Abstract
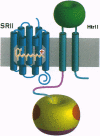
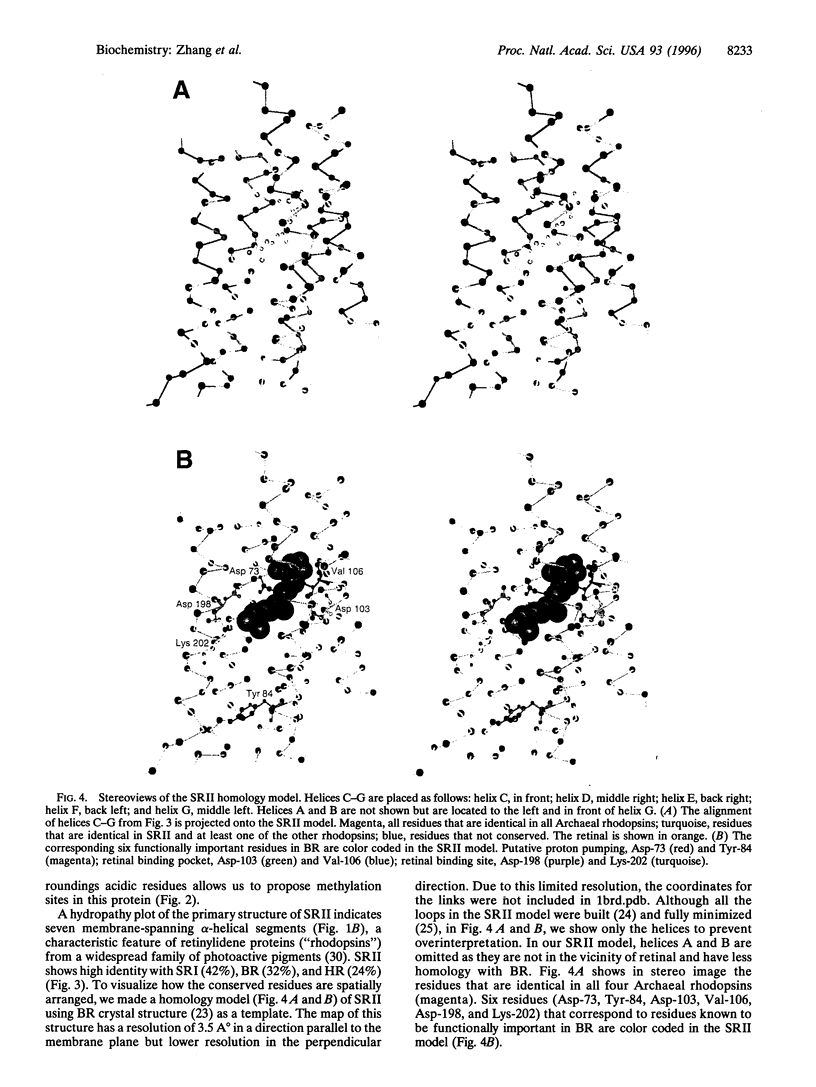
Recently, a large family of transducer proteins in the Archaeon Halobacterium salinarium was identified. On the basis of the comparison of the predicted structural domains of these transducers, three distinct subfamilies of transducers were proposed. Here we report isolation, complete gene sequences, and analysis of the encoded primary structures of transducer gene htrII, a member of family B, and its blue light receptor gene (sopII) of sensory rhodopsin II (SRII). The start codon ATG of the 714-bp sopII gene is one nucleotide beyond the termination codon TGA of the 2298-bp htrII gene. The deduced protein sequence of HtrII predicts a eubacterial chemotaxis transducer type with two hydrophobic membrane-spanning segments connecting sizable domains in the periplasm and cytoplasm. HtrII has a common feature with HtrI, the sensory rhodopsin I transducer; like HtrI, HtrII possesses a hydrophilic loop structure just after the second transmembrane segment. The C-terminal 299 residues (765 amino acid residues total) of HtrII show strong homology to the signaling and methylation domain of eubacterial transducer Tsr. The hydropathy plot of the primary structure of SRII indicates seven membrane-spanning alpha-helical segments, a characteristic feature of retinylidene proteins ("rhodopsins") from a widespread family of photoactive pigments. SRII shows high identity with SRI (42%), bacteriorhodopsin (BR) (32%), and halorhodopsin (24%). The crucial positions for retinal binding sites in these proteins are nearly identical, with the exception of Met-118 (numbering according to the mature BR sequence), which is replaced by Val in SRII. In BR, residues Asp-85 and Asp-96 are crucial in proton pumping. In SRII, the position corresponding to Asp-85 in BR is conserved, but the corresponding position of Asp-96 is replaced by an aromatic Tyr. Coexpression of the htrII and sopII genes restores SRII phototaxis to a mutant (Pho81) that contains a deletion in the htrI/sopI and insertion in htrII/sopII regions. This paper describes the first example that both HtrI and HtrII exist in the same halobacterial cell, confirming that different sensory rhodopsins SRI and SRII in the same organism have their own distinct transducers.
Full text
PDF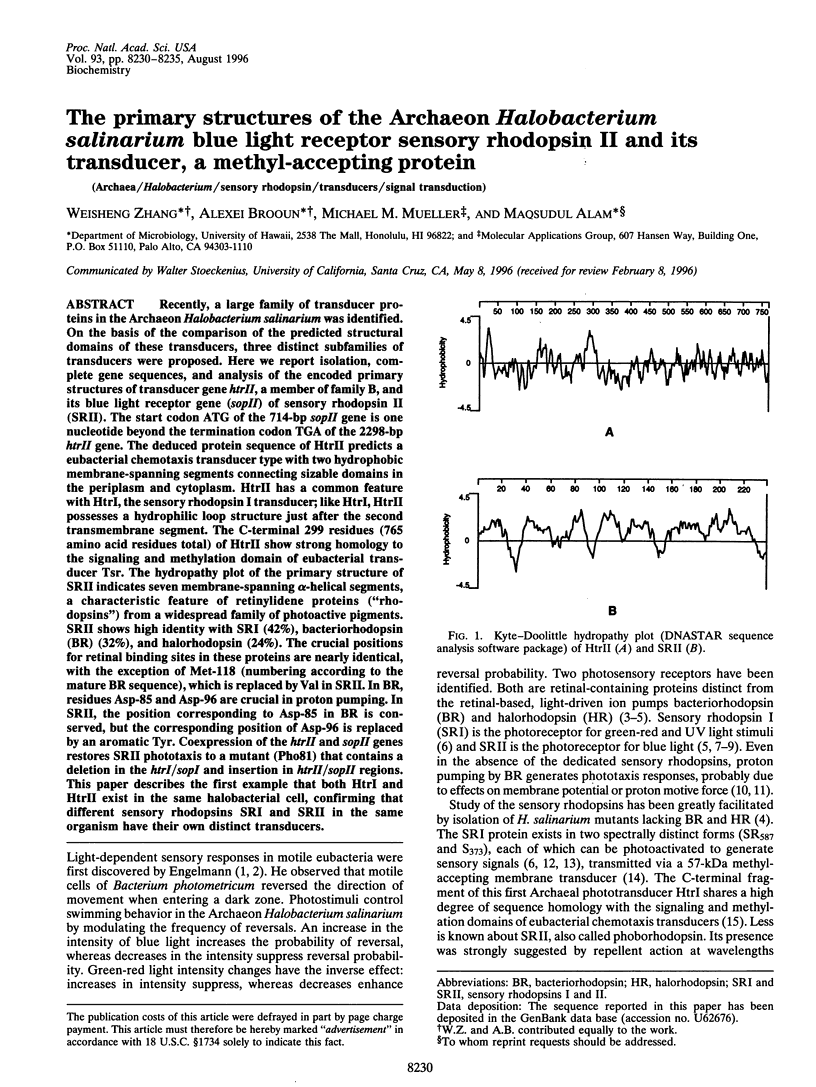




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibikov S. I., Grishanin R. N., Kaulen A. D., Marwan W., Oesterhelt D., Skulachev V. P. Bacteriorhodopsin is involved in halobacterial photoreception. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9446–9450. doi: 10.1073/pnas.90.20.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Spudich J. L. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Stoeckenius W., Szundi I., Perozo E., Olson K. D., Spudich J. L. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980 Aug;143(2):809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D., Dahlquist F. W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci U S A. 1980 May;77(5):2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S. W., Lam W. L., Charlebois R. L., Schalkwyk L. C., Doolittle W. F. Transformation methods for halophilic archaebacteria. Can J Microbiol. 1989 Jan;35(1):148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- Dencher N. A., Hildebrand E. Sensory transduction in Halobacterium halobium: retinal protein pigment controls UV-induced behavioral response. Z Naturforsch C. 1979 Sep-Oct;34(9-10):841–847. doi: 10.1515/znc-1979-9-1030. [DOI] [PubMed] [Google Scholar]
- Engström P., Hazelbauer G. L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980 May;20(1):165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E., Krah M., Marwan W., Oesterhelt D. The methyl-accepting transducer protein HtrI is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. EMBO J. 1993 Aug;12(8):2999–3005. doi: 10.1002/j.1460-2075.1993.tb05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh D. A., Farrens D. L., Subramaniam S., Khorana H. G. Hydrophobic amino acids in the retinal-binding pocket of bacteriorhodopsin. J Biol Chem. 1993 Sep 25;268(27):20305–20311. [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Hildebrand E., Dencher N. Two photosystems controlling behavioural responses of Halobacterium halobium. Nature. 1975 Sep 4;257(5521):46–48. doi: 10.1038/257046a0. [DOI] [PubMed] [Google Scholar]
- Jung K. H., Spudich J. L. Protonatable residues at the cytoplasmic end of transmembrane helix-2 in the signal transducer HtrI control photochemistry and function of sensory rhodopsin I. Proc Natl Acad Sci U S A. 1996 Jun 25;93(13):6557–6561. doi: 10.1073/pnas.93.13.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. Bacteriorhodopsin as a model for proton pumps. Nature. 1995 Jun 8;375(6531):461–463. doi: 10.1038/375461a0. [DOI] [PubMed] [Google Scholar]
- Levitt M. Accurate modeling of protein conformation by automatic segment matching. J Mol Biol. 1992 Jul 20;226(2):507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- Levitt M. Protein folding by restrained energy minimization and molecular dynamics. J Mol Biol. 1983 Nov 5;170(3):723–764. doi: 10.1016/s0022-2836(83)80129-6. [DOI] [PubMed] [Google Scholar]
- Marwan W., Oesterhelt D. Signal formation in the halobacterial photophobic response mediated by a fourth retinal protein (P480). J Mol Biol. 1987 May 20;195(2):333–342. doi: 10.1016/0022-2836(87)90654-1. [DOI] [PubMed] [Google Scholar]
- Rath P., Spudich E., Neal D. D., Spudich J. L., Rothschild K. J. Asp76 is the Schiff base counterion and proton acceptor in the proton-translocating form of sensory rhodopsin I. Biochemistry. 1996 May 28;35(21):6690–6696. doi: 10.1021/bi9600355. [DOI] [PubMed] [Google Scholar]
- Rose I. A., Hanson K. R., Wilkinson K. D., Wimmer M. J. A suggestion for naming faces of ring compounds. Proc Natl Acad Sci U S A. 1980 May;77(5):2439–2441. doi: 10.1073/pnas.77.5.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel R., Scharf B., Gautel M., Kleine K., Oesterhelt D., Engelhard M. The primary structure of sensory rhodopsin II: a member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Sundberg S. A., Manor D., Spudich J. L. Properties of a second sensory receptor protein in Halobacterium halobium phototaxis. Proteins. 1986 Nov;1(3):239–246. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Takahashi T., Spudich J. L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984 Dec 6;312(5994):509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994 Dec 2;79(5):747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Mochizuki Y., Kamo N., Kobatake Y. Evidence that the long-lifetime photointermediate of s-rhodopsin is a receptor for negative phototaxis in Halobacterium halobium. Biochem Biophys Res Commun. 1985 Feb 28;127(1):99–105. doi: 10.1016/s0006-291x(85)80131-5. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Yan B., Mazur P., Derguini F., Nakanishi K., Spudich J. L. Color regulation in the archaebacterial phototaxis receptor phoborhodopsin (sensory rhodopsin II). Biochemistry. 1990 Sep 11;29(36):8467–8474. doi: 10.1021/bi00488a038. [DOI] [PubMed] [Google Scholar]
- Tomioka H., Takahashi T., Kamo N., Kobatake Y. Flash spectrophotometric identification of a fourth rhodopsin-like pigment in Halobacterium halobium. Biochem Biophys Res Commun. 1986 Sep 14;139(2):389–395. doi: 10.1016/s0006-291x(86)80003-1. [DOI] [PubMed] [Google Scholar]
- Wolff E. K., Bogomolni R. A., Scherrer P., Hess B., Stoeckenius W. Color discrimination in halobacteria: spectroscopic characterization of a second sensory receptor covering the blue-green region of the spectrum. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7272–7276. doi: 10.1073/pnas.83.19.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Cline S. W., Doolittle W. F., Spudich J. L. Transformation of a bop-hop-sop-I-sop-II-Halobacterium halobium mutant to bop+: effects of bacteriorhodopsin photoactivation on cellular proton fluxes and swimming behavior. Photochem Photobiol. 1992 Oct;56(4):553–561. doi: 10.1111/j.1751-1097.1992.tb02200.x. [DOI] [PubMed] [Google Scholar]
- Yao V. J., Spudich E. N., Spudich J. L. Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J Bacteriol. 1994 Nov;176(22):6931–6935. doi: 10.1128/jb.176.22.6931-6935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao V. J., Spudich J. L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Brooun A., McCandless J., Banda P., Alam M. Signal transduction in the archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci U S A. 1996 May 14;93(10):4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]