SUMMARY
The mitosis-specific phosphorylation of Histone H3 at Thr3 (H3T3ph) plays an important role in chromosome segregation by recruiting Aurora B. H3T3 phosphorylation is catalyzed by Haspin, an atypical protein kinase, whose kinase domain is intrinsically active without phosphorylation at the activation loop. We report here the molecular basis for Haspin inhibition during interphase and its reactivation in M phase. We identify a conserved basic segment that autoinhibits Haspin during interphase. This autoinhibition is neutralized when Cdk1 phosphorylates the N terminus of Haspin to recruit Polo-like kinase (Plk1/Plx1), which in turn further phosphorylates multiple sites at the Haspin N terminus. While Plx1, but not Aurora B, is critical for H3T3 phosphorylation in Xenopus egg extracts, Plk1 and Aurora B both promote this modification in human cells. Thus, M phase-specific H3T3 phosphorylation is governed by the combinatorial action of mitotic kinases that neutralizes Haspin autoinhibition through a mechanism dependent on multisite phosphorylation.
INTRODUCTION
Phosphorylation of histone H3 is recognized as a hallmark of mitosis. Histone H3 phosphorylation at Thr3 (H3T3ph) acts as a mitotic ligand for Survivin (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010), a subunit of the chromosomal passenger complex (CPC), which plays multiple essential roles during mitosis and meiosis (Carmena et al., 2012b). H3T3ph promotes CPC localization on mitotic chromatin, particularly at the centromere. Enrichment of the CPC on chromatin locally activates its kinase subunit Aurora B by promoting autophosphorylation, leading to downstream phosphorylation of a variety of substrates (De Antoni et al., 2012; Funabiki and Wynne, 2013; Kelly et al., 2010; Wang et al., 2012). While dephosphorylation of H3T3ph at the exit from M phase is required for proper chromosome decondensation and nuclear envelope formation (Kelly et al., 2010), the molecular mechanisms that limit H3T3ph to M phase remain unclear.
Mitotic H3T3 phosphorylation is catalyzed by Haspin (Dai et al., 2005), which is an atypical protein kinase in several regards. For example, in most kinases, the highly conserved DFG (Asp-Phe-Gly) motif anchors the N-terminal portion of the activation segment and coordinates the catalytic magnesium (Nolen et al., 2004), but in Haspin it is changed into DYT (Asp-Tyr-Thr). Crystal structure analysis of the Haspin kinase domain revealed that it exhibits an intrinsically active conformation in the absence of a phosphorylated activation loop, assisted by several unique insertions at its N-terminal and C-terminal lobes (Eswaran et al., 2009; Villa et al., 2009).
How can H3T3 phosphorylation be limited to M phase if the Haspin kinase domain is intrinsically active? Here, we reveal that Haspin activity is autoinhibited during interphase by a conserved basic segment adjacent to its kinase domain, and that the multisite phosphorylation of its N-terminal region by Cdk1 and Polo-like kinase (Plx1 in Xenopus egg extracts or Plk1 in human cells) in M phase neutralizes its autoinhibition.
RESULTS
Plx1 stimulates H3T3 phosphorylation
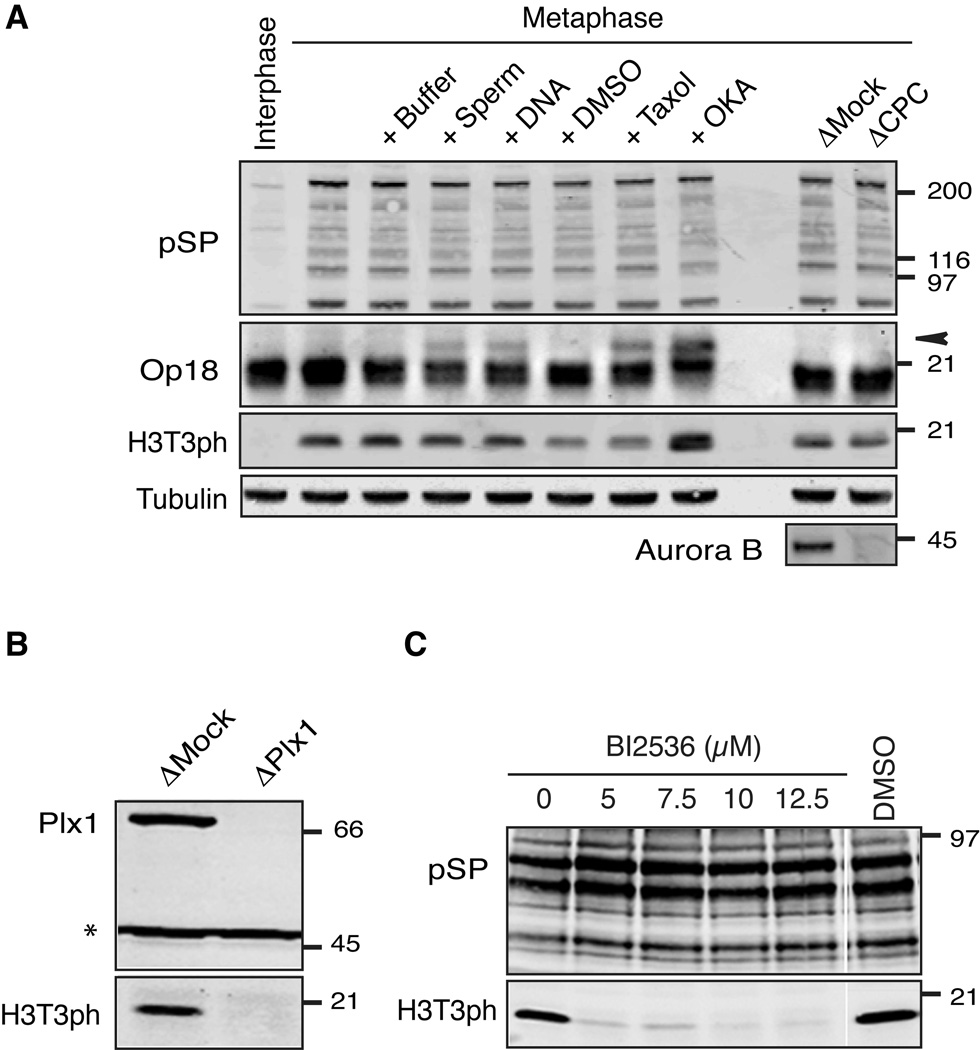
It has been reported that Aurora B-dependent phosphorylation of Haspin is important for H3T3 phosphorylation on mitotic chromosomes in human tissue culture cells (Wang et al., 2011). However, even though Aurora B activity is generally suppressed in meiotic metaphase II-arrested Xenopus egg extracts (CSF extracts) (Kelly et al., 2010; Kelly et al., 2007), histone H3 stockpiled in these extracts is highly phosphorylated at Thr3 (Figure 1A). While Aurora B activity is stimulated by addition of chromatin or taxol to extracts, resulting in Op18 hyperphosphorylation (Gadea and Ruderman, 2006; Kelly et al., 2007; Tseng et al., 2010), these treatments did not change levels of H3T3ph (Figure 1A). Lack of stimulation was not due to H3T3 phosphorylation being saturated in metaphase extracts, as adding the phosphatase inhibitor okadaic acid enhanced H3T3ph. Additionally, depletion of the CPC, including Aurora B, did not affect the level of H3T3ph (Figure 1A), suggesting that the mechanism responsible for stimulating phosphorylation of H3T3 in Xenopus egg extract is independent of Aurora B.
Figure 1. Plx1 stimulates H3T3 phosphorylation.
(A) H3T3 phosphorylation is not dependent on Aurora B in Xenopus egg extracts. Aurora B activity in metaphase Xenopus egg extracts was stimulated by adding DNA, sperm chromosomes or taxol. Maximum levels of substrate phosphorylation were revealed by okadaic acid (OKA). The CPC was depleted from metaphase extracts by anti-INCENP antibodies. Western blot analysis of total extracts is shown. The hyperphosphorylated form of Op18 is indicated by an arrowhead. Anti-phosphoSP antibody (pSP) was used to monitor M phase-specific Cdk1 substrate phosphorylation.
(B) Metaphase egg extracts were depleted with either an anti-Plx1 antibody (ΔPlx1) or a control IgG (ΔMock). Western blot analysis of extract proteins using anti-Plx1 and anti-H3T3ph antibodies is shown. Asterisk indicates a cross-reacting antigen used as a loading control.
(C) The Plx1 inhibitor BI2536 was added to metaphase egg extracts at the indicated concentrations. Western blot analysis of extract proteins using indicated antibodies is shown.
See also Figure S1.
The mobility of Xenopus Haspin (xHaspin) in polyacrylamide gels was highly reduced after incubation with metaphase egg extracts, but not with interphase extracts (Figure S1A), and this mobility shift depended on phosphorylation (Figure S1B). We found that Plx1 greatly contributes to xHaspin modification and H3T3 phosphorylation in metaphase extracts. Depleting Plx1 from Xenopus egg extracts (ΔPlx1 extracts) effectively reduced the level of H3T3ph (Figure 1B) and the mobility shift of xHaspin (Figure S1C). Similarly, the Polo inhibitor BI2536 (Steegmaier et al., 2007) reduced the level of H3T3ph (Figure 1C), indicating that xHaspin hyperphosphorylation and maintenance of H3T3ph in metaphase extracts depend on Plx1 activity. Polo-like kinases appear to support H3T3ph by an indirect mechanism, since recombinant Plk1 failed to phosphorylate H3 N terminus in vitro (Figure S1D). Taken together, these results suggest that Plx1 controls xHaspin activity via a phosphorylation-dependent mechanism.
xHaspin-Plx1 interaction is critical for H3T3 phosphorylation
Effective substrate phosphorylation by Plx1/Plk1 typically requires binding of its Polo-box-domain (PBD) to a phosphorylated PBD-binding motif (S[pS/pT]) on its substrates (Elia et al., 2003a; Elia et al., 2003b). xHaspin contains three potential PBD-binding motifs that overlap with the Cdk1 substrate consensus (S[S/T]P). While the conservation of the first motif (ST121P) is limited to Xenopus laevis and tropicalis, the second motif (ST206P) appears to be conserved in vertebrate Haspin homologs (Figure S2A). Since the third motif (SS486P) is not conserved even in Xenopus tropicalis, we did not examine it further.
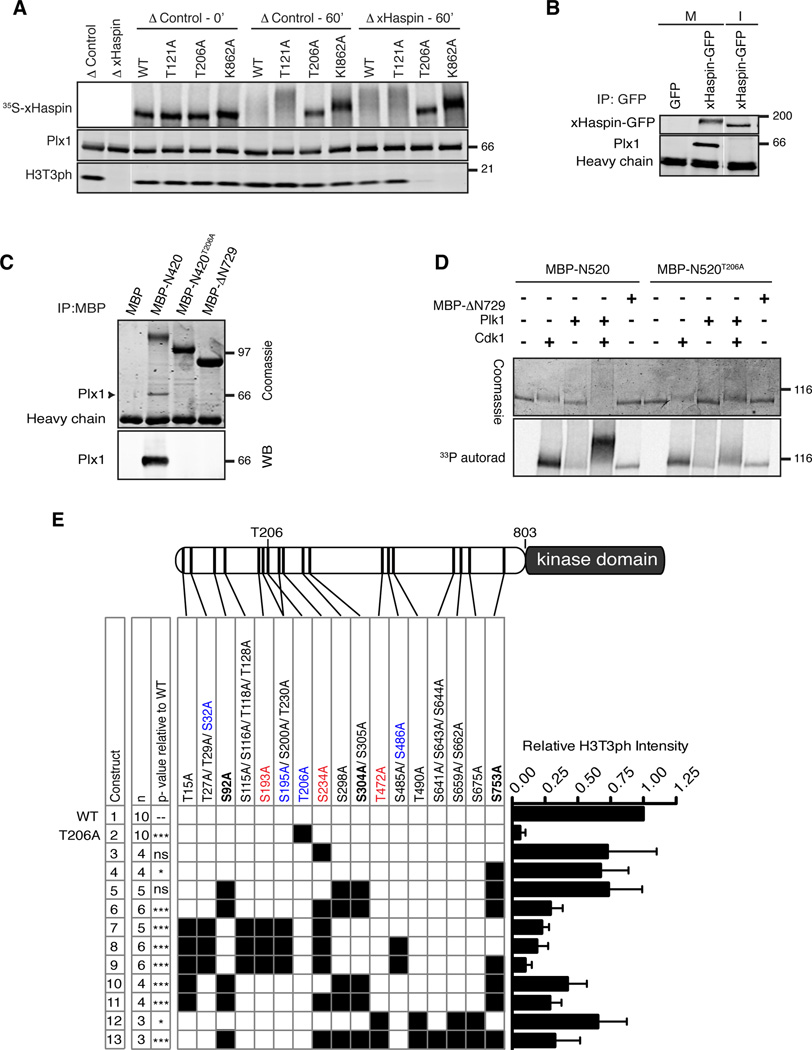
In order to test the possibility that Plx1 associates with the PBD-binding motif to directly phosphorylate and activate xHaspin, we established an experimental system where xHaspin can be immunodepleted from egg extracts, and wild-type or mutant xHaspin added back (Figure 2A). As reported previously (Kelly et al., 2010), H3T3ph activity was effectively depleted following immunodepletion with an antibody against the N terminus of xHaspin. When 35S-labeled xHaspin, translated in reticulocyte lysates, was added to xHaspin-depleted (ΔxHaspin) metaphase extracts, the exogenous xHaspin was hyperphosphorylated and H3T3ph was restored (Figure 2A). Mutating a critical lysine residue required for ATP-recognition in the xHaspin kinase domain to alanine (K862A) abolished its activity towards H3T3 and partially reduced the M phase-specific modification of xHaspin, indicating that some of M phase-specific modifications are autophosphorylation. Mutating T206, but not T121 to alanine abolished both xHaspin’s hyperphosphorylation and its capacity to rescue H3T3ph in ΔxHaspin extracts, demonstrating that T206 is critical for xHaspin activity in Xenopus egg extracts (Figures 2A and 2E).
Figure 2. H3T3 phosphorylation in metaphase extracts requires Plx1- docking and multisite phosphorylation at the Haspin N terminus.
(A) Wild-type (WT) or indicated mutants of full-length xHaspin, translated and labeled with 35S in reticulocyte lysates, were added to control or ΔxHaspin metaphase extracts. Total extracts were analyzed by autoradiography (35S-xHaspin) and Western blotting (anti-Plx1 and anti-H3T3ph).
(B) GFP or xHaspin-GFP were translated from exogenously added mRNA in metaphase egg extracts, which were then released into interphase (I) by calcium addition or maintained in metaphase (M). GFP-fused proteins were immunopurified using anti-GFP antibody beads. The bead fraction was probed with anti-GFP or anti-Plx1 antibodies.
(C) Purified MBP, MBP-xHaspin-N420 with or without a T206A mutation, or MBP-xHaspin-ΔN729 were incubated with metaphase egg extracts, followed by immunopurification using anti-MBP antibody beads. Coomassie staining (top) and a Western blot of the bead fraction using an anti-Plx1 antibody are shown.
(D) Recombinant MBP-xHaspin-N520 (with or without T206A) was subjected to in vitro kinase reactions γ-33P-ATP and a combination of purified Cdk1-cyclin B, Plk1 and MBP-xHaspin-ΔN729. Coomassie staining (top) and autoradiography, corresponding to MBP-xHaspin N520 proteins, are shown.
(E) Quantitation of H3T3ph levels in metaphase ΔxHaspin extracts supported by various xHaspin phosphorylation site mutants. Within the schematic of xHaspin, black bars point to the relative location of the residue in the N terminus. Constructs are numbered 1 through 13 on the left hand side. A black square in the matrix indicates the presence of the indicated serine/threonine-to-alanine mutation for a given construct. Phosphorylation sites matching to substrate consensus sequences for Plk1 and Cdk1 are marked in red and blue, respectively. Residues whose phosphorylation depends on T206 are bolded (see Table S1). Constructs were synthesized and labeled with 35S in reticulocyte lysates and added to metaphase ΔxHaspin for 60min before assessing the H3T3ph levels by Western blot. The graph on the right displays the mean and SEM of relative H3T3ph levels, divided by the input amount of 35S-xHaspin proteins and standardized to the wild-type. “n” represents number of independent experiments. P-values relative to xHaspin-WT are reported for all constructs and they were determined using a one-way ANOVA followed by Bonferroni post-hoc test. ***, p < 0.001; **, p < 0.01; *, p , <0.05; ns= not significant.
See also Figure S2.
Multiple lines of evidence support the idea that the M phase-specific T206 phosphorylation of xHaspin acts as a priming modification for Plx1 docking and subsequent Plx1-dependent phosphorylation of xHaspin. First, xHaspin interacts with Plx1 in metaphase extracts but not in interphase extracts (Figure 2B). This metaphase interaction with Plx1 can be recapitulated by the N-terminal 420 amino acid residues of xHaspin tagged with maltose-binding protein (MBP-xHaspin-N420) in a manner dependent on T206, but not with the kinase domain that lacks 729 N-terminal residues (MBP-xHaspin-ΔN729) (Figure 2C). Second, phosphorylation of T206 by Cdk1-Cyclin B can directly promote binding to the PBD (Figure S2B). Third, Plk1 alone can only poorly phosphorylate MBP-xHaspin-N520 (the N-terminal 520 residues tagged with MBP) in a purified system, but this reaction was greatly enhanced by Cdk1 in a T206-dependent manner (Figure 2D). Finally, M phase-specific phosphorylation of T206 in egg extracts was confirmed by a phospho-specific antibody recognizing the phosphorylated T206 residue (Figure S2C) and by mass spectrometry (MS) (Figure S2D and Table S1). Altogether, these data suggest that Cdk1-dependent phosphorylation of xHaspin at T206 directly recruits Plx1, which phosphorylates and activates xHaspin.
Phosphorylation at multiple sites in the N terminus of xHaspin is required for its mitotic activation
How does the Plx1 activate xHaspin? Although the activation loop of Haspin harbors an evolutionarily conserved potential Polo-substrate motif (Ser1054 in xHaspin), mutation of this serine to alanine did not alter xHaspin’s capacity to phosphorylate H3T3ph in Xenopus egg extracts (Figures S3A–B), consistent with a mechanism of Haspin activation independent of phosphorylation at its activation loop (Eswaran et al., 2009; Villa et al., 2009).
To identify the phosphorylation sites that are responsible for Haspin activation, we employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) on full-length or parts of xHaspin purified from metaphase egg extracts, and on recombinant N-terminal MBP-xHaspin-N520 incubated with purified Cdk1-Cyclin B and Plk1. We found that xHaspin is phosphorylated at more than 30 sites, the majority of which are located in the N terminus (Figure 2E and Table S1). Comparing the phosphorylation profiles of wild-type xHaspin, xHaspin T206A and the kinase dead mutant xHaspinK862A, we identified at least two residues (S304, Y307) whose phosphorylation seems to depend on T206, and two residues (S92 and S753) that require both T206 and xHaspin kinase activity. Given that S92 is phosphorylated in vitro by Cdk1/Plk1 on xHaspin-N520 (which lacks the kinase domain), these residues may be phosphorylated by both Cdk1/Plx1 and xHaspin. Alternatively, Plx1-dependent activation of Haspin may stimulate autophosphorylation at these residues, or conversely Haspin autophosphorylation may facilitate Plx1-dependent phosphorylation at these sites.
We next asked which phosphorylation sites are important for xHaspin activation in egg extract. Alanine mutations at five phosphorylation sites (S92, S298, S304, S305, S753) including those whose phosphorylation depended on T206 in metaphase extracts, did not significantly affect its capacity to complement H3T3 phosphorylation in metaphase ΔxHaspin egg extracts (construct #5, Figures 2E, S2E and S2F). This is likely due to contribution of T206-dependent phosphorylation at other sites that were not identified by the MS analysis. Indeed, adding another alanine mutation at S234, which fits the canonical Polo substrate consensus (Alexander et al., 2011; Santamaria et al., 2011) and was phosphorylated on xHaspin-N520 in vitro by Cdk1/Plk1, showed significant impact on H3T3ph level and the xHaspin mobility shift (#6), while S234A mutation alone did not (#3). Interestingly, mutating 13 N-terminal phosphorylation sites (#7), including S234, but not the rest of residues mutated in construct #6, also failed to fully support H3T3 phosphorylation. Among those 13 residues mutated in construct #7, T15A reduced the H3T3ph level when added to construct #5 (#10), indicating that T15 may not be essential but can contribute to xHaspin activity. In addition, mutating five residues (T472, T490, S659, S662, S675), which appear to be phosphorylated in a manner dependent on xHaspin kinase activity (Table S1) and were mutated in neither construct #7 nor construct #6, mildly but significantly reduced H3T3ph level (#12). Altogether these data demonstrate that phosphorylation at multiple residues in its N terminus collaborates to activate xHaspin.
xHaspin is inhibited by a basic segment adjacent to its kinase domain
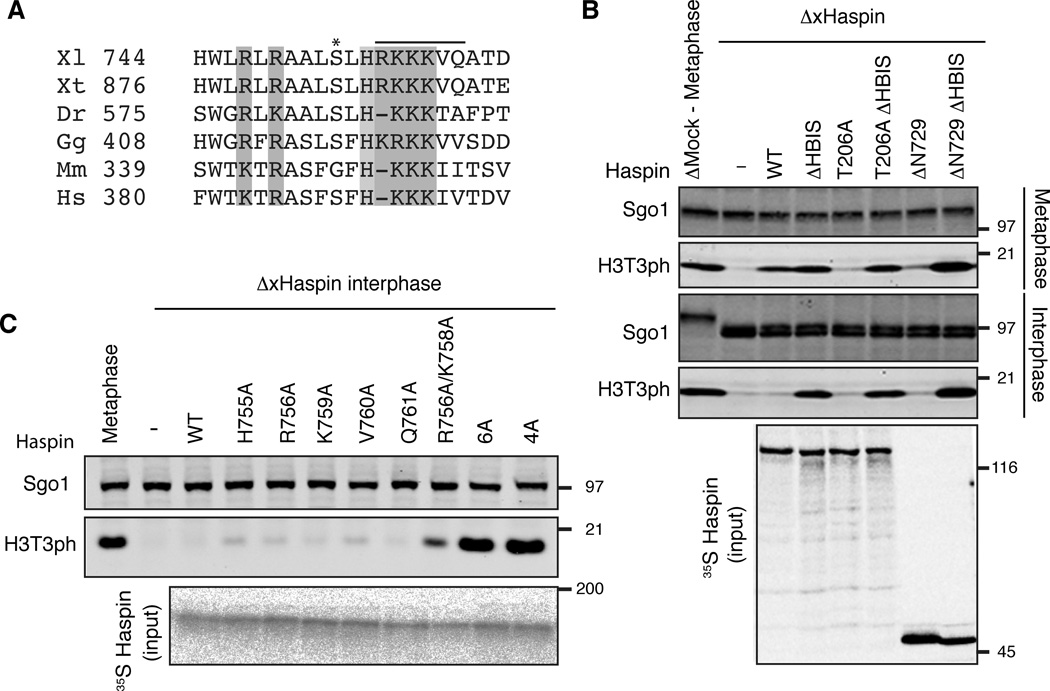
The requirement of N-terminal phosphorylation for Haspin activation raises the possibility that the Plx1-dependent phosphorylation neutralizes the action of a hypothetical autoinhibitory module within the xHaspin N terminus. If this were the case, removal of such an inhibitory module should stimulate xHaspin’s kinase activity in a Plx1-independent manner. However, deleting as much as the first 729 amino acid residues of xHaspin (xHaspin-ΔN729) abolished, rather than activated, the H3T3 kinase activity of xHaspin in Xenopus egg extracts (Figure S3B–D), indicating that this construct still harbors the inhibitory module. Within the remaining N-terminal domain of xHaspin-ΔN729, we found an evolutionarily conserved segment, enriched with basic residues (Figures 3A and S3B). Based on results described below, we named this region the Haspin Basic Inhibitory Segment or HBIS.
Figure 3. The HBIS inhibits xHaspin activity in interphase Xenopus egg extracts.
(A) Multisequence alignment showing conservation of the HBIS. A line indicates the amino acids that are deleted in ΔHBIS. An asterisk indicates S753, which is phosphorylated in metaphase egg extracts. Xl, Xenopus laevis; Xt, Xenopus tropicalis; Dr, Danio rerio; Gg, Gallus gallus, Mm, Mus musculus; Hs, Homo sapiens.
(B) Full-length xHaspin (WT), xHaspin-ΔHBIS, xHaspinT206A, xHaspinT206A ΔHBIS mutant protein or N-terminally truncated xHaspin (ΔN729) with or without an HBIS deletion (ΔHBIS) were labeled with 35S in reticulocyte lysates and then incubated with metaphase (top) or interphase (middle) ΔxHaspin extracts. Western blot analyses (top and middle) using anti-H3T3ph and anti-Sgo1 (as a loading control) antibodies, and an autoradiography of the input samples are shown.
(C) Full-length xHaspin proteins with indicated point mutation(s) within the HBIS were translated and 35S-labeled in reticulocyte lysates before incubation with interphase ΔxHaspin extracts. Multiple point mutations are introduced in 6A mutant (R756A, K757A, K758A, K759A, V760A, Q761A) and 4A mutant (R756A, K757A, K758A, K759A).
See also Figure S3.
To test if the HBIS negatively regulates Haspin activity, we deleted the RKKKVQ sequence in the HBIS from xHaspin (ΔHBIS) and assessed its impact on H3T3 kinase activity in metaphase and interphase egg extracts. Deletion of the HBIS from xHaspin-ΔN729 not only restored its capacity to phosphorylate H3T3 in metaphase ΔxHaspin extracts, but also promoted H3T3 phosphorylation in interphase extracts (Figure 3B). Deleting the HBIS also bypassed the requirement for T206 in H3T3 phosphorylation (Figure 3B). Point mutation analysis further confirmed the importance of multiple basic residues within the HBIS for its inhibitory action (Figure 3C). These results indicate that the HBIS is critical to inactivate Haspin during interphase.
The HBIS directly autoinhibits xHaspin
To understand the mechanism behind HBIS-mediated Haspin inhibition, we initially explored the possibility that the HBIS recruits an inhibitory factor in egg extracts. The sequence similarity between the HBIS and the basic nuclear localization signal (NLS) (Lange et al., 2007) prompted us to test if nuclear import factors contribute to this inhibition. Intriguingly, Importin β (also known as karyopherin β) interacted with xHaspin specifically in interphase and dissociated from it during M phase (Figure S4A) in a Plx1-dependent manner (Figure S4B). However, forced dissociation of Importin β from xHaspin by adding excess amounts of the Importin β binding (IBB) domain of Importin α (Weis et al., 1996) did not promote H3T3 phosphorylation in interphase extracts (Figure S4C), indicating that Importin β does not act as an inhibitor of xHaspin.
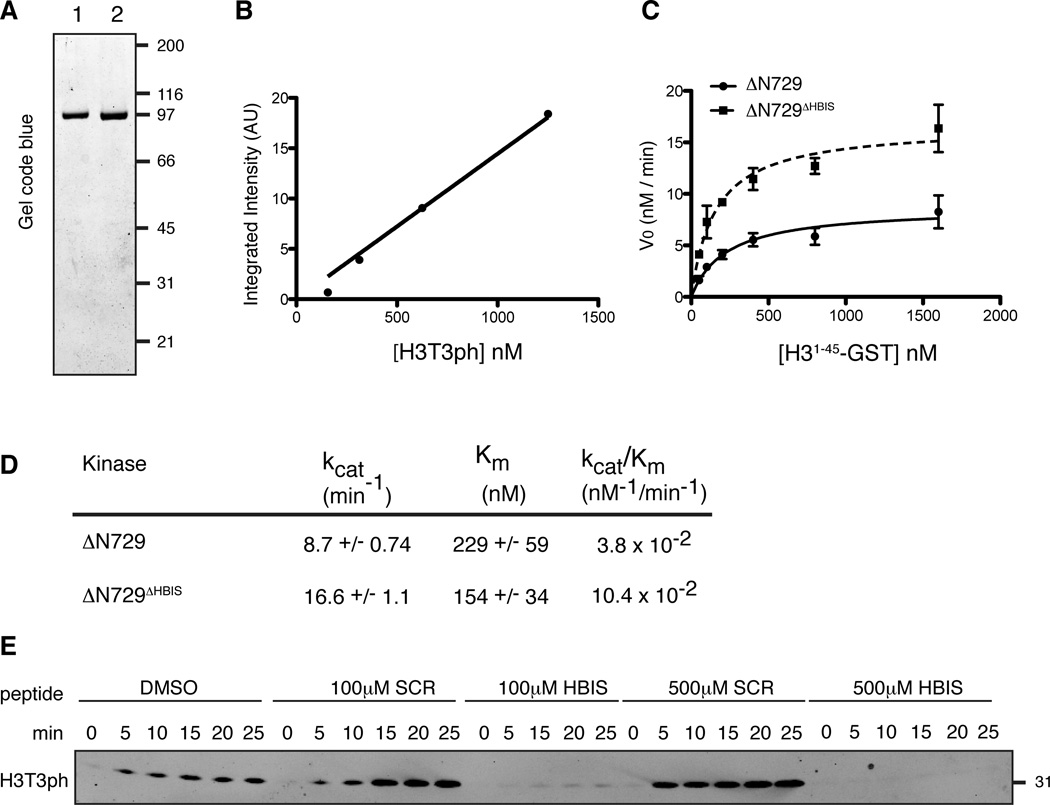
We next tested whether the HBIS directly inhibits Haspin activity. The monomeric fractions of bacterially produced MBP-xHaspin-ΔN729 and MBP-xHaspin-ΔN729ΔHBIS were purified using affinity chromatography and gel filtration chromatography (Figure 4A). We set up a Western blot-based in vitro kinase assay to compare the kinase activity of the two enzymes and extract quantitative kinetic information. The absolute levels of generated H3T3ph were quantified by Western blot under conditions where anti-H3T3ph signal intensity shows a linear relationship with the amount of H3T3ph (Figure 4B). We measured initial velocities (V0) of phosphorylation by MBP-xHaspin-ΔN729 and MBP-xHaspin-ΔN729ΔHBIS as a function of substrate concentration (H31–45-GST) in the presence of saturating amounts of ATP (750 µM). While removal of the HBIS had only a modest effect on the Km, the Vmax increased by two-fold (Figures 4C and D), consistent with a noncompetitive mode of inhibition rather than competitive inhibition. MBP-xHaspin-ΔN729ΔHBIS exhibited a two-fold increase in kcat and a three-fold increase in its catalytic efficiency (kcat/Km) compared to xHaspin-ΔN729 (Figure 4D).
Figure 4. The HBIS directly inhibits xHaspin.
(A) Gel code blue staining of purified MBP-xHaspin-ΔN729 (1) and MBP-xHaspin-ΔN729ΔHBIS (2).
(B) Signal intensity of Western blots using the anti-H3T3ph antibody (A.U.) was plotted against the amount of H3T3ph (loaded). The signal intensity showed a linear relationship to the input H3T3ph.
(C) Initial velocities of H3T3 phosphorylation reaction using 1 nM MBP-xHaspin-ΔN729 or MBP-xHaspin-ΔN729ΔHBIS at various H31–45-GST concentration are shown. Triplicate data sets were fit to the Michaelis-Menten equation using the nonlinear least square fit method. Error bars represent standard error of the mean.
(D) Km and kcat values were obtained after fitting the data in C to the Michaelis-Menten equation.
(E) Either HBIS- or SCR-peptides were added at the indicated concentration to the in vitro kinase reaction with 1 nM xHaspin-ΔN729 and 200 nM H31–45-GST. Substrate phosphorylation was detected by Western blots using anti-H3T3ph antibody.
See also Figure S4.
Furthermore, adding a synthetic 21 amino acid peptide containing the HBIS to the kinase reaction resulted in strong inhibition of H3T3 phosphorylation (Figure 4E). Since a control peptide with an identical amino acid composition, but scrambled sequence (SCR peptide), did not exert such an inhibitory effect, we concluded that the HBIS-peptide directly inhibits xHaspin in a sequence-specific manner. Altogether, these data demonstrate that the HBIS directly inhibits xHaspin kinase activity.
Phosphorylation of the xHaspin N terminus controls binding to the HBIS
How can Polo-dependent phosphorylation neutralize the inhibitory action of the HBIS? Our MS-analysis of full-length xHaspin proteins revealed that S753 within the HBIS is phosphorylated in mitosis in a T206-dependent manner (Figure 3A and Table S1). However, the S753A mutation, by itself or in combination with other phosphorylation site mutations, showed only a mild effect on the xHaspin’s capacity to complement H3T3ph in ΔxHaspin extracts (Figures 2E, S2E and S2F), indicating that the phosphorylation within the HBIS may contribute but is not a sole mechanism to neutralize the inhibitory action of HBIS.
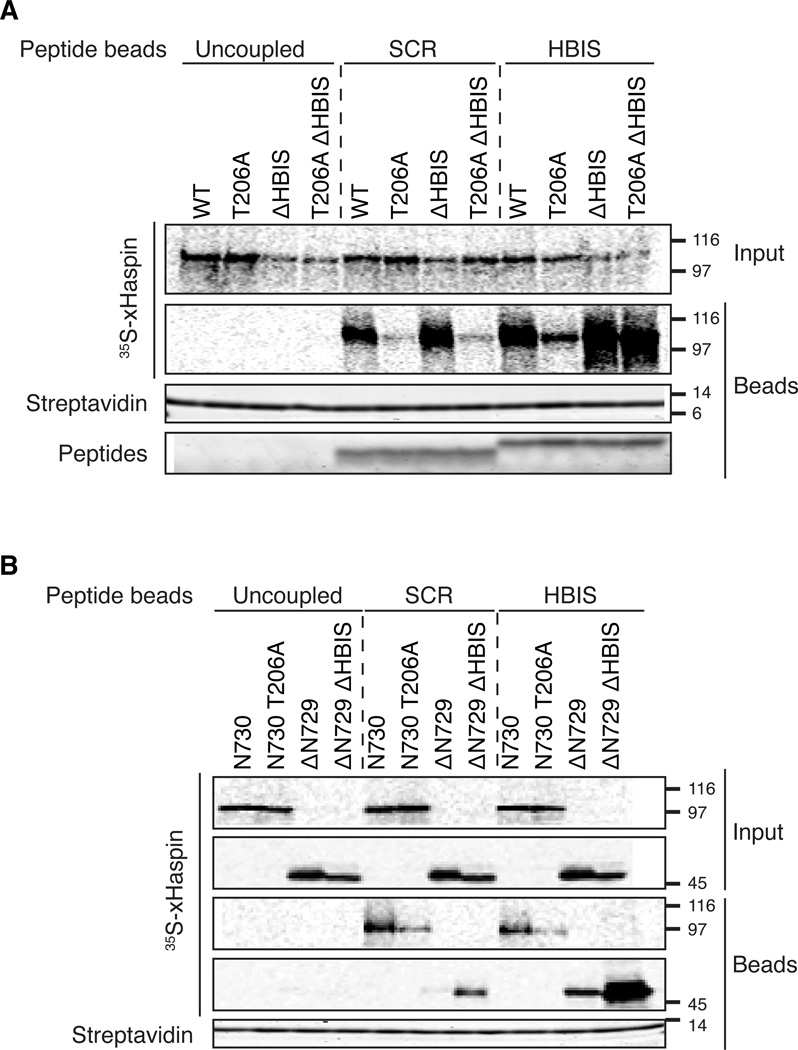
We hypothesized that in M phase, negative charges imparted by phosphorylation of the xHaspin N terminus would interact with basic residues in the HBIS, displacing it to relieve inhibition of the kinase. If this were the case, the HBIS would be expected to bind the xHaspin kinase domain in a sequence-dependent manner, while its interaction with the N terminus would hinge on the presence of phosphorylations. To test this hypothesis, we first examined the modes of interaction between the HBIS-peptide and full-length xHaspin. 35S-labeled full-length xHaspin wild-type, T206A, ΔHBIS, or the T206A ΔHBIS double mutant, were incubated with metaphase extracts containing magnetic beads coated with either HBIS- or SCR-peptides. While wild-type xHaspin copurified with both peptides (with some preference for the HBIS-peptide), the T206A mutant, which prevents phosphorylation of the xHaspin N terminus, bound poorly to both peptides. This suggests that the interaction of full-length xHaspin with the HBIS is largely dependent on T206-mediated phosphorylation and does not require rigorous sequence specificity. Similar to wild-type xHaspin, xHaspin-ΔHBIS binds both SCR-peptides and HBIS-peptides. However, the T206A-ΔHBIS double mutant showed robust binding to HBIS- but not SCR-peptides, suggesting that deleting the HBIS in a T206A mutant liberates a sequence-specific HBIS-peptide binding site (Figure 5A). These data are consistent with a model in which xHaspin can bind the HBIS in two modes: 1) phospho-dependent binding, and 2) HBIS sequence-dependent, phospho-independent binding.
Figure 5. xHaspin binds the HBIS through two distinct mechanisms.
(A) 35S-labeled full length xHaspin wild-type (WT), a T206A mutant (T206A), a mutant lacking the HBIS (ΔHBIS) or a construct with both mutations (T206A ΔHBIS) were pre-incubated in metaphase egg extract. This extract was incubated for 60 min with uncoupled control beads, or beads coated in the HBIS- or SCR-peptide. Input and bead fractions (treated with phosphatase to reduce the mobility shifts) were analyzed by autoradiography.
(B) 35S-labeled xHaspin N terminus (N730), the N terminus with a T206A mutation (N730T206A), xHaspin kinase domain (ΔN729) or a kinase domain mutant without the HBIS (ΔN729ΔHBIS) were processed as in A.
See also Figure S5.
To examine whether these two modes of binding are supported by distinct domains of xHaspin, the N-terminal 730 amino acids of xHaspin with or without a T206A mutation (xHaspin-N730 and xHaspin-N730T206A), and the C-terminal kinase domain with or without the HBIS deletion (xHaspin-ΔN729 and xHaspin-ΔN729ΔHBIS) were subjected to a peptide-binding assay in egg extracts as above. The xHaspin kinase domain (xHaspin-ΔN729) showed weak binding to HBIS-peptides, but deletion of the HBIS dramatically enhanced this binding (Figure 5B), suggesting that an HBIS binding site had become available. In contrast, the xHaspin N terminus (xHaspin-N730) bound to both SCR peptides and HBIS-peptides in a T206-dependent manner. Binding of xHaspin-N730 to these peptides was also sensitive to Polo inhibition (Figure S5), further confirming that this interaction is mediated by Polo-dependent phosphorylation.
Thus, while the HBIS can bind to the kinase domain of xHaspin, it can also interact with the N-terminus of xHaspin upon Polo-dependent phosphorylation. Without Polo-dependent phosphorylation, the majority of xHaspin exists in a configuration where its HBIS segment interacts with the kinase domain, rendering it unable to interact with exogenous HBIS-peptide. These results are consistent with the model in which phosphorylation relieves the inhibitory interaction between the HBIS and the kinase domain, thus stimulating xHaspin kinase activity.
Plk1 directly stimulates Haspin activity
To test if Plk1/Plx1-dependent phosphorylation of Haspin directly stimulates the H3T3 kinase activity, GFP-tagged full-length xHaspin and the kinase inactive K862A mutant were isolated from interphase egg extracts (Figure S6A). They were then incubated with Cdk1-Cyclin B and/or Plk1 and the kinetics of H3T3 phosphorylation was measured (Figure S6B). While xHaspin purified from interphase egg extracts exhibit a substantial phosphorylation activity towards H3T3, the phosphorylation kinetics is significantly enhanced by Plk1 or Cdk1-Cyclin B + Plk1, but not Cdk1-Cyclin B alone.
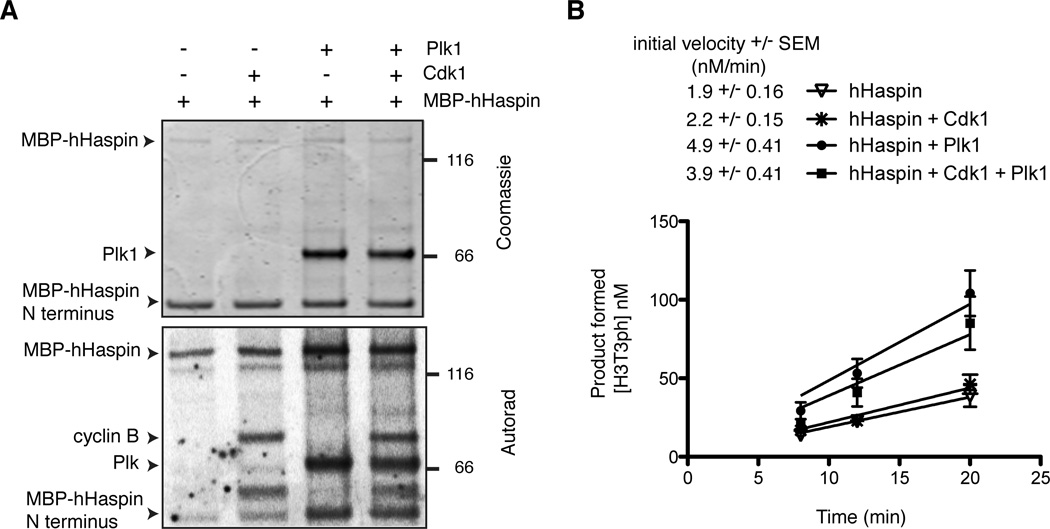
To rule out the possibility that Plk1 indirectly activated Haspin by affecting additional factor(s) that copurified with xHaspin from interphase egg extracts, we repeated the above experiment using full-length Haspin purified from bacteria. For technical reasons, MBP-fused human Haspin (hHaspin) was used instead of xHaspin. Plk1 (with or without Cdk1-Cyclin B) phosphorylated MBP-hHaspin in vitro and stimulated H3T3 phosphorylation kinetics (Fig. 6). The apparent lack of requirement for Cdk1 in this assay may be due to the large excess of Plk1 over recombinant hHaspin in the reaction or a possible contribution of hHaspin autophosphorylation. Altogether, these data demonstrate that Plk1-mediated phosphorylation can directly activate Haspin in the absence of additional factors.
Figure 6. Plk1 directly stimulates the H3T3 kinase activity of hHaspin in the absence of other contributors.
(A) A Coomassie staining and an autoradiograph of the in vitro kinase reaction involving MBP-hHaspin, Cdk1 and Plk1. Bands migrating ~45 kD are degradation products of MBP-hHaspin, containing MPB and a portion of hHaspin N terminus. The Cdk1/Cyclin B components were below the level of detection by Coomassie staining.
(B) A kinetic analysis of the H3T3 kinase activity using the pre-phosphorylated MBP-hHaspin (with or without Plk1/Cdk1) shown in A. The production of H3T3ph was determined by quantitative Western blots using H31–45-GST as a substrate. The initial velocities and the reaction curves of the various conditions are shown. Bars represent SEM of 3 independent experiments.
See also Figure S6.
Plk1 and Aurora B both support H3T3 phosphorylation in human cells
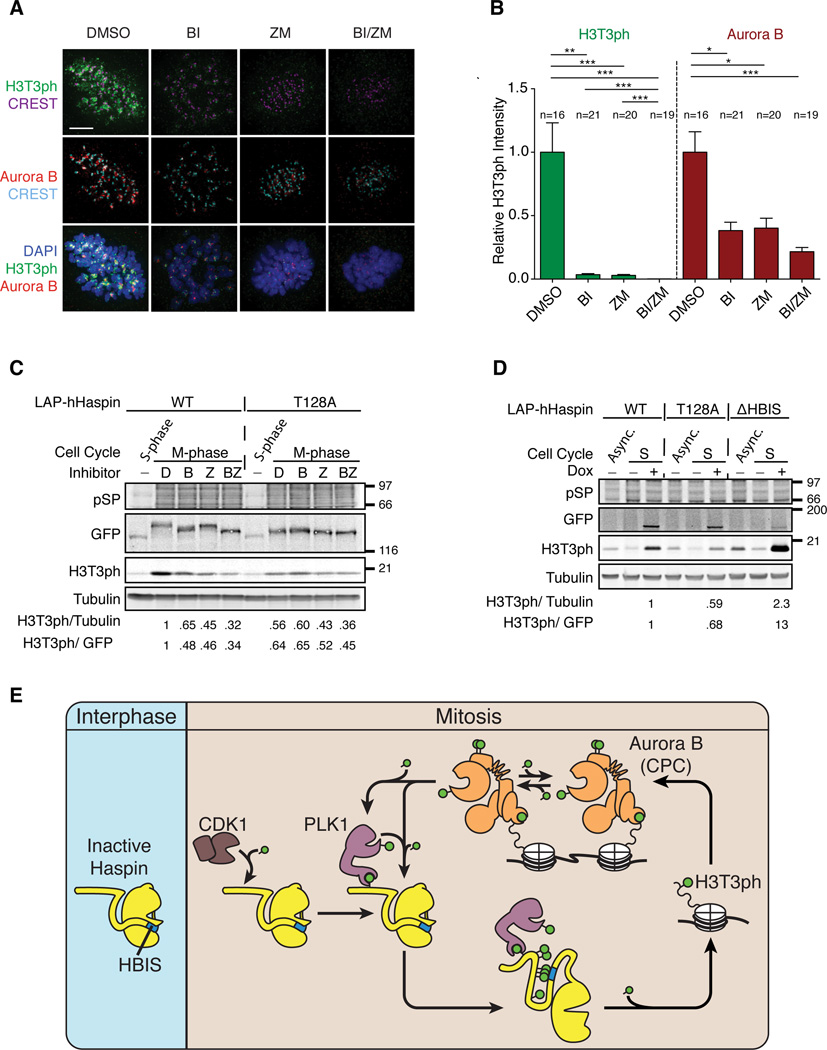
To test whether polo-dependent Haspin activation is conserved in human somatic cells, mitotic RPE1-hTERT and HeLa cells were collected, and treated with either the Plk1 inhibitor BI2536, the Aurora B inhibitor ZM447439 (Ditchfield et al., 2003), or both inhibitors in the presence of MG132 and nocodazole. In both cell lines, BI2536 greatly diminished H3T3 phosphorylation without significantly affecting phosphorylation of the Aurora B substrate, H3S10 (Figures S7A and B). Consistent with published observations (Wang et al., 2011), the Aurora B inhibitor ZM447439 reduced phosphorylation of both H3T3 and H3S10. Simultaneous treatment with BI2536 and ZM447439 showed an additive effect on reducing H3T3ph levels, indicating that both Aurora B and Plk1 collaborate to promote H3T3 phosphorylation in human tissue culture cells during mitosis.
We further tested the effect of ZM447439 and BI2536 on the chromosomal distribution and levels of H3T3ph in mitotic RPE1-hTERT cells. In control cells, H3T3ph was particularly enriched on chromosomal regions near centromeres. Consistent with Western blot analysis, ZM447439 and BI2536 both dramatically reduced the H3T3ph signal (Figures 7A, B and S7C). Quantification of H3T3ph levels indicates that the effects of ZM447439 and BI2536 on total chromosomal H3T3ph levels are comparable (Figures 7B and S7C). Simultaneous treatment with ZM447439 and BI2536 further decreased the H3T3ph signal to background levels, suggesting that activation of Haspin is largely under the control of Aurora B and Plk1. Consistent with published studies (Salimian et al., 2011; Wang et al., 2011), ZM447439 and BI2536 both reduced the centromeric enrichment of Aurora B (Figures 7C–D), while addition of both inhibitors further displaced Aurora B from the centromere. These results indicate that hHaspin requires both Aurora B and Plk1 for robust H3T3ph in human mitotic cells.
Figure 7. Plk1 and Aurora B activities are both required for H3T3ph in human mitotic cells.
(A and B) RPE1-hTERT cells were synchronized in mitosis for 3 hr with nocodazole, collected by mitotic shake-off and transferred to media containing MG132 and nocodazole for 1hr. Cells were then treated with either DMSO, 100 nM BI2536 (BI), 2 μM ZM447439 (ZM) or both inhibitors (BI/ZM) for 3 hr, collected by mitotic shake-off, processed for indirect immunofluorescence using the indicated antibodies.
(A) Representative maximum projections. Bar, 5 μm.
(B) Integrated intensity was calculated in Metamorph on deconvolved images and was standardized to the average intensity of the control. Mean and SEM are shown. ***, p < 0.001; **, p < 0.01; *, p , <0.05.
(C) DLD1 T-REx Flp-In cell lines expressing either LAP-hHaspin (WT) or LAP-hHaspinT128A were synchronized in mitosis with nocodazole for 16 hr, then collected by mitotic shake-off and transferred to media containing MG132 and nocodazole for 1 hr. Cells were then treated with inhibitors as A and B. The ratios of H3T3ph/ GFP and H3T3ph/ tubulin are reported relative to LAP-hHaspin (WT) + DMSO. LAP-hHaspin was detected by anti-GFP antibodies.
(D) DLD1 T-REx Flp-In cell lines expressing LAP-hHaspin (WT), LAPhHaspinT128A or LAP-hHaspinΔHBIS were synchronized in S-phase by double thymidine block and processed for Western blot analysis. The ratios of H3T3ph/ GFP and H3T3ph/ tubulin are reported relative to LAP-hHaspin (WT).
(E) Schematic of the activation mechanism of Haspin and the chromatinassociated feed back loop. During interphase, Haspin kinase activity is inhibited by the HBIS. Upon entry into mitosis, activated Cdk1 phosphorylates the N terminus of Haspin (T206 in Xenopus and T128 in human). This is recognized by Plk1, which further phosphorylates the Haspin N terminus. The phosphorylated N terminus interacts with the HBIS, displacing it from the kinase domain to activate Haspin and promote H3T3 phosphorylation. H3T3ph is then recognized by the CPC, resulting in cluster-mediated autophosphorylation and autoactivation of Aurora B. Activated Aurora B phosphorylates the activation loop of Plk1 to stimulate its activity and also directly phosphorylates Haspin to promote H3T3 phosphorylation.
See also Figure S7.
hHaspin activity is regulated by polo-dependent activation and HBIS-dependent autoinhibition in human cells
To evaluate the conservation of the Polo-dependent activation and the HBIS-dependent autoinhibition of Haspin in human cells, LAP-tagged full-length cDNA encoding either wild-type hHaspin, hHaspinT128A (equivalent to xHaspinT206A) or hHaspinΔHBIS was integrated at a single, predetermined locus into DLD-1 cells (Girdler et al., 2006). Expression of these constructs was induced with doxycycline after synchronizing cells in either mitosis or interphase. LAP-hHaspin expressed during mitosis showed a large gel mobility shift (Figure 7C). In the cell line expressing wild-type LAP-hHaspin, BI2536 reduced the mobility shift and the H3T3ph level. ZM44739 also greatly reduced H3T3ph level, but showed a milder effect on the mobility shift. In the T128A mutant cell line, the mobility shift of the LAP-hHaspinT128A was greatly reduced, but insensitive to BI2536. The H3T3ph level was lower in the T128A cell line than in the wild-type LAP-hHaspin cell line, which was further reduced by ZM44739, but not by BI2536. These results are consistent with the model that T128 supports Plk1-dependent, but not Aurora-dependent, activation of Haspin.
To demonstrate that the HBIS is required to autoinhibit hHaspin activity in interphase, we synchronized DLD1 cells in S-phase and compared the level of H3T3ph after expression of LAP-hHaspin wild-type, T128A or ΔHBIS (deleting KKKIV of the HBIS shown in Figure 3A). Even under identical induction conditions, the protein level of hHaspinΔHBIS was reproducibly lower than wild-type hHaspin or the T128A mutant (Figure 7D). This phenomenon was observed in several independent clones in both DLD1 and HeLa cell lines (data not shown), suggesting that deletion of the HBIS reduced the hHaspin protein level for unknown reasons (data not shown). Despite its relatively low abundance, DLD1 cells expressing hHaspin-ΔHBIS had significantly higher levels of H3T3ph in interphase than those expressing wild-type hHaspin or the T128A mutant. This result suggests that the autoinhibitory function of the HBIS is conserved in human cells.
DISCUSSION
In this study, we revealed the mechanistic basis for cell cycle-dependent activation of the H3T3 kinase Haspin. In interphase, Haspin is autoinhibited by the HBIS that interacts with its kinase domain. When Cdk1 is activated upon entry into M phase and phosphorylates a specific priming site (T206 in Xenopus or T128 in human) at the N terminus of Haspin, Plk1 docks on this priming phosphorylation site and subsequently phosphorylates multiple residues on the N terminus of Haspin, a cascade of events which neutralizes the action of HBIS and stimulates the kinase activity of Haspin (Figure 7E).
The Mechanism of Haspin autoinhibition in interphase
xHaspin is autoinhibited by the HBIS located immediately upstream of the kinase domain. Given that a synthetic HBIS-peptide can inhibit purified kinase in vitro, we propose that the HBIS directly inhibits kinase activity. The HBIS-peptide inhibits the purified MBP-xHaspin-ΔN729, even though this construct harbors the HBIS. This suggests that the kinase domain dynamically switches between HBIS-bound and –unbound state in this isolated context. Indeed, MBP-xHaspin-ΔN729 can phosphorylate H3T3 in a purified system, albeit with slower kinetics (Figure 4), while equivalent constructs in egg extracts failed to support H3T3 phosphorylation (Figures 3B and S3B–D). This difference is not due to the MBP-tag or nature of the substrates (H31–45-GST in vitro v.s. an H3-H4 complex in extract) (data not shown). We speculate that a counteracting phosphatase(s) that acts on xHaspin and/or histone H3 sets a higher threshold level of kinase activity to achieve robust H3T3 phosphorylation in egg extracts. It is also possible that egg extracts contain a factor(s) that modulates the folding and/or binding affinity between the HBIS and the kinase domain.
Removal of the HBIS doubles the Vmax of Haspin without significantly affecting its Km towards H3T3, consistent with the model in which HBIS acts as an allosteric, rather than a competitive inhibitor. The contribution of multiple basic residues in the HBIS in a redundant manner suggests a charge-based interaction with the kinase domain. However, the highly conserved acidic triplets in the xHaspin activation loop (Asp707, Glu708, Asp709 in human) are not involved in HBIS binding, as mutating these residues to alanine did not significantly reduce binding to the HBIS-peptide (C.G., M.S.W., and H.F., unpublished data). Future studies on the structure of the kinase domain-HBIS complex should provide more detailed mechanistic insights into the nature of HBIS autoinhibition.
Reactivation of Haspin in mitosis by phosphorylation
How could Polo and/or Aurora B-dependent phosphorylation activate Haspin? In the case of calcium/calmodulin-dependent kinase II (CaMKII), phosphorylation of its autoinhibitory module activates CaMKII kinase activity (Rellos et al., 2010; Rosenberg et al., 2005). S753 within the Xenopus HBIS is phosphorylated in a Polo-dependent manner in metaphase egg extracts, but only partially, if any, contributes to neutralization of the HBIS. In hHaspin, in addition to S389 (the counterpart of xHaspin S753), an adjacent S387 within the HBIS is also phosphorylated during mitosis (Wang et al., 2011). Since S387 falls within the substrate consensus motif for Aurora kinases (RX[S/T][no Pro]) (Alexander et al., 2011; Hengeveld et al., 2012), this phosphorylation may contribute to the Aurora B-dependent activation of Haspin in human cells.
Cdk1-Cyclin B and Plx1/Plk1 phosphorylate the xHaspin N terminus to promote binding of this region to the HBIS (Figures 2, 5 and S5). We speculate that this phosphorylation-dependent interaction is important to neutralize the action of the HBIS. Consistent with this hypothesis, mutating multiple phosphorylation sites in the N terminus of xHaspin greatly reduced its activity in metaphase egg extracts. Included in these phosphorylation sites is S234, which is proximal to the polo-docking site at T206, and may correspond to the known phosphorylated site in hHaspin, S151 (Wang et al., 2011). S234A mutation alone did not show a significant impact on Haspin activity, but its importance was revealed when combined with mutations at other phosphorylation sites. S234 phosphorylation may facilitate phosphorylation of additional sites on the Haspin N terminus, either by Polo, other kinases or Haspin itself. Indeed, the gel mobility shift of the catalytically inactive xHaspin K862A mutant was substantially reduced (Figure 2A), and phosphorylation of S753, for example, was sensitive to both T206 and Haspin activity (Table S1). In addition, the residues (T472, T490, S659, S662, S675) whose phosphorylation depends on Haspin activity appear to contribute to further Haspin activation. Thus, xHaspin activation may be mediated by a sequence of phosphorylation events, beginning with Cdk1-Cyclin B-dependent recruitment of Polo to T206, partial activation of Haspin by Polo-mediated phosphorylation of residues such as S234, and finally full activation of Haspin by Polo- and/or Haspin-dependent phosphorylation of residues in the N terminus. Some of these phosphorylation sites may affect Haspin activity without directly interacting with the HBIS. In the future, it will be interesting to determine if individual residues serve distinct functions in Haspin activation.
Functional significance of Haspin activation by Polo and Aurora B
Multiple mitotic kinases are known to form a complex signaling network on chromosomes and kinetochores (Funabiki and Wynne, 2013), but it was unclear how stimulation of the network is restricted to mitosis. Our study links the major cell cycle driver Cdk1 and this mitotic kinase network (Figure 7E). A great majority of H3T3ph signal depends on either Aurora B activity or Plk1 activity in human cells, indicating that full activation of hHaspin may require simultaneous phosphorylation of hHaspin by both kinases. We propose that spatiotemporal control of H3T3 phosphorylation is dictated by Cdk1/Plk1-dependent and Aurora B-dependent phosphorylation of Haspin.
Aurora B activation is mediated by autophosphorylation (Sessa et al., 2005), which is induced by local enrichment at the specific intracellular structure (Kelly et al., 2007). Therefore, recruitment of Aurora B to the centromere by H3T3ph and by Bub1-dependent H2A phosphorylation (Yamagishi et al., 2010) provides spatial information to which H3T3 phosphorylation (and Aurora B) is restricted on mitotic chromosomes. Aurora B-dependent activation of hHaspin in human cells creates a positive feedback loop to further enforce this centromeric enrichment of H3T3ph and Aurora B (Wang et al., 2011). This enrichment of H3T3ph at the centromere is important for recruiting Aurora B and controlling kinetochore functions such as activation of the spindle assembly checkpoint and regulation of kinetochore-microtubule attachments (De Antoni et al., 2012; Wang et al., 2010; Wang et al., 2012; Yamagishi et al., 2010).
In contrast, Plk1/Plx1 couples Haspin activation to the Cdk1-Cyclin B activity that peaks at mitosis, since Plk1-dependent phosphorylation depends on a priming phosphorylation on the substrate (Elia et al., 2003b), rather than local enrichment of Plk1 (Liu et al., 2012; Tan and Kapoor, 2011). The cascade from Cdk1 to Plk1 to Haspin to Aurora B generates additional feedback since Aurora B contributes to Plk1 activation by phosphorylating its activation loop (Carmena et al., 2012a) (Figure 7). Consistent with the requirement of Plk1 activity for its own kinetochore localization (Nishino et al., 2006; Zhou et al., 2003), Aurora B activity also positively controls Plk1 localization (Carmena et al., 2012a). Thus, together with Bub1 (Yamagishi et al., 2010), Plk1 at the kinetochore further promotes Aurora B enrichment at the centromere by activating Haspin. The presence of multiple positive feedback loops may locally augment the activity of these kinases.
Since H3T3 phosphorylation can dissociate the transcription factor TFIID, which recognizes the neighboring trimethylation at lysine 4 (H3K4me3) (Varier et al., 2010), requirement of multisite phosphorylation for Haspin activation may help spatiotemporally restrict H3T3 to obviate the risk of transcriptional misregulation. However, it is possible that phosphorylation can be induced in interphase under specific conditions, as has been implied in plant cells (Houben et al., 2007). It will be interesting to see whether Haspin can be activated outside of mitosis to regulate transcription.
EXPERIMENTAL PROCEDURES
Xenopus egg extract and Immunodepletions
Meiosis metaphase II-arrested CSF extracts were prepared using a method previously described (Murray, 1991). Immunodepletion was executed following the established protocols (Kelly et al., 2010; Sampath et al., 2004).
Extract Reconstitution Reactions using 35S-labeled proteins
To complement ΔxHaspin extracts by full-length or mutant xHaspin, the proteins were produced using the transcription-coupled translation rabbit reticulocyte lysate system (Promega). The reactions were incubated for 2.5 hr at 30°C in the presence of 35S-methionine. The samples were diluted 1:10 into either CSF or interphase extracts and incubated for 60–90 min.
Imaging and quantification
Cells were imaged with Deltavision Image Restoration microscope system (Applied Precision/GE Healthcare), mounted on an Olympus IX-70 microscope and fitted with a 100x/1.40 UPLSAPO objective lens and a CoolSnap QE CCD camera (Photometrics) in the Rockefeller Bioimaging Facility. Images were deconvolved using SoftWoRx (Applied Precision) and exported to Metamorph (Universal Imaging) for analysis and quantification. Representative images are maximum projections. Integrated intensity for each channel was determined after applying a threshold to remove background. Statistical significance was determined by unpaired Student’s t-test.
HBIS-peptides
HBIS (biotin- HWLRLRAALSLHRKKKVQATD) or SCR (biotin-ARDQKLWSKARTHVAHLKLLR) peptides (>95% pure) were synthesized by the Rockefeller University Proteomics Resource Center. Peptide bead pulldowns were performed similar to (Kelly et al., 2010).
Supplementary Material
Highlights.
The histone H3T3 kinase Haspin is autoinhibited by a basic segment during interphase
Priming phosphorylation of Haspin by Cdk1 during M phase recruits Polo-like kinase
Polo-dependent multisite phosphorylation of Haspin neutralizes the autoinhibition
Polo and Aurora B act together to support H3T3 phosphorylation in human cells
ACKNOWLEDGEMENTS
We thank A. Desai, R. Gassmann, J. Higgins, A. Losada, S. Taylor and K. Weis for reagents, T. Tong at the Rockefeller University Bio-Imaging Resource Center for helping with imaging analyses, J. Fernandez, M. Tesic Mark and H. Molina at the Proteomics Resource Center for MS analysis, M. Good, R. Subramanian, D. Patnaik, and M. Unciuleac for their assistance with setting up a quantitative kinetic assay for Haspin, S. Giunta, T. Maniar, D. Wynne, and C. Zierhut for critical reading of the manuscript, and members of the Funabiki lab for discussions. This work was supported by the grant from National Institute of Health to H. F. (R01-GM075249).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, one table and references.
REFERENCES
- Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JR, Ehrenberger T, Ivins F, Sessa F, Hudecz O, Nigg EA, et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal. 2011;4:ra42. doi: 10.1126/scisignal.2001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Pinson X, Platani M, Salloum Z, Xu Z, Clark A, Macisaac F, Ogawa H, Eggert U, Glover DM, et al. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS biology. 2012a;10:e1001250. doi: 10.1371/journal.pbio.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012b;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes & development. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Maffini S, Knapp S, Musacchio A, Santaguida S. A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. The Journal of cell biology. 2012;199:269–284. doi: 10.1083/jcb.201205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. The Journal of cell biology. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003a;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003b;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JM, Knapp S. Structure and functional characterization of the atypical human kinase haspin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 2013;122:135–158. doi: 10.1007/s00412-013-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea BB, Ruderman JV. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4493–4498. doi: 10.1073/pnas.0600702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdler F, Gascoigne KE, Eyers PA, Hartmuth S, Crafter C, Foote KM, Keen NJ, Taylor SS. Validating Aurora B as an anti-cancer drug target. J Cell Sci. 2006;119:3664–3675. doi: 10.1242/jcs.03145. [DOI] [PubMed] [Google Scholar]
- Hengeveld RC, Hertz NT, Vromans MJ, Zhang C, Burlingame AL, Shokat KM, Lens SM. Development of a chemical genetic approach for human aurora B kinase identifies novel substrates of the chromosomal passenger complex. Molecular & cellular proteomics : MCP. 2012;11:47–59. doi: 10.1074/mcp.M111.013912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A, Demidov D, Caperta AD, Karimi R, Agueci F, Vlasenko L. Phosphorylation of histone H3 in plants--a dynamic affair. Biochim Biophys Acta. 2007;1769:308–315. doi: 10.1016/j.bbaexp.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. The Journal of biological chemistry. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. The Journal of cell biology. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Cell Cycle Extracts. Meth Cell Biol. 1991;36:573–597. [PubMed] [Google Scholar]
- Nishino M, Kurasawa Y, Evans R, Lin SH, Brinkley BR, Yu-Lee LY. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Current biology : CB. 2006;16:1414–1421. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Molecular cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Rellos P, Pike AC, Niesen FH, Salah E, Lee WH, von Delft F, Knapp S. Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS biology. 2010;8:e1000426. doi: 10.1371/journal.pbio.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, Black BE. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Current biology : CB. 2011;21:1158–1165. doi: 10.1016/j.cub.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatininduced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Sillje HH, Korner R, Nigg EA. The Plk1-dependent phosphoproteome of the early mitotic spindle. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M110.004457. M110 004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, Stukenberg PT, Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Molecular cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase1, inhibits tumor growth in vivo. Current biology : CB. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Tan L, Kapoor TM. Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16675–16680. doi: 10.1073/pnas.1106748108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng BS, Tan L, Kapoor TM, Funabiki H. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell. 2010;18:903–912. doi: 10.1016/j.devcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varier RA, Outchkourov NS, de Graaf P, van Schaik FM, Ensing HJ, Wang F, Higgins JM, Kops GJ, Timmers HT. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. The EMBO journal. 2010;29:3967–3978. doi: 10.1038/emboj.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa F, Capasso P, Tortorici M, Forneris F, de Marco A, Mattevi A, Musacchio A. Crystal structure of the catalytic domain of Haspin, an atypical kinase implicated in chromatin organization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20204–20209. doi: 10.1073/pnas.0908485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ulyanova NP, Daum JR, Patnaik D, Kateneva AV, Gorbsky GJ, Higgins JM. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. The Journal of cell biology. 2012;199:251–268. doi: 10.1083/jcb.201205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ulyanova NP, van der Waal MS, Patnaik D, Lens SM, Higgins JM. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Current biology : CB. 2011;21:1061–1069. doi: 10.1016/j.cub.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K, Ryder U, Lamond AI. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. The EMBO journal. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell. 2003;5:127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.