for gene expression studies and analysis of biological pathways, as it allows fast, extremely sensitive, and highly reproducible quantification of mRNA levels [3-5]. However, some factors including RNA stability, RNA extraction, retrotransciption efficiency, PCR steps, and amounts of RNA added into the reaction may negatively affect the accuracy and reliability of the results obtained from RT-qPCR [6,7]. Several procedures have been developed for normalization of the variations from sample to sample among which, the most common and reliable method is to normalize the total amounts of RNA or a single internal reference gene, known as housekeeping gene [8,9]. An ideal reference gene is expected to be stable in terms of expression level across various experimental conditions such as developmental stages, tissue types, experimental treatments, and external stimuli [10]. However, recent studies have shown that many commonly used reference genes are not suitable for RT-qPCR, as their expression might be altered by some experimental conditions [11-13]. Therefore, it is essential to screen and select appropriate reference gene(s) with a constant level of expression under certain experimental conditions for valid interpretation of expression data [14].
To date, several mathematical models and software programs have been developed which allows selection of the most stable reference genes [5,7,13,14]. However, it has been repeatedly reported that different software programs may give different rankings of reference genes [15-17]. This may be explained by the fact that the routine programs use different algorithms, thus they would not be expected to yield identical results. Recently, a rank aggregation method was developed to combine the ordered lists of genes, obtained from different software programs, to a consensus ranking of reference genes [15-20].
The objective of this study was to select from a panel of nine commonly used reference genes the most stable genes for normalization of gene expression data in the domestic goat, C. hircus. Six different software programs were used to identify the most stable reference genes across the four studied tissues under chromium treatment. Additionally, a rank aggregation method was used to provide a consensus ranking by combining the ranking results obtained from six different programs. The variant experimental conditions include four different tissue types (liver, visceral fat, subcutaneous fat, and longissimus lumborum (LL) muscles) as well as two dietary conditions based on presence or absence of the supplementary trivalent chromium (Cr+3). Trivalent chromium has been known as an essential element for normal metabolism of proteins, lipids, carbohydrates, and nucleic acids in human and husbandry animals such as cows, sheep, and goats[21,22]. A large body of evidence has also confirmed the positive effects of chromium on various biological properties of domestic animals including immunity against viral diseases [23], live body weight, daily weight gain, dressing percentage, longissimus muscle area, nutrient digestibility, etc. [21,22,24,25]. Importantly, chromium has been suggested to alter the expression level of a variety of genes in human and laboratory animals [26-28]. Therefore, we used the trivalent chromium as food supplementation to evaluate the effects of external agents on expression stability of the nine candidate genes.
Materials and Methods
Ethics Statement
All experiments with animals were performed according to the recommendations in the Guide for the Care and Use of Laboratory Animals of the Research Station of Department of Animal Science, University of Tehran, Iran. The protocols were approved by the Animal Care and Use Committee of the University of Tehran Institutional Animal Care and Use Committee and included in a Research project.
Animal husbandry and experimental design
This study was conducted at the Research Station of Department of Animal Science, College of Agriculture and Natural Resources, University of Tehran, Iran. Twenty-four, 4 to 5-months old male goat kids belonging to the native Iranian breed, Mahabadi, were selected for the experiment. All procedures of immunity and nutrition were conducted under protocols approved by this station.
The kids were weighed (BW=22 ± 2 kg) and allocated randomly to one of the two following dietary treatments: standard diet plus 0 and 1 mg chromium per day as chromium-methionine (Availa®Cr 1000, Zinpro Corporation, USA). The standard diet was balanced and prepared using NRC computer software (Table 1). The kids were individually penned for 100 days (10 days for adaptation and 90 days for feeding period), with access to enough water, and provided with the prepared diets twice a day (08:00 h and 17:00 h). The kids were weighed before the morning feeding meal triweekly (i.e. after 14-16 hours of starvation) throughout the experimental period to determine changes in their body weight.
Table 1. Ingredient and chemical composition of basal standard diet fed to goat kids.
| Ingredient | % of DM |
|---|---|
| Alfalfa hay | 16.49 |
| Corn silage | 8.32 |
| Wheat straw | 5.19 |
| Barley grain | 51 |
| Wheat bran | 9.09 |
| Canola meal | 4.55 |
| Soybean meal | 2.21 |
| Calcium carbonate | 1.3 |
| Mineral-vitamin supplement a | 0.91 |
| Sodium bicarbonate | 0.78 |
| Salt | 0.52 |
| Nutrient fractions | |
| DM (%) | 80.78 |
| CP (% of DM) | 13.5 |
| Ether extract (% of DM) | 2.6 |
| NDF (% of DM) | 36.6 |
| Ash (% of DM) | 9 |
| ME (Mcal/kgDM) | 36.6 |
| Calcium (% of DM) | 0.89 |
| Phosphorus (% of DM) | 0.49 |
| Chromium (% of DM) | 0.83 |
a Containing per kg DM: calcium, 195 g; phosphor, 80 g; magnesium, 21000 mg; sodium, 50 g; manganese, 2200 mg; iron, 3000 mg; copper, 300 mg; iodine, 120 mg; cobalt, 100 mg; zinc, 300 mg; selenium, 1.1 mg; antioxidant, 2500 mg; vitamin A, 600,000 IU; Vitamin D3, 200,000 IU; vitamin E, 200 mg.
Slaughtering and tissue sampling
After feeding on the prepared diets for 90 days, the kids were transferred to the departmental abattoir, where they were kept for 12 h with free access to water. They were then slaughtered by decapitation and tissue samples from liver, visceral fat, subcutaneous fat, and longissimus lumborum muscle were taken from the corpses. The samples were immediately frozen in liquid nitrogen and transferred to the laboratory, where they were maintained at -80 °C until used.
Total RNA isolation, clean up and cDNA synthesis
Total RNA was extracted according to the method of Chomczynski and Sacchi (2006) using Trizol Reagent (Invitrogen Co., Carlsbad,CA, USA) [29]. The extracted RNA was then treated with RNase-free DNase I (TaKaRa, Shuzo, Kyoto, Japan). RNA concentrations were estimated by Nanodrop spectrophotometry at 260 nm and their purities were checked by determining the absorption ratios at 260/280 nm. The quality of extracted RNA was assessed by electrophoresis at 1% agarose-gel containing Ethidium Bromide. First-strand cDNA was synthesized from 100 ng of total RNA using an oligo (dT) primer, random hexamers and a commercially available kit (AccuPower® RocketScript™ RT PreMix) according to manufacturer’s instructions.
Selection of reference genes and primer designing
Nine classical reference genes including glyceraldehyd-3-phosphate dehydrogenase (GAPDH), beta-actin (ACTB), heat shock protein-90 (HSP-90), aminolevulinate synthase 1 (ALAS1), 18S ribosomal RNA (18S rRNA), ribosomal protein S18 (RPS18), hydroxyl methyl bilane synthase (HMBS), acetyl coenzyme A carboxylase alpha (ACAC-α), and beta-2-microglobulin (B2M) were considered as candidate genes. The nucleotide sequences of all candidate genes (except for B2M) belonging to the domestic goat (Capra hircus) were obtained from the GenBank database (http://www.ncbi.nlm.nih.gov). For B2M gene, only one sequence was available for the relative species, Ovis aries. Primer pairs were designed from these sequences (optimal Tm at 59.8°C and GC% between 45-50%) using primer3Plus online software [30] and checked using OligoAnalyzer 3.1 (http://eu.idtdna.com/analyzer/applications/oligoanalyzer/), OligoCalc [31], and PrimerBLAST [32] (Table 2).
Table 2. Sequence and some characterization of specific primer pairs for the nine genes used for selection of reference gene in goat kids.
| Gene Name | Accession number | Sense primer sequence 5ʹ→3ʹ | Anti-sense primer sequence 5ʹ→3ʹ | Length (bp) | Tm (°C) |
|---|---|---|---|---|---|
| GAPDH | AJ431207.1 | GGCACAGTCAAGGCAGAGAA | TCTCGCTCCTGGAAGATGGT | 71 | 59.8 |
| ACTB | HQ993072.1 | GCAAGGACCTTTACGCCAAC | CTTGATCTTCATCGTGCTGGG | 116 | 59.8 |
| HSP-90 | AF548366.1 | GCCCGAGATAGAAGACGTTG | AGTCGTTGGTCAGGCTCTTG | 197 | 59.8 |
| ALAS | AB232536.1 | ATGTGGCCCACGAGTTTGG | CTTGTGCTGGCGATGTACC | 178 | 59.8 |
| 18S rRNA | DQ149973.1 | TAATCCCGCCGAACCCCATT | GGTGTGTACAAAGGGCAGG | 125 | 59.8 |
| RPS18 | EF564275.1 | ATGCAGAATCCACGCCAATAC | GGCCCGAATCTTCTTCAGG | 147 | 59.8 |
| HMBS | AB232537.1 | CTTGCCAGAGAAGAGTGTGG | CAGCCGTGTGTTGAGGTTTC | 115 | 59.8 |
| ACAC | DQ370054.1 | CGCTATGGAAGTCGGCTGTG | CAGGAAGAGGCGGATGGGAA | 105 | 59.8 |
| B2M | DQ386890.1 | TGTCCCACGCTGAGTTCACT | TGAGGCATCGTCAGACCTTGA | 137 | 59.8 |
Two step real-time RT-PCR
Real-time Quantitative PCR was performed using SYBR Green I technology on iQ5 System (BioRad, USA). The reactions consisted of 1x SYBR Green PCR Master Mix (SYBR biopars, GUASNR, Iran), 300 nm of each specific forward and reverse primers, 10 ng of cDNA, and nuclease free water to a final volume of 20 µL.
The cycling conditions were as follows: cDNA was denatured at 94 °C for 3 min, followed by 35 cycles of 94 °C for 15 s and 59.8 °C for 15 s (gain set at 10 for SYBR Green). All samples were amplified in triplicate from the same RNA preparation and the mean value was considered. Two biological replications were used for each plate. The real-time RT-qPCR efficiency was assessed for each gene based on the slope of a linear regression model [33]. The bulks of each cDNA sample were used as PCR template in a range of 10-fold dilution series. The corresponding real-time RT-qPCR efficiencies were calculated based on the slope of the standard curve using the following equation: (E = 10 -1/slope - 1) [34].
Determination of genes expression stability
Six popular software programs including geNorm version 2.3 [15], NormFinder version 0.953 [10] and BestKeeper version 1 [35], qBasePlus version 2.3 (Biogazelle, Ghent, Belgium) and GenEx version 5 (NormFinder and GeNorm) were used to validate the most stable reference genes for different tissues taken from goat kids. In general, all programs are based on the principle that the expression of reference genes should be stable under all experimental conditions and tissue types studied.
Statistical Method for Rank Aggregation
The RankAggreg package of R software was used to combine the stability measurements obtained from the six software and establish a consensus rank of reference genes [36]. The term stability refers to the variation of the Ct values for a given gene across different samples or experimental conditions. Transcripts with the lowest stability measurement would usually yield the best reference gene. Six matrix of rank-ordered reference genes, according to the different stability measurements (NormFinder stability value, M values by qBasePlus, geNorm (qbase plus) and GenEx (geNorm), standard deviation of BestKeeper and GenEx (NormFinder) were used as input for this statistical package (for each of the studied tissues separately and all tissues together). An unweighted rank aggregation was applied by using BruteAggreg function of the package. This function performs rank aggregation using the brute force approach.
The aim of rank aggregation is to find an aggregated ranking that minimizes the distance to each of the ranked lists in the input set. The distance among ordered lists is calculated using the Spearman foot rule function. In summary Brute force approach was used to account all ranking lists and find the list with the minimum Spearman foot rule distance according to the Brute Aggreg function.
The permutation function of the gtools package was used to generate all possible ordered lists. This approach is suggested when the size of the ranking list is smaller than10. RankAggreg function is another function of this package that performs rank aggregation via the Cross-Entropy Monte Carlo algorithm. This algorithm is usually recommended when the size of the ranking list is larger than 10 [36]. Since, our size of the ranking list was near to 10 (9), we used RankAggreg function additionally to validate the consensus rank of reference genes resulting from the brute force approach. Because GenEx (geNorm) yields the same M stability value as the two most stable genes, two consensus lists of reference genes were created by altering the position of the two most stable genes. The consensus ranking with the lower score was selected.
Results
In this study, after a detailed literature review, nine reference genes, including GAPDH, ACTB, HSP-90, ALAS1, 18S rRNA, RPS18, HMBS, ACAC-α and B2M were used to select a suitable reference gene for expression analyses in different tissues of domestic goats. These genes are among the most common ones frequently used as reference genes for normalization of RT-qPCR data across animal taxa. To confirm reproducibility of real-time PCR, standard error of means (SEM) was determined (Table 3). We found a wide variation in the averages of the cycle threshold (CT) values for the nine reference genes where it ranged between 23.71 to 33.17 in liver, 24.18 to 30.86 in visceral fat, 22.33 to 27.72 in subcutaneous fat, and 23.42 to 30.84 in longissimus lumborum muscle tissues (these values has been visualized in Figure 1 for each studied tissue and in Figure 2 for all tissues together). The highest and lowest expression levels were detected for 18S rRNA in subcutaneous fat and for ALAS in liver, respectively. Although, the expression of the studied genes showed some degrees of differences across different tissues and dietary treatments, the most stability was recorded for HSP-90 (Figure 2). This was implied by the fact that HSP-90 was nearly always among the most three stable genes ranked by the six programs for all tissues (Tables 4-7). The GAPDH, RPS18 and ACTB genes, in contrast, were among the most unstable genes in term of expression level (Tables 4-7). Table 8 shows the ranking of the nine candidate genes by considering all sample tissues as a single unit (Information S1).
Table 3. The calculated mean the cycle threshold (Ct) values and their SEM for the nine reference genes in different tissues.
| Reference gene | Treatment | Tissue |
|||
|---|---|---|---|---|---|
| Liver | Visceral Fat | Subcutaneous Fat | LL Muscle | ||
| HSP-90 | Control | 30.47 ± 0.067 | 29.06 ± 0.116 | 27.11 ± 0.179 | 30.83 ± 0.009 |
| Chromium | 30.13 ± 0.055 | 30.13 ± 0.283 | 27.33 ± 0.223 | 28.81 ± 0.087 | |
| ALAS | Control | 31.37 ± 0.378 | 28.63 ± 0.167 | 26.58 ± 0.104 | 30.01 ± 0.032 |
| Chromium | 30.15 ± 0.544 | 29.91 ± 0.196 | 27.56 ± 0.095 | 28.50 ± 0.046 | |
| B2M | Control | 32.10 ± 0.237 | 25.70 ± 0.012 | 26.82 ± 0.038 | 29.60 ± 0.179 |
| Chromium | 29.43 ± 0.450 | 27.79 ± 0.153 | 26.46 ± 0.144 | 25.67 ± 0.058 | |
| RPS18 | Control | 28.85 ± 0.381 | 24.23 ± 0.023 | 25.62 ± 0.012 | 27.53 ± 0.095 |
| Chromium | 28.58 ± 0.029 | 25.63 ± 0.026 | 23.96 ± 0.023 | 25.65 ± 0.061 | |
| GAPDH | Control | 23.82 ± 0.061 | 26.14 ± 0.020 | 22.83 ± 0.286 | 26.42 ± 0.251 |
| Chromium | 25.72 ± 0.017 | 26.90 ± 0.185 | 23.94 ± 0.023 | 23.54 ± 0.066 | |
| ACAC | Control | 30.12 ± 0.009 | 27.99 ± 0.153 | 26.36 ± 0.092 | 29.01 ± 0.156 |
| Chromium | 30.04 ± 0.061 | 30.35 ± 0.294 | 26.58 ± 0.078 | 29.76 ± 0.346 | |
| ACTB | Control | 31.27 ± 0.785 | 28.94 ± 0.393 | 25.91 ± 0.110 | 28.07 ± 0.014 |
| Chromium | 29.43 ± 0.012 | 29.27 ± 0.147 | 27.38 ± 0.081 | 23.74 ± 0.026 | |
| 18s rRNA | Control | 26.71 ± 0.153 | 25.27 ± 0.003 | 23.44 ± 0.058 | 25.85 ± 0.084 |
| Chromium | 26.97 ± 0.101 | 24.70 ± 0.300 | 23.09 ± 0.306 | 25.87 ± 0.052 | |
| HMBS | Control | 28.12 ± 0.222 | 27.33 ± 0.196 | 24.69 ± 0.052 | 27.31 ± 0.121 |
| Chromium | 26.64 ± 0.534 | 28.35 ± 0.274 | 25.75 ± 0.081 | 26.12 ± 0.064 | |
Figure 1. The distribution of gene expression levels of nine candidate reference genes analyzed for four different tissues (liver (a), visceral fat (b), subcutaneous muscle (c), and longissimus muscle (d)) in pooled CT value.
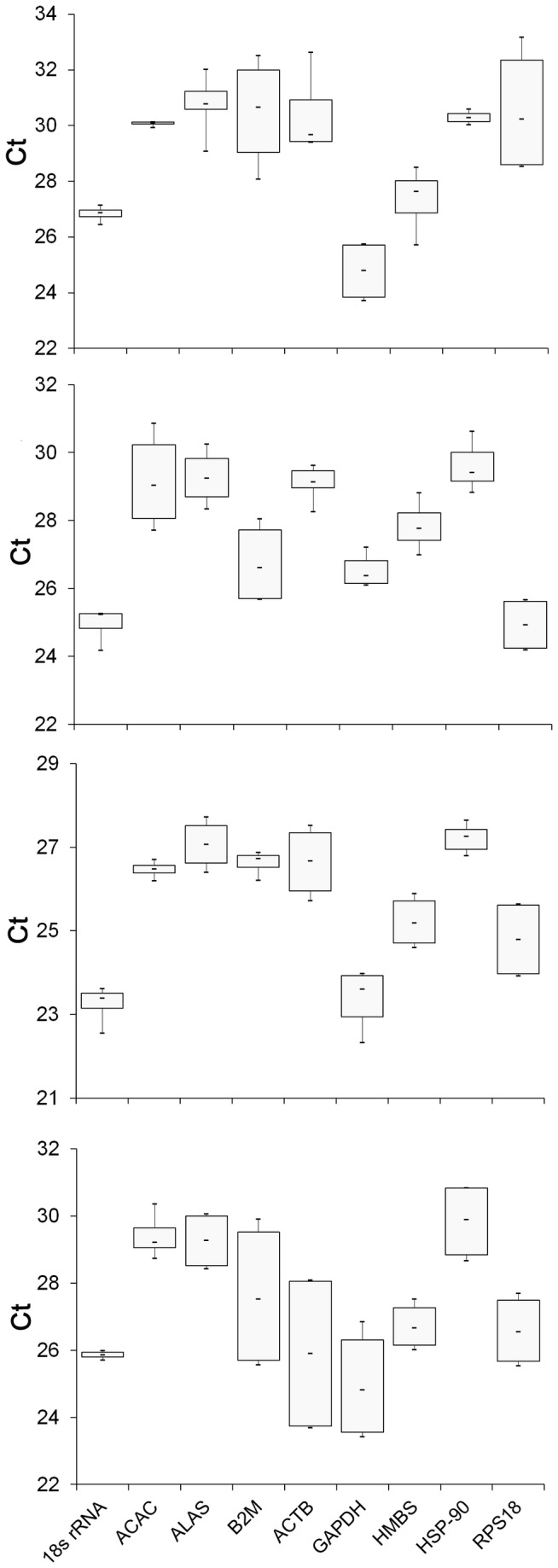
Figure 2. The distribution of gene expression levels of nine candidate reference genes in pooled CT value.
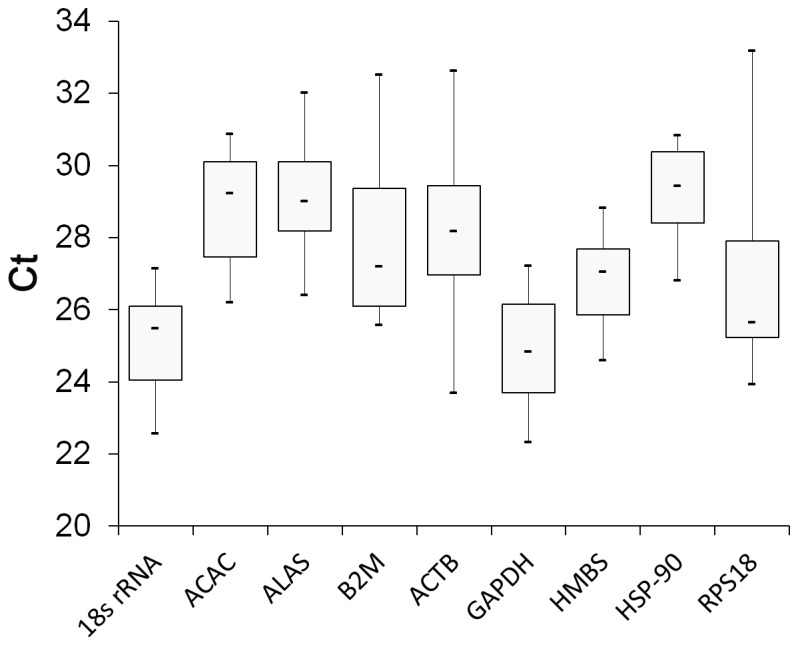
The variations are related to data taken together from four different tissues (liver, visceral fat, subcutaneous muscle, and longissimus muscle).
Table 4. Results of ranking of nine candidate reference genes obtained using six different software programs and rank aggregation of candidate reference genes for liver.
| Rank Position | GenEx (NormFinder) | GenEx (geNorm) | NormFinder | BestKeeper | geNorm | qBasePlus | Consensus |
|---|---|---|---|---|---|---|---|
| 1 | HSP-90 | HSP-90 | HSP-90 | ACAC | ACACa | ALAS | HSP-90 |
| 2 | ACAC | ACAC | ACAC | HSP-90 | HSP-90 | HMBS | ACAC |
| 3 | HMBS | 18s rRNA | HMBS | 18s rRNA | 18s rRNA | HSP-90 | 18s rRNA |
| 4 | ALAS | ALAS | ALAS | ALAS | ALAS | ACTB | ALAS |
| 5 | 18s rRNA | HMBS | 18s rRNA | HMBS | HMBS | ACAC | HMBS |
| 6 | ACTB | ACTB | ACTB | GAPDH | ACTB | 18s rRNA | ACTB |
| 7 | B2M | B2M | B2M | ACTB | B2M | B2M | B2M |
| 8 | RPS18 | RPS18 | RPS18 | B2M | RPS18 | RPS18 | RPS18 |
| 9 | GAPDH | GAPDH | GAPDH | RPS18 | GAPDH | GAPDH | GAPDH |
a ACAC was the first in consensus list if alter the position of the two most stable genes in the geNorm list.
Table 8. Results of ranking of nine candidate reference genes obtained using six different software programs and rank aggregation of candidate reference genes for all studied tissues.
| Rank Position | GenEx (NormFinder) | GenEx (geNorm) | NormFinder | BestKeeper | geNorm | qBasePlus | Consensus |
|---|---|---|---|---|---|---|---|
| 1 | ALAS | HSP-90 | ALAS | HMBS | HSP-90 | ALAS | ALAS |
| 2 | HSP-90 | ALAS | HSP-90 | 18s rRNA | ALAS | HSP-90 | HSP-90 |
| 3 | HMBS | ACAC | HMBS | HSP-90 | HMBS | HMBS | HMBS |
| 4 | 18s rRNA | 18s rRNA | ACAC | ALAS | ACAC | 18s rRNA | 18s rRNA |
| 5 | ACAC | HMBS | 18s rRNA | ACAC | B2M | ACAC | ACAC |
| 6 | B2M | GAPDH | B2M | GAPDH | 18s rRNA | B2M | B2M |
| 7 | GAPDH | B2M | GAPDH | ACT-B | GAPDH | GAPDH | GAPDH |
| 8 | ACT-B | ACT-B | ACT-B | B2M | ACT-B | ACT-B | ACT-B |
| 9 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 |
Table 6. Results of ranking of nine candidate reference genes obtained using six different software programs and rank aggregation of candidate reference genes for subcutaneous fat.
| Rank Position | GenEx (NormFinder) | GenEx (geNorm) | NormFinder | BestKeeper | geNorm | qBasePlus | Consensus |
|---|---|---|---|---|---|---|---|
| 1 | ALAS | HSP-90 | ALAS | HMBS | HSP-90 | ALAS | ALAS |
| 2 | HSP-90 | ALAS | HSP-90 | 18s rRNA | ALAS | HSP-90 | HSP-90 |
| 3 | HMBS | ACAC | HMBS | HSP-90 | HMBS | HMBS | HMBS |
| 4 | 18s rRNA | 18s rRNA | ACAC | ALAS | ACAC | 18s rRNA | 18s rRNA |
| 5 | ACAC | HMBS | 18s rRNA | ACAC | B2M | ACAC | ACAC |
| 6 | B2M | GAPDH | B2M | GAPDH | 18s rRNA | B2M | B2M |
| 7 | GAPDH | B2M | GAPDH | ACTB | GAPDH | GAPDH | GAPDH |
| 8 | ACTB | ACTB | ACTB | B2M | ACTB | ACTB | ACTB |
| 9 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 |
The BestKeeper software uses a pairwise correlation analysis of all reference genes and calculates the geometric mean of the best suited ones. BestKeeper uses two most important criteria including the stability (coefficient of variance (CV) and standard deviation (SD)) and coefficient of correlation to the BestKeeper index for evaluating the stability of reference genes. The genes with the lowest CV and SD are considered as the most stable genes [37,38]. Results based on BestKeeper showed that the most stable gene in both liver and subcutaneous fat under chromium treatment was ACAC. But in visceral fat and longissimus lumborum muscles 18srRNA was identified as the most stable gene. HMBS and 18srRNA were found to have a remarkably stable expression in all studied tissues together.
GeNorm, the first reported software, is based on pairwise comparison model and calculates the M value, where M is the average pairwise variation of an individual gene to other genes, of all candidate genes and use geometric averaging across a matrix of reference genes [15,17]. Genes are ranked according to their expression stability by a repeated process of stepwise exclusion of the least stably expressed genes [39]. Via geNorm, M values of all reference genes were less than 1.5 indicating that these candidate genes have stable expression levels. Among nine reference genes, the most stable genes in liver were ACAC and HSP-90, in visceral fat were HSP-90 and HMBS and in LL muscles were HMBS and GAPDH. But in subcutaneous fat and all studied tissues together, ALAS and HSP-90 were identified as most stable reference genes.
The qBasePlus evaluates the stability of the applied reference genes by calculating two quality measures: the coefficient of variation (CV) and the geNorm stability M-value. Both values are only meaningful, or can be calculated only if multiple reference genes are quantified. The reference genes with the lower quality values would have the higher stability [40]. The algorithm of qBasePlus for calculation of relative quantities selecting different reference genes and specific efficiencies has four steps: 1) Calculation of the average Ct value for all replicates of the same gene/sample combination within a given run, 2) transformation of mean Ct value into relative quantity using the gene specific PCR efficiency, 3) calculation of the normalization factor and 4) calculation of the normalized relative quantity for gene of interest for each sample [40]. Our results of qBasePlus revealed that the most stable gene in liver, subcutaneous fat and all studied tissues together under chromium treatment was ALAS, whereas in visceral fat and LL muscle tissues was HSP-90.
NormFinder software program employs an ANOVA-based model to estimate overall reference gene stability; but also considers variations between sample subgroups. The software calculates a stability value for all candidate reference genes. The stability value is based on the combined estimate of intra- and inter-group expression variations of the studied genes [41,42]. Using NormFinder, the top-ranked reference gene was ALAS in visceral and subcutaneous fat tissues and all studied tissues together. NormFinder also indicates HSP-90 and ACAC as the best stable reference genes for normalizing calculations in Liver and LL muscles, respectively.
GenEx (http://www.multid.se/) has the advantage of incorporating both NormFinder and geNorm in the software. Thus, users can get both of these algorithms on which to base their choice of reference genes in one software installation [43]. These algorithms detect the most stably expressed genes in an experimental setup. Optimal number of reference gene was selected using pairwise variation analysis integrated in geNorm algorithm implanted in GenEx [44]. The accumulated standard deviation (Acc. SD), as an indicator for the optimal number of reference genes, was determined using GenEx software. For each sample, the normalization factor based on n reference genes was calculated as the geometric average of the n raw reference gene quantities [45]. The results of GenEx (NormFinder) shown that the most stable genes in liver, visceral and subcutaneous fat, LL muscles and all studied tissues together were HSP-90, RPS-18, ALAS, ACAC and ALAS, respectively. But in GenEx (geNorm) were HSP-90, ALAS, HSP-90, ALAS and HSP-90.
We found significant differences in ranking patterns of reference genes obtained from different software programs (Tables 4-7). These discrepancies are probably related to differences in the algorithm each program uses in gene ranking. To provide a consensus ranking, a rank aggregation method was used to combine the ranking patterns of all six software. The RankAggreg package provides two functions for combining the ordered lists: the BruteAggreg function for k<10 and the RankAggreg function for k>10, where k is the size of the ranking list. In this study, the use of both functions yielded the same ranking list suggesting that the consensus ranks of genes were robust to the methods used. Additionally, the ranking lists were consistent when we altered the position of the two most stable genes in the geNorm list except for liver tissue. The results of rank aggregation method revealed that the two most stably expressed genes under chromium treatment were HSP-90 and ACAC in liver, HSP-90 and HMBS in visceral fat, ALAS and HSP-90 in subcutaneous muscles, and ACAC and HSP-90 in longissimus muscles (Tables 4-7). The three genes, GAPDH, RPS18 and ACTB showed the least stability in expression level across all studied tissues (Tables 4-7). The two candidate genes, ALAS and HSP-90 were nearly always the most stably expressed genes when different software programs were applied for the selection of suitable reference gene in all studied tissues (Tables 4-7). Therefore, it is not surprising that the rank aggregation method ranked the two genes as the most stable ones when compared to other candidate genes (Table 8).
Discussion
Goats are of excellent economical and historical importance in Iran. Strong evidences suggest that the Fertile Crescent (stretching from the southern Levant in south eastern Turkey and northern Syria to the high Zagros Mountain pastures of Iran) is the center of domestication of both sheep and goat [46]. In addition to especial importance of goats as sources of milk, pelt, and meat, they have been long considered as a model species in a wide range of biological and medicinal studies [1,2]. Therefore, gene expression profiles of this animal in different tissues at the mRNA and protein levels would be extremely valuable for further elucidation of the molecular mechanisms involved in different biological pathways.
The RT-qPCR technique is one of the most common methods to understand gene expression profiles in different biological systems, which is an important step in identifying gene function. In this regard, the importance of reference genes to accurately analyze expression of a particular gene is well known. The best reference genes are expected to undergo a constant, unregulated expression in all experimental conditions and analyzed tissues. However, it has been clearly demonstrated that no universal reference gene exists that express stably in all experimental conditions [47,48]. To date, there are a few studies that use RT-qPCR as a technique for gene expression analyses in domestic goats. In most of these studies, the reference genes are usually selected based on literature review and experience in other organisms rather than empirical evidence in support of their efficacy. But it is demonstrated that the most appropriate reference gene for animals of interest would not expect to be the same as found in other organisms, even if the animals are closely relative [49,50]. Therefore, it is generally proposed that the reference genes need to be validated for each species and for each specific experimental condition.
Here, to improve the gene expression analysis by RT-qPCR in the domestic goats, we evaluated the suitability of nine candidate reference genes in four different tissues and under two different nutritional diets (presence or absence of chromium supplementation). Then, their expression was analyzed by using six different software programs: GeNorm, GenEx (GeNorm), qBasePlus, NormFinder, GenEx (NormFinder) and BestKeeper.
The six software programs produced similar results, but the rankings were not identical and, in some cases, were substantially different, especially in the top ranked genes. Our results in this part suggest that different computer software might introduce a different ranking of reference genes. For example, the two genes, 18s rRNA and ACTB were among the least stable genes of visceral fat when the two programs GeNorm and NormFinder were applied, while, the program BestKeeper ranked these genes as the most stable ones (Table 5). Generally, the two programs GeNorm and NormFinder showed the most consistency compared with other studied software in term of ranking of the candidate reference genes (Tables 4-7). Difference in ranking results obtained from different software programs is a well-known criterion because the programs typically use different algorithms to determine gene expression stability [17-20]. The programs GeNorm, GenEx (GeNorm) and qBasePlus rank the reference genes according to a stability value (M). This value represents the mean pairwise variation between a candidate reference gene and all the other studied genes. The lowest M value indicates genes with the most stable expression. The genes are then ranked using stepwise elimination of the least stable genes [15,17]. qBasePlus also calculates a coefficient of variation (CV) for each gene as a stability measurement [17]. The NormFinder and GenEx (NormFinder) fit the data to a mathematical model, which allows comparison of intra- and intergroup variation and calculation of expression stability [10,17]. BestKeeper [33] uses repeated pairwise correlation analysis to determine the optimal reference genes. Therefore, it is not surprising that these algorithms differ in the ranking of the best reference genes.
Table 5. Results of ranking of nine candidate reference genes obtained using six different software programs and rank aggregation of candidate reference genes for visceral fat.
| Rank Position | GenEx (NormFinder) | GenEx (geNorm) | NormFinder | BestKeeper | geNorm | qBasePlus | Consensus |
|---|---|---|---|---|---|---|---|
| 1 | RPS18 | ALAS | ALAS | 18s rRNA | HSP-90 | HSP-90 | HSP-90 |
| 2 | ALAS | HMBS | HSP-90 | ACTB | HMBS | HMBS | HMBS |
| 3 | HSP-90 | HSP-90 | GAPDH | GAPDH | ALAS | ALAS | ALAS |
| 4 | HMBS | RPS18 | HMBS | HMBS | RPS18 | RPS18 | RPS18 |
| 5 | GAPDH | GAPDH | RPS18 | HSP-90 | GAPDH | GAPDH | GAPDH |
| 6 | B2M | B2M | ACTB | ALAS | ACTB | ACTB | ACTB |
| 7 | ACTB | ACAC | B2M | RPS18 | B2M | B2M | B2M |
| 8 | ACAC | ACTB | ACAC | B2M | ACAC | ACAC | ACAC |
| 9 | 18s rRNA | 18s rRNA | 18s rRNA | ACAC | 18s rRNA | 18s rRNA | 18s rRNA |
Although our study was not [10,17] designed to measure the effect of chromium supplementation on individual reference gene expression, it appears that chromium affects the stability rankings of some specific genes. This implies that the expression of a given reference gene may vary with experimental conditions and should be tested in a set of conditions. In addition, our results revealed that the appropriate reference gene for a given tissue type may differ from those of other tissues. For example, the gene GAPDH was between the two most stable genes according to ranking results obtained from the GeNorm program for liver. However, in longissimus muscles, GAPDH was the worst candidate as reference gene (Tables 4 & 7). All of these findings strongly confirm the previously reported fact that different tissues may differ in the expression stability of the reference genes.
Table 7. Results of ranking of nine candidate reference genes obtained using six different software programs and rank aggregation of candidate reference genes for longissimus muscle.
| Rank Position | GenEx (NormFinder) | GenEx (geNorm) | NormFinder | BestKeeper | geNorm | qBasePlus | Consensus |
|---|---|---|---|---|---|---|---|
| 1 | ACAC | ALAS | ACAC | ACAC | HMBS | HSP-90 | ACAC |
| 2 | HSP-90 | GAPDH | HSP-90 | B2M | GAPDH | ACAC | HSP-90 |
| 3 | B2M | ACTB | B2M | HSP-90 | ALAS | ALAS | ALAS |
| 4 | ALAS | HMBS | ALAS | 18s rRNA | HSP-90 | HMBS | B2M |
| 5 | HMBS | ACAC | HMBS | ALAS | ACAC | GAPDH | HMBS |
| 6 | 18s rRNA | HSP-90 | 18s rRNA | HMBS | 18s rRNA | 18s rRNA | 18s rRNA |
| 7 | GAPDH | B2M | GAPDH | GAPDH | B2M | B2M | GAPDH |
| 8 | ACTB | 18s rRNA | ACTB | ACTB | ACTB | ACTB | ACTB |
| 9 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 | RPS18 |
We used rank aggregation method to combine the ordered lists of reference genes provided by different programs to a consensus rank. Results of rank aggregation using RankAggreg package offer that the most appropriate candidates as reference genes are HSP-90 for liver and visceral fat, ALAS for subcutaneous fat and ACAC for longissimus muscle (Tables 4-7). Generally, HSP-90 seems to be the most suitable candidate as reference gene across all studied tissues as it was nearly always among the two most stable genes (Tables 4-7). The suitability of HSP-90 as reference gene in expression studies has been previously confirmed in some other organisms [51]. Compared to the other reference genes examined, the three candidate genes GAPDH, ACTB, and RPS18 showed the least expression stability across all the studied tissues and are not appropriate enough for use as reference gene in gene expression analysis studies (Tables 4-7). Our results are in contrast to many other studies in which, GAPDH and ACTB have been reported to show high expression stability in different organisms (for examples see 52-54).
To our knowledge, this is the first study which investigates different candidate reference genes for gene expression analyses in four different tissues of the domestic goat (C. hicus) and seems to be useful in guiding researchers performing gene expression analyses in different breeds of this animal. Although, our results did not clarify any reference gene with constant expression level across all studied tissues, they highlighted HSP-90 as the most stably expressed gene that can be used for normalization of expression data in different tissues of the goat.
Supporting Information
Threshold Cycle (Ct) of housekeeping genes in four tissues and original data of ranking (HKGs) via six softwares.
(XLS)
Acknowledgments
The authors would like to appreciate Mr. H. Mousapour and Dr. S. A. Salami for their great helps with this work and their technical assistance.
Funding Statement
This study was conducted at Research Station of Department of Animal Science, College of Agriculture and Natural Resources, University of Tehran, Iran. All authors have equal contributions to study design, data collection and analysis, and writing the manuscript. No external funding sources except exists for this study. Someone else had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dong Y, Xie M, Jiang Y, Xiao N, Du X et al. (2013) Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat Biotechnol, 31: 135–41. PubMed: 23263233. [DOI] [PubMed] [Google Scholar]
- 2. Fulton LK, Clarke MS, Farris HE (1994) The goat as a model for biomedical research and teaching. Ilar Journal 36: 21-29. doi: 10.1093/ilar.36.2.21. [DOI] [Google Scholar]
- 3. Everaert BR, Boulet GA, Timmermans J-P, Vrints CJ (2011) Importance of suitable reference gene selection for quantitative real-time PCR: special reference to mouse myocardial infarction studies. PLOS ONE 6: e23793. doi: 10.1371/journal.pone.0023793. PubMed: 21858224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan C, Ma J, Guo Q, Li X, Wang H et al. (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLOS ONE 8: e56573. doi: 10.1371/journal.pone.0056573. PubMed: 23437174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang X, Ding L, Sandford AJ (2005) Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol 6: 4. doi: 10.1186/1471-2199-6-4. PubMed: 15720708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han X, Lu M, Chen Y, Zhan Z, Cui Q et al. (2012) Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLOS ONE 7: e43084. doi: 10.1371/journal.pone.0043084. PubMed: 22912794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Teste M-A, Duquenne M, François JM, Parrou J-L (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10: 99. doi: 10.1186/1471-2199-10-99. PubMed: 19874630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29: 23-39. doi: 10.1677/jme.0.0290023. PubMed: 12200227. [DOI] [PubMed] [Google Scholar]
- 9. Kadegowda AK, Bionaz M, Thering B, Piperova LS, Erdman RA et al. (2009) Identification of internal control genes for quantitative polymerase chain reaction in mammary tissue of lactating cows receiving lipid supplements. J Dairy Sci 92: 2007-2019. doi: 10.3168/jds.2008-1655. PubMed: 19389958. [DOI] [PubMed] [Google Scholar]
- 10. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245-5250. doi: 10.1158/0008-5472.CAN-04-0496. PubMed: 15289330. [DOI] [PubMed] [Google Scholar]
- 11. Bémeur C, Ste-Marie L, Desjardins P, Hazell AS, Vachon L et al. (2004) Decreased β-actin mRNA expression in hyperglycemic focal cerebral ischemia in the rat. Neurosci Lett 357: 211-214. doi: 10.1016/j.neulet.2003.12.081. PubMed: 15003287. [DOI] [PubMed] [Google Scholar]
- 12. Lee JH, Fitzgerald JB, DiMicco MA, Grodzinsky AJ (2005) Mechanical injury of cartilage explants causes specific time-dependent changes in chondrocyte gene expression. Arthritis and Rheumatism 52: 2386-2395. doi: 10.1002/art.21215. [DOI] [PubMed] [Google Scholar]
- 13. Ohl F, Jung M, Xu C, Stephan C, Rabien A et al. (2005) Gene expression studies in prostate cancer tissue: which reference gene should be selected for normalization? J Mol Med (Berl) 83: 1014-1024. doi: 10.1007/s00109-005-0703-z. PubMed: 16211407. [DOI] [PubMed] [Google Scholar]
- 14. Gabrielsson BG, Olofsson LE, Sjögren A, Jernås M, Elander A et al. (2005) Evaluation of reference genes for studies of gene expression in human adipose tissue. Obes Res 13: 649-652. doi: 10.1038/oby.2005.72. PubMed: 15897472. [DOI] [PubMed] [Google Scholar]
- 15. Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J et al. (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Molecular Breeding 23: 607-616. doi: 10.1007/s11032-009-9259-x. [DOI] [Google Scholar]
- 16. Lardizábal MN, Nocito AL, Daniele SM, Ornella LA, Palatnik JF et al. (2012) Reference genes for real-time PCR quantification of microRNAs and messenger RNAs in rat models of hepatotoxicity. PLOS ONE 7: e36323. doi: 10.1371/journal.pone.0036323. PubMed: 22563491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10: 4. doi: 10.1186/1471-2229-10-4. PubMed: 20056000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beekman L, Tohver T, Dardari R, Léguillette R (2011) Evaluation of suitable reference genes for gene expression studies in bronchoalveolar lavage cells from horses with inflammatory airway disease. BMC Mol Biol 12: 5. doi: 10.1186/1471-2199-12-5. PubMed: 21272375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim I, Yang D, Tang X, Carroll JL (2011) Reference gene validation for qPCR in rat carotid body during postnatal development. BMC Res Notes 4: 440. doi: 10.1186/1756-0500-4-440. PubMed: 22023793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klie M, Debener T (2011) Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res Notes 4: 518. doi: 10.1186/1756-0500-4-518. PubMed: 22123042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anderson RA (1994) Stress effects on chromium nutrition of humans and farm animals. Nothingham University Press, Nothingam, UK: pp. 267-274. [Google Scholar]
- 22. Emami A, Zali A, Ganjkhanlou M, AFJANI AA (2012) Nutrient digestibility, carcass characteristics and plasma metabolites in kids fed diets supplemented with chromium methionine. Online. Journal Animal and Feed Research (OJAFR) 2: 127-132. [Google Scholar]
- 23. Haldar S, Mondal S, Samanta S and Ghosh TK (2009) Effects of dietary chromium supplementation on glucose tolerance and primary antibody response against peste des petits ruminants in dwarf Bengal goats ( Capra hircus). Animal : an international journal of animal bioscience 3: 209-217 [DOI] [PubMed]
- 24. El-Monem UA and El-Hamid AA (2008) Effect of chromium picolinate supplementation on growth performance, carcass traits, biochemical parameters and blood constituents of growing lambs under the summer Egyptian conditions. 2nd International Scientific Conference on Small Ruminant Production, Cairo, Egypt, 14-16 February 2008. Egyptian Association for Sheep and Goat; pp. 95-104. [Google Scholar]
- 25. Pechova A, Podhorský A, Lokajova E, Pavlata L, Illek J (2002) Metabolic effects of chromium supplementation in dairy cows in the peripartal period. Acta Veterinaria Brno 71: 9-None. doi: 10.2754/avb200271010009. [DOI] [Google Scholar]
- 26. Maples NL, Bain LJ (2004) Trivalent chromium alters gene expression in the mummichog (Fundulus heteroclitus). Environmental Toxicology and Chemistry 23: 626-631. doi: 10.1897/03-130. PubMed: 15285355. [DOI] [PubMed] [Google Scholar]
- 27. Permenter MG, Lewis JA, Jackson DA (2011) Exposure to nickel, chromium, or cadmium causes distinct changes in the gene expression patterns of a rat liver derived cell line. PLOS ONE 6: e27730. doi: 10.1371/journal.pone.0027730. PubMed: 22110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sadeghi M, Najafpanah MJ (2013) Analysis of Immune-Relevant Genes Expressed in Spleen of Capra hircus Kids Fed with Trivalent Chromium. Biological Trace Element Research: 1-6. PubMed: 24078326. [DOI] [PubMed] [Google Scholar]
- 29. Chomczynski P, Sacchi N (2006) The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat Protoc 1: 581-585. doi: 10.1038/nprot.2006.83. PubMed: 17406285. [DOI] [PubMed] [Google Scholar]
- 30. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R et al. (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35: W71-W74. doi: 10.1093/nar/gkl806. PubMed: 17485472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35: W43-W46. doi: 10.1093/nar/gkm234. PubMed: 17452344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S et al. (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. doi: 10.1186/1471-2105-13-134. PubMed: 22708584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45-e45. doi: 10.1093/nar/29.9.e45. PubMed: 11328886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Radonić A, Thulke S, Mackay IM, Landt O, Siegert W et al. (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313: 856-862. doi: 10.1016/j.bbrc.2003.11.177. PubMed: 14706621. [DOI] [PubMed] [Google Scholar]
- 35. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509-515. doi: 10.1023/B:BILE.0000019559.84305.47. PubMed: 15127793. [DOI] [PubMed] [Google Scholar]
- 36. Pihur V, Datta S, Datta S (2009) RankAggreg, an R package for weighted rank aggregation. BMC Bioinformatics 10: 62. doi: 10.1186/1471-2105-10-S1-S62. PubMed: 19228411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonefeld BE, Elfving B, Wegener G (2008) Reference genes for normalization: a study of rat brain tissue. Synapse 62: 302-309. doi: 10.1002/syn.20496. PubMed: 18241047. [DOI] [PubMed] [Google Scholar]
- 38. Mehta D, Menke A, Binder EB (2010) Gene expression studies in major depression. Curr Psychiatry Rep 12: 135-144. doi: 10.1007/s11920-010-0100-3. PubMed: 20425299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ponton F, Chapuis M-P, Pernice M, Sword GA, Simpson SJ (2011) Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster . J Insect Physiol 57: 840-850. doi: 10.1016/j.jinsphys.2011.03.014. PubMed: 21435341. [DOI] [PubMed] [Google Scholar]
- 40. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. doi: 10.1186/gb-2007-8-2-r19. PubMed: 17291332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Condori J, Nopo-Olazabal C, Medrano G, Medina-Bolivar F (2011) Selection of reference genes for qPCR in hairy root cultures of peanut. BMC Res Notes 4: 392. doi: 10.1186/1756-0500-4-392. PubMed: 21985172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Uddin MJ, Cinar MU, Tesfaye D, Looft C, Tholen E et al. (2011) Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues. BMC Res Notes 4: 441. doi: 10.1186/1756-0500-4-441. PubMed: 22023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neville MJ, Collins JM, Gloyn AL, McCarthy MI, Karpe F (2011) Comprehensive Human Adipose Tissue mRNA and MicroRNA Endogenous Control Selection for Quantitative Real-Time-PCR Normalization. Obesity (Silver Spring) 19: 888-892. doi: 10.1038/oby.2010.257. PubMed: 20948521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Becker C, Riedmaier I, Reiter M, Tichopad A, Pfaffl MW et al. (2011) Changes in the miRNA profile under the influence of anabolic steroids in bovine liver. Analyst 136: 1204-1209. doi: 10.1039/c0an00703j. PubMed: 21212882. [DOI] [PubMed] [Google Scholar]
- 45. Swijsen A, Nelissen K, Janssen D, Rigo J-M, Hoogland G (2012) Validation of reference genes for quantitative real-time PCR studies in the dentate gyrus after experimental febrile seizures. BMC Res Notes 5: 685. doi: 10.1186/1756-0500-5-685. PubMed: 23237195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zeder MA, Hesse B (2000) The initial domestication of goats (Capra hircus) in the Zagros Mountains 10,000 years ago. Science 287: 2254-2257. doi: 10.1126/science.287.5461.2254. PubMed: 10731145. [DOI] [PubMed] [Google Scholar]
- 47. Cankorur-Cetinkaya A, Dereli E, Eraslan S, Karabekmez E, Dikicioglu D et al. (2012) A novel strategy for selection and validation of reference genes in dynamic multidimensional experimental design in yeast. PLOS ONE 7: e38351. doi: 10.1371/journal.pone.0038351. PubMed: 22675547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lallemant B, Evrard A, Combescure C, Chapuis H, Chambon G et al. (2009) Reference gene selection for head and neck squamous cell carcinoma gene expression studies. BMC Mol Biol 10: 78. doi: 10.1186/1471-2199-10-78. PubMed: 19650912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kosir R, Acimovic J, Golicnik M, Perse M, Majdic G et al. (2010) Determination of reference genes for circadian studies in different tissues and mouse strains. BMC Mol Biol 11: 60. doi: 10.1186/1471-2199-11-60. PubMed: 20712867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Podevin N, Krauss A, Henry I, Swennen R, Remy S (2012) Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol Breed 30: 1237-1252. doi: 10.1007/s11032-012-9711-1. PubMed: 23024595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ovesná J, Kučera L, Vaculová K, Štrymplová K, Svobodová I et al. (2012) Validation of the beta-amy1 transcription profiling assay and selection of reference genes suited for a RT-qPCR assay in developing barley caryopsis. PLOS ONE 7: e41886. doi: 10.1371/journal.pone.0041886. PubMed: 22860024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bogaert L, Van Poucke M, De Baere C, Peelman L, Gasthuys F et al. (2006) Selection of a set of reliable reference genes for quantitative real-time PCR in normal equine skin and in equine sarcoids. BMC Biotechnol 6: 24. doi: 10.1186/1472-6750-6-24. PubMed: 16643647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seol D, Choe H, Zheng H, Jang K, Ramakrishnan PS et al. (2011) Selection of reference genes for normalization of quantitative real-time PCR in organ culture of the rat and rabbit intervertebral disc. BMC Res Notes 4: 162. doi: 10.1186/1756-0500-4-162. PubMed: 21615931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7: 33. doi: 10.1186/1471-2199-7-33. PubMed: 17026756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Threshold Cycle (Ct) of housekeeping genes in four tissues and original data of ranking (HKGs) via six softwares.
(XLS)


