Abstract
The genome of bacteria is organized and compacted by the action of nucleoid-associated proteins. These proteins are often present in tens of thousands of copies, binding with low specificity along the genome. DNA-bound proteins thus potentially act as roadblocks to the progression of machinery that moves along the DNA. In this study we investigate the effect of HU, one of the key proteins involved in shaping the bacterial nucleoid, on DNA helix stability by mechanically unzipping single dsDNA molecules. Our study demonstrates that 1) individually bound HU proteins have no observable effect on DNA helix stability, whereas 2) side-by-side bound HU proteins within filaments increase DNA helix stability. As the stabilizing effect is small compared to the power of DNA based motor enzymes, our results suggest that HU alone does not provide substantial hindrance to their progression in vivo.
Keywords: Biochemistry, DNA, Molecular biology, proteins
Introduction
The dimensions of an unrestrained bacterial genome largely exceed that of the bacterial cell itself. Therefore the genomic DNA of bacteria needs to be compacted and functionally organized by architectural proteins. (1-3) In E. coli approximately ten different nucleoid-associated proteins (NAPs) are likely involved in this process. Two main types of architectural properties can be noted: bridging of two sites/regions along DNA (e.g. H-NS) and bending of DNA (e.g. HU, IHF). (1-3) Under certain experimental conditions a DNA stiffening mode is observed in addition to one of these two modes. In the case of H-NS like proteins switching between bridging and stiffening appears to depend on magnesium ions (4) and may correspond to a switch between binding in trans and in cis. (5) The HU protein bends DNA at low protein concentrations, promoting compaction, and stiffens DNA at high concentrations (supplementary figure S1). (6-8) The affinity of HU for short regular B-DNA substrates is in the micromolar range, whereas affinities in the nanomolar range have been reported for binding to pre-distorted DNA substrates. (9-11)
DNA transactions, such as transcription, replication and repair, all take place on a genome decorated with many different proteins (a large fraction of which are NAPs). (12) The speed and efficiency of the motor enzymes RNA polymerase and DNA polymerase translocating along DNA can be affected by the presence of DNA bound proteins. Transcription and replication may come to a halt if a protein's off rate is so low that it acts as a roadblock. (13) The effect of nucleoid-associated proteins on transcription elongation and replication has not been investigated directly. Earlier, we argued, based on force measurements that DNA-DNA bridges mediated by H-NS do not interfere with transcription. (14) In this study, we investigate the effects of HU on DNA helix stability with implications for both the aforementioned processes. To this purpose we apply single-molecule DNA-unzipping force analysis to single HU-DNA complexes. This study provides insight into the strength of binding of the HU protein in its two binding modes, and dissects the role of DNA sequence composition.
Results & Discussion
Unzipping of single DNA molecules yields a characteristic sequence-dependent unzipping force landscape with GC-rich regions requiring higher forces for unzipping than AT-rich regions. (15) Binding of a protein to a specific site along the DNA results in specific changes in this landscape. (16, 17) For instance, force-induced dissociation of a eukaryotic nucleosome from a specific positioning sequence is manifested in a complex force landscape reflecting the disruption of specific nucleosomal protein-DNA interactions. (18, 19) The force required for dissociation of nucleosomes is on the order of several tens of pNs. In addition to yielding information on dissociation pathways and kinetics under force, unzipping experiments also permit mapping of protein binding locations along DNA.
In this study we investigated the generic effect of the binding of the nucleoid-associated protein HU on the force required for DNA unzipping and specific effects related to DNA sequence composition. DNA unzipping experiments can provide detailed insight into the binding patterns of HU (and their stability against applied force) along DNA, thus deepening and extending observations on global mechanical effects as seen in DNA stretching experiments (20) (supplementary figure S2). In control force-extension experiments we first reproduced the bimodal binding behaviour as seen in our previous magnetic tweezers studies (7) (data not shown). Up to a concentration of 400 nM the apparent persistence length is reduced (yielding DNA compaction). This behaviour is attributed to the binding of individual HU proteins. At higher concentrations (400 -1600 nM) the apparent persistence length increases. This corresponds to the ‘DNA stiffening’ mode in which HU proteins are bound closely side-by-side. (7) Next, we constructed 750 bp DNA substrates suitable for DNA unzipping (16, 18, 19) and carried out DNA unzipping experiments over a range of concentrations (from 0 – 1600 nM HU) under the same conditions as used in our previous studies. (7) The speed of unzipping (~75 bp/s) is on the order of the progression rate found in vivo for RNA polymerase, (21) but an order of magnitude lower than the rate of replication. (22)
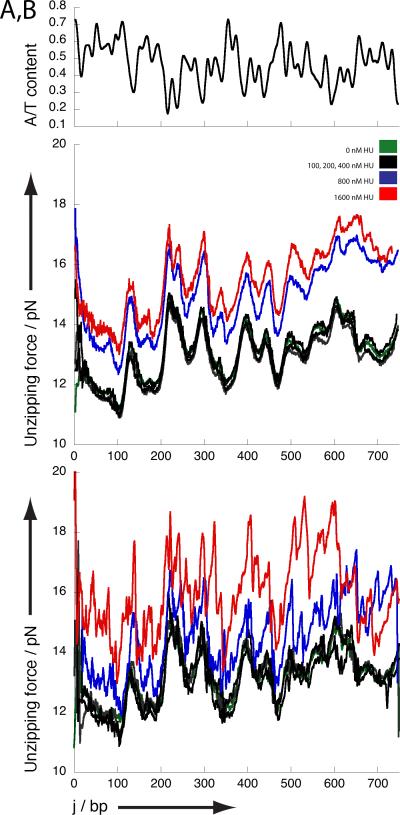
Under these conditions DNA unzipping curves characteristic of bare DNA are obtained in the absence of protein (figure 1a): the local force required for unzipping ranges between 11 and 15 pN depending on AT-content. Regions of low AT/high GC-content require higher unzipping forces than regions of high AT/low GC content. Following addition of 100, 200 or 400 nM HU the average unzipping curves do not change substantially. However, when the amount of HU is increased beyond 400 nM corresponding to the DNA stiffening regime, the average force required for unzipping increases by ~2 pN.
Fig 1. Unzipping of DNA and HU-DNA complexes.
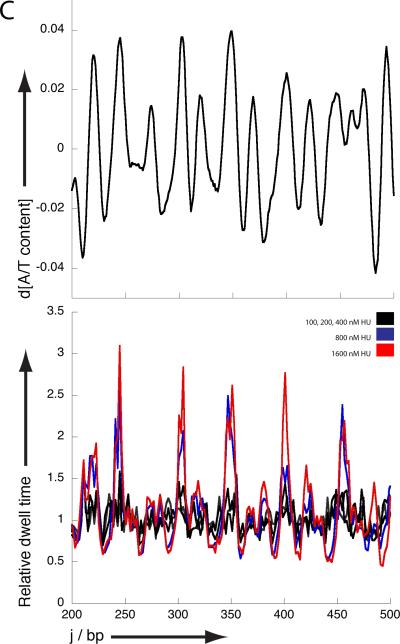
Unzipping traces for HU DNA complexes. A) Average unzipping force landscape obtained at different HU concentrations. Note that the unzipping forces at 800 and 1600 nM HU increase by ~ 2 pN compared to bare DNA and HU concentrations up to 400 nM.; B) Typical unzipping force landscape obtained at each HU concentration. Note that in the traces obtained at 800 and 1600 nM additional peaks are observed. Figures A and B also show the A/T content along the unzipped DNA fragment.; C) Relative dwell time along the unzipped DNA fragment indicating how much longer a certain position is maintained within an HUDNA complex compared to bare DNA. Figure C shows the derivative of A/T content along the unzipped fragment. Data obtained at 0 nM HU (green), 100, 200 and 400 nM HU (black) overlaps. Data obtained at 800 nM and 1600 nM HU are shown in blue and red respectively.
If we next focus on individual unzipping traces rather than on the averages (figure 1b), it is evident that the traces obtained at concentrations up to 400 nM are essentially identical to the bare DNA traces. This indicates that individual HU molecules sparsely bound along the DNA are dissociated very easily. However, distinct features are present along the unzipping trace (peaks superimposed on the average raised DNA baseline) in the concentration regime where filaments are formed (i.e. HU concentrations higher than 400 nM). The occurrence of these features is correlated with regions of high AT content directly following regions of low AT content (characterized by high unzipping force). These features are expected to translate into long dwell times at the locations of the maxima in a plot of the derivative of AT content. Indeed, sites with long dwell times perfectly match the location of such maxima (figure 1c). This suggests that HU release from such regions requires more energy than elsewhere along the DNA, which is in line with the reported in vivo propensity of HU to bind AT-rich DNA. (23) The large drop in force after rupture suggests gaps are present in the HU-DNA filaments following AT-rich regions. It is tempting to speculate that HU filament formation initiates at AT-rich regions and that the observed irregularities within the filament arise from improper phasing between independently nucleated filaments. Note that under these conditions no individual proteins can be seen to dissociate from the filament extremity due to their close packing.
The average difference in unzipping force between bare DNA and the HUDNA filament at the unzipping rate used is about 2 pN. As the dissociation rate of the individual bound proteins at lower concentrations is too fast to be measured, this value reflects the energy required to disrupt cooperative HU-HU interactions within HU-DNA filaments. (7, 24) In individual traces high force peaks (up to 5 pN above the reference trace) corresponding to rupture events are observed. Our current studies do not permit the determination of a consensus sequence in the DNA related to these events. We speculate that structural features such as sequence-dependent stiffness promote the formation of stable filaments, so that a number of subunits held together via cooperative interactions dissociate at once.
What are the implications of our experimental observations for in vivo collisions between HU and the transcription and replication machinery? The applied rate of unzipping of 75 bp/s is in the range that is physiologically relevant for both types of processes. The high off rate of HU (leading to undetectable events in our experiments) suggests that neither of the two types of machinery is affected by the sparse binding of individual HU molecules along the genome. Assuming a binding site of 9 bp a maximum of 8 HU proteins is force to dissociate from the DNA by the applied force. This condition is only encountered in the filament and results in a 2 pN increase in unzipping force along the DNA. At low DNA coverage no force peaks are observed along the DNA suggesting an off rate >> 8 s-1. Although local filament formation may occur in vivo in regions of high AT content, sparse binding is physiologically likely the most relevant. Nevertheless, even such filaments have only a minor effect on DNA duplex stability in our experiments and thus on progression of enzymes moving along DNA. Transcription has even been observed to occur through eukaryotic nucleosomes,(25, 26) which have much higher characteristic rupture forces. (18, 19) Also for bacteria it has been previously suggested that DNA regions bridged by H-NS are not a barrier to the progression of RNA polymerase. (14) However, HU filaments (as well as H-NS bridged regions) likely do act as barriers to diffusion of proteins involved in target search, stressing the importance of target localization via multiple association/dissociation events. One also needs to keep in mind that the situation in vivo is more complex and involves cooperative action of different nucleoid-associated proteins. This suggests that, locally, more stable complexes composed of different NAPs can be formed that affect the progression of DNA-based motor enzymes and thus NAPs may play direct regulatory roles.
Experimental section
Biological materials
The DNA construct used in unzipping experiments was designed as two separate segments as described. (16, 18, 19) A 1.1 kb anchoring segment was prepared by PCR from plasmid pRL574 (27) using a digoxigenin (dig)-labeled primer and then digested with BstXI (NEB) to produce a ligatable overhang. The 0.8 kb unzipping fragment was prepared by PCR from plasmid 601 (28) using a biotin-labeled primer and then digested with BstXI and dephosphorylated using CIP (NEB) to introduce a nick into the final DNA template. A 2.2 kb DNA construct was prepared by PCR from plasmid pRL574 using primers with a biotin label and a digoxigenin (dig) label for force-extension measurements. HU was purified as described before. (7)
Single-molecule sample preparation
The sample preparation for the optical trapping experiments was essentially the same as that previously described. (16) HU was diluted to concentrations from 50 – 1600 nM in 20 mM HEPES (pH 7.9), 60 mM KCl and 1 mM DTT. (7) First, the glass surface of the sample chamber was functionalized with antidigoxigenin. Next, the surface was passivated with Blotting Grade Blocker (Biorad). The digoxigenin end-labeled DNA construct was introduced into the sample chamber, followed by binding of streptavidin-coated 0.48 μm polystyrene microspheres to the biotinylated extremity of the DNA construct. Finally, HU solutions of different concentrations were flowed into the sample chamber and incubated for 30 minutes before data acquisition. Unzipping experiments were carried out in HU solution.
Single-molecule data collection
Data were collected using a single beam optical trap as described previously (16). Experiments were conducted at constant loading rate (8.2 ± 1.1 pN/s) as achieved via computer-controlled feedback. Data were low-pass filtered to 5 kHz, digitized at ~ 15 kHz, and then further averaged to 100 Hz. The tether length used to calculate base pair position was Gaussian filtered with σ = 0.01. Experiments were conducted at a room temperature of 23 °C. The acquired data signals were converted into force and number of base pairs unzipped as previously described. (16)
Data analysis
The unzipping landscape for each of the individual traces collected at 0 nM HU was aligned to a theoretical unzipping landscape for the used DNA sequence using a SIMPLEX procedure, improving positional accuracy and precision to a few base pair. (19) Parameters used to create the theoretical unzipping curve that best fit our experimental data were energies of 1.02 and 2.9 kT for AT and GC pairs respectively. Average AT-content was calculated using a 9 bp moving window. Traces collected in the presence of HU were aligned to the average of 37 aligned experimental reference traces. Dwell times for all positions j along the unzipped DNA fragment were determined using a 2 bp window. The calculated dwell times were averaged over all obtained unzipping traces at each HU concentration. Relative dwell times were calculated using the bare DNA unzipping trace as reference. Average data is obtained by averaging multiple unzipping traces/dwell time plots (0 nM, N=57; 100 nM, N=76; 200 nM, N=43; 400 nM, N= 87, 800 nM, N=130; 1600 nM, N=54). The apparent persistence length for DNA and HU-DNA complexes was estimated by fitting to the WLC-model. (29)
Supplementary Material
Acknowledgments
We thank members of the Dame and Wang labs for critical reading of the manuscript. R.T.D. is financially supported by the Netherlands Organization for Scientific Research [VIDI grant 864.08.001] and M.D.W. by an NIH grant (GM059849) and an NSF grant (MCB-0820293).
Abbreviations
- HU
Histone-like protein from strain U93
REFERENCES
- 1.Dame RT. The role of nucleoid-associated proteins in the organizations and compaction of bacterial chromatin. Mol Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 2.Dame RT, Kalmykowa OJ, Grainger DC. Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in gram negative bacteria. PLoS Genet. 2011;7:e1002123. doi: 10.1371/journal.pgen.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luijsterburg MS, White MF, van Driel R, Dame RT. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit Rev Biochem Mol Biol. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiggins PA, Dame RT, Noom MC, Wuite GJ. Protein-mediated molecular bridging: a key mechanism in biopolymer organization. Biophys J. 2009;97:1997–2003. doi: 10.1016/j.bpj.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dame RT, Goosen N. HU: promoting or counteracting DNA compaction? FEBS Lett. 2002;529:151–156. doi: 10.1016/s0014-5793(02)03363-x. [DOI] [PubMed] [Google Scholar]
- 7.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc Natl Acad Sci U S A. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skoko D, Wong B, Johnson RC, Marko JF. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 9.Wojtuszewski K, Hawkins ME, Cole JL, Mukerji I. HU binding to DNA: evidence for multiple complex formation and DNA bending. Biochemistry. 2001;40:2588–2598. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 10.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. The EMBO journal. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinson V, Takahashi M, Rouviere-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. Journal of molecular biology. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 12.Vora T, Hottes AK, Tavazoie S. Protein occupancy landscape of a bacterial genome. Mol Cell. 2009;35:247–253. doi: 10.1016/j.molcel.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein IJ, Greene EC. Molecular traffic jams on DNA. Annual review of biophysics. 2013;42:241–263. doi: 10.1146/annurev-biophys-083012-130304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 15.Essevaz-Roulet B, Bockelmann U, Heslot F. Mechanical separation of the complementary strands of DNA. Proc Natl Acad Sci U S A. 1997;94:11935–11940. doi: 10.1073/pnas.94.22.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch SJ, Shundrovsky A, Jantzen BC, Wang MD. Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys J. 2002;83:1098–1105. doi: 10.1016/S0006-3495(02)75233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch SJ, Wang MD. Dynamic force spectroscopy of protein- DNA interactions by unzipping DNA. Physical review letters. 2003;91 doi: 10.1103/PhysRevLett.91.028103. [DOI] [PubMed] [Google Scholar]
- 18.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- 20.Dame RT. Single-molecule micromanipulation studies of DNA and architectural proteins. Biochem Soc Trans. 2008;36:732–737. doi: 10.1042/BST0360732. [DOI] [PubMed] [Google Scholar]
- 21.Gotta SL, Miller OL, Jr., French SL. rRNA transcription rate in Escherichia coli. J Bacteriol. 1991;173:6647–6649. doi: 10.1128/jb.173.20.6647-6649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirose S, Hiraga S, Okazaki T. Initiation site of deoxyribonucleotide polymerization at the replication origin of the Escherichia coli chromosome. Molecular & general genetics : MGG. 1983;189:422–431. doi: 10.1007/BF00325904. [DOI] [PubMed] [Google Scholar]
- 23.Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee AS, Luscombe NM. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012;40:3524–3537. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sagi D, Friedman N, Vorgias C, Oppenheim AB, Stavans J. Modulation of DNA conformations through the formation of alternative high-order HU-DNA complexes. Journal of molecular biology. 2004;341:419–428. doi: 10.1016/j.jmb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol. 2010;17:745–752. doi: 10.1038/nsmb.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer DA, Gelles J, Sheetz MP, Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352:444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 28.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of molecular biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 29.Bouchiat C, Wang MD, Allemand J, Strick T, Block SM, Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.