Abstract
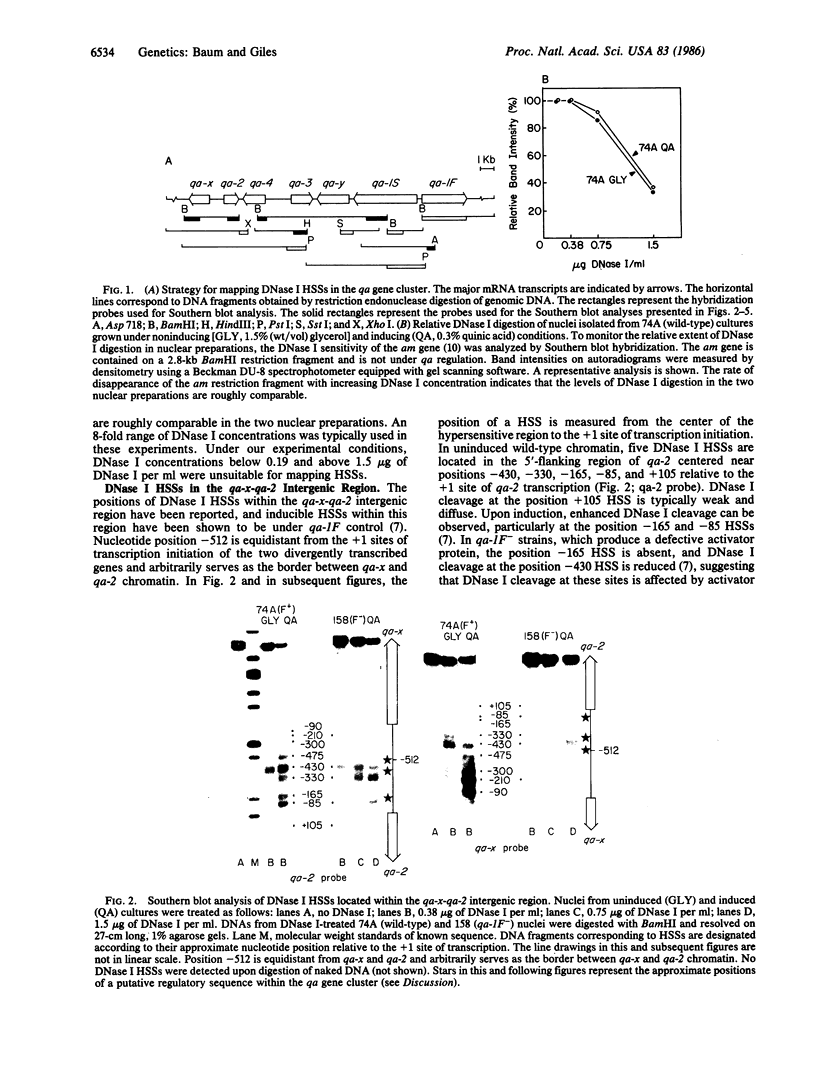
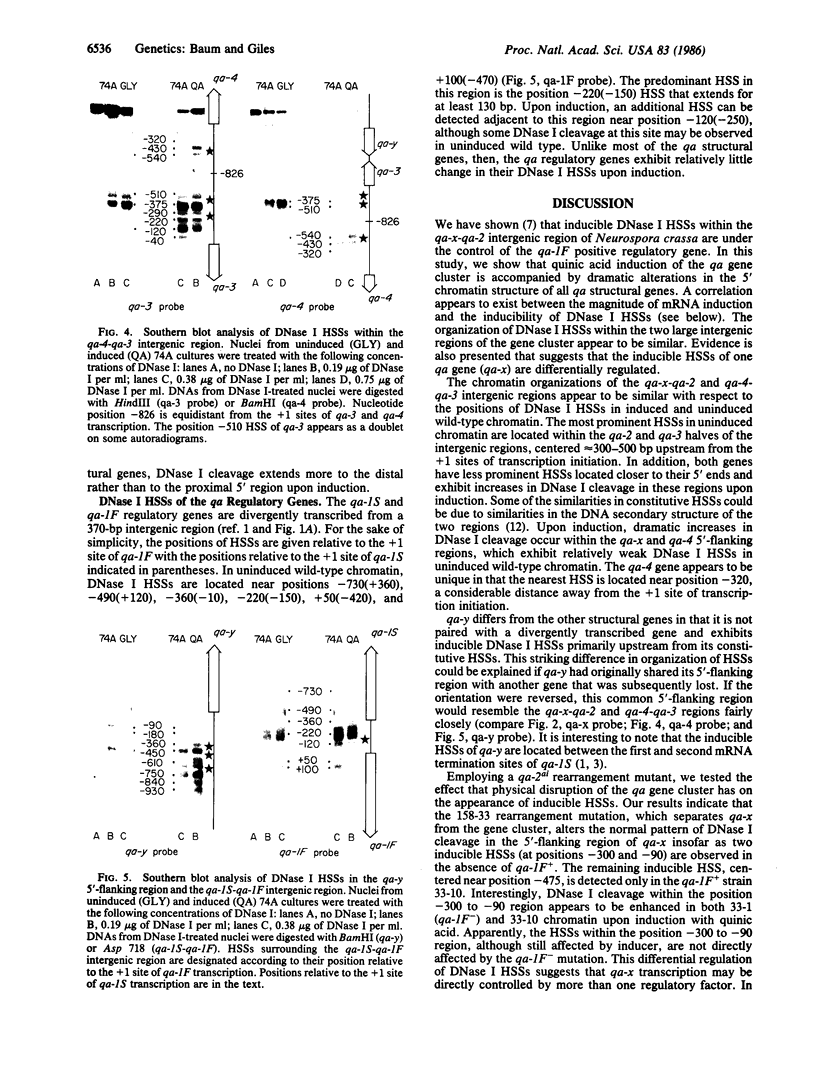
DNase I hypersensitive regions were mapped within the 17.3-kilobase qa (quinic acid) gene cluster of Neurospora crassa. The 5'-flanking regions of the five qa structural genes and the two qa regulatory genes each contain DNase I hypersensitive sites under noninducing conditions and generally exhibit increases in DNase I cleavage upon induction of transcription with quinic acid. The two large intergenic regions of the qa gene cluster appear to be similarly organized with respect to the positions of constitutive and inducible DNase I hypersensitive sites. Inducible hypersensitive sites on the 5' side of one qa gene, qa-x, appear to be differentially regulated. Employing these and previously published data, we have identified a conserved sequence element that may mediate the activator function of the qa-1F regulatory gene. Variants of the 16-base-pair consensus sequence are consistently found within DNase I-protected regions adjacent to inducible DNase I hypersensitive sites within the gene cluster.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baum J. A., Giles N. H. Genetic control of chromatin structure 5' to the qa-x and qa-2 genes of Neurospora. J Mol Biol. 1985 Mar 5;182(1):79–89. doi: 10.1016/0022-2836(85)90029-4. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Geever R. F., Case M. E., Tyler B. M., Buxton F., Giles N. H. Point mutations and DNA rearrangements 5' to the inducible qa-2 gene of Neurospora allow activator protein-independent transcription. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7298–7302. doi: 10.1073/pnas.80.23.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever R. F., Murayama T., Case M. E., Giles N. H. Rearrangement mutations on the 5' side of the qa-2 gene of Neurospora implicate two regions of qa-1F activator-protein interaction. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3944–3948. doi: 10.1073/pnas.83.11.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Baum J., Geever R., Huiet L., Patel V., Tyler B. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol Rev. 1985 Sep;49(3):338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiet L. Molecular analysis of the Neurospora qa-1 regulatory region indicates that two interacting genes control qa gene expression. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1174–1178. doi: 10.1073/pnas.81.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird J. H., Keighren M. A., Kinsey J. A., Eaton M., Fincham J. R. Cloning of the am (glutamate dehydrogenase) gene of Neurospora crassa through the use of a synthetic DNA probe. Gene. 1982 Dec;20(3):387–396. doi: 10.1016/0378-1119(82)90207-4. [DOI] [PubMed] [Google Scholar]
- Lohr D., Hopper J. E. The relationship of regulatory proteins and DNase I hypersensitive sites in the yeast GAL1-10 genes. Nucleic Acids Res. 1985 Dec 9;13(23):8409–8423. doi: 10.1093/nar/13.23.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V. B., Giles N. H. Autogenous regulation of the positive regulatory qa-1F gene in Neurospora crassa. Mol Cell Biol. 1985 Dec;5(12):3593–3599. doi: 10.1128/mcb.5.12.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V. B., Schweizer M., Dykstra C. C., Kushner S. R., Giles N. H. Genetic organization and transcriptional regulation in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5783–5787. doi: 10.1073/pnas.78.9.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechtman M. G., Yanofsky C. Structure of the trifunctional trp-1 gene from Neurospora crassa and its aberrant expression in Escherichia coli. J Mol Appl Genet. 1983;2(1):83–99. [PubMed] [Google Scholar]
- Tyler B. M., Geever R. F., Case M. E., Giles N. H. Cis-acting and trans-acting regulatory mutations define two types of promoters controlled by the qa-1F gene of Neurospora. Cell. 1984 Feb;36(2):493–502. doi: 10.1016/0092-8674(84)90242-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]








