Abstract
In clinical diagnostics, both array comparative genomic hybridization (array CGH) and single nucleotide polymorphism (SNP) genotyping have proven to be powerful genomic technologies utilized for the evaluation of developmental delay, multiple congenital anomalies, and neuropsychiatric disorders. Differences in the ability to resolve genomic changes between these arrays may constitute an implementation challenge for clinicians: which platform (SNP vs array CGH) might best detect the underlying genetic cause for the disease in the patient? While only SNP arrays enable the detection of copy number neutral regions of absence of heterozygosity (AOH), they have limited ability to detect single-exon copy number variants (CNVs) due to the distribution of SNPs across the genome. To provide comprehensive clinical testing for both CNVs and copy-neutral AOH, we enhanced our custom-designed high-resolution oligonucleotide array that has exon-targeted coverage of 1860 genes with 60 000 SNP probes, referred to as Chromosomal Microarray Analysis – Comprehensive (CMA-COMP). Of the 3240 cases evaluated by this array, clinically significant CNVs were detected in 445 cases including 21 cases with exonic events. In addition, 162 cases (5.0%) showed at least one AOH region >10 Mb. We demonstrate that even though this array has a lower density of SNP probes than other commercially available SNP arrays, it reliably detected AOH events >10 Mb as well as exonic CNVs beyond the detection limitations of SNP genotyping. Thus, combining SNP probes and exon-targeted array CGH into one platform provides clinically useful genetic screening in an efficient manner.
Keywords: array CGH, SNP, uniparental disomy, absence of heterozygosity, medically actionable variants
Introduction
Variation in the human genome consists of two major types: (1) single nucleotide variation, in the form of DNA base-pair substitutions and short indels, and (2) structural variation affecting many base pairs, including inversions, translocations, insertions, deletions, and duplications resulting in copy number variation (CNV).1 Advances in genome-wide analytical techniques, such as array comparative genomic hybridization (array CGH) and single nucleotide polymorphism (SNP) genotyping, can be used to detect CNVs. The number of identified CNVs has increased dramatically as the resolution of the detection technologies has improved.2, 3, 4, 5, 6, 7 Rapidly, CNV detection has become an integral part of genetic studies of disease susceptibility, delineation of novel genomic disorders,8, 9, 10 and analysis of data from genome-wide association studies.11, 12
Both array CGH and SNP genotyping are routinely utilized by clinicians for the evaluation of patients with developmental delay (DD)/intellectual disability (ID), multiple congenital anomalies (MCA),13 and neuropsychiatric disorders.14 Genomic resolution by both array platforms (SNP and array CGH) used in clinical laboratories allows for the detection of genomic gains and losses of ∼400 kb in size.15 Custom-designed oligonucleotide array CGH with greater resolution of the human genome, enabling detection of single-exon CNVs for clinically relevant genes, has also been implemented clinically.16, 17 This approach increases diagnostic yield, but also increases the likelihood of detecting variants of uncertain clinical significance.
While single-exon resolution throughout the genome is not feasible for SNP arrays, SNP arrays show higher sensitivity for the detection of low-level mosaic aneuploidies and chimerism18 and offer the ability to detect copy number neutral regions of absence of heterozygosity (AOH).19 Consanguinity can be revealed by AOH, because multiple regions of AOH are expected to be present in individuals from inbred populations, representing chromosomal segments that are identical by descent after transmission through parental lineages. The size and number of AOH blocks correlate with the degree of parental relatedness.20 Researchers and clinicians are also using the location of homozygous regions for mapping information in consanguineous families to identify autosomal recessive disease-causing genes.
When confined to a single chromosome, AOH regions may indicate uniparental disomy (UPD).21, 22 Several mechanisms can lead to UPD, resulting in isodisomy if both transmitted homologs are identical, heterodisomy if both homologs from one parent are present or segmental isodisomy if part of the chromosome is isodisomic and the other part is heterodisomic.23, 24 Although the true prevalence of UPD is not known, it is expected to be at least ∼1 in 3500 live births based upon information available in the ‘pre-genome analyses era'.25 UPD is a well-known mechanism leading to human disease if a chromosome containing imprinted genes is involved or if a recessive disease-causing mutation is present.25, 26
We now show that, by combining SNPs with our exon-targeted oligonucleotide array, detection for both copy number variation and copy-neutral AOH are optimized and enabled in a convenient, single genomic assay.
MATERIALS AND METHODS
Patients and sample preparation
A total of 3240 patient samples referred to the Medical Genetics Laboratory (http://www.bcm.edu/geneticlabs/) at Baylor College of Medicine from October 2010 to March 2012 were analyzed by the Chromosomal Microarray Analysis – Comprehensive (CMA-COMP) array. As an independent experimental SNP platform and quality assurance measure, 59 consecutive cases in which an AOH event (>10 Mb) was identified by the CMA-COMP array were also analyzed on an Illumina SNP array (Illumina Inc., San Diego, CA, USA).
All studies were performed on patient DNA extracted from peripheral blood using the Puregene DNA blood kit (Gentra, Minneapolis, MN, USA) according to the manufacturer's instructions.
CMA-COMP
Microarray design
The custom-designed 400K CMA-COMP array used in this study was manufactured by Agilent Technologies, Inc. (Santa Clara, CA, USA), and contains ∼280 000 oligonucleotide probes targeting 1860 genes at the exon level of genome resolution (<500 bp), with an average backbone coverage of one probe per 30 kb, an improved design of a previously described version by Boone et al,16 and 60 000 SNP probes in duplicate.
Analysis
DNA was digested with AluI and RsaI in order to allow detection of the SNPs located at the enzymes' recognition sites. The remaining procedures of DNA labeling and hybridization were performed according to the manufacturers' instructions, with minor modifications.27
For detailed array analysis, see Supplementary I.
Reporting
CNVs were reported and classified into three categories: (1) abnormal, (2) unclear clinical significance, and (3) likely benign. Abnormal CNVs include aneuploidy, known microdeletion/duplication syndromes, genomic imbalances larger than 2 Mb, other known genomic disorders (eg, the 1.5-Mb CMT1A duplication) and copy number changes involving pathogenic single genes. In cases where a pathogenic CNV is identified that may be associated with a presymptomatic condition, we report these incidental findings to the referring physician in order to facilitate adequate counseling and prompt medical attention as needed,28 consistent with current ACMG recommendations. CNVs of unclear clinical significance include those smaller than 2 Mb in size that have not been correlated with a clinical phenotype. CNVs are classified as likely benign if they are polymorphic in the normal population as determined by reviews of recent literature. Evaluation of potential medically actionable variants includes careful consideration of the size of the CNV, variant allele frequency, gene(s) involved, whether it represents a de novo or inherited event, and the reported phenotypic clinical findings in the child or other relevant family members.29, 30
Additionally, AOH segments greater than 10 Mb in size were reported.
SNP genotyping platform
SNP array analysis was performed on the Illumina Infinium HD assay platform using HumanOmni1-Quad BeadChip (Illumina Inc.) with 200 ng of genomic DNA according to the manufacturer's instructions. The GenomeStudio software (Illumina, Inc.) was used for data processing and analysis. AOH regions larger than 10 Mb were used for comparison to the CMA-COMP results.
Results
Comparison of AOH calling by CMA-COMP and Illumina SNP array
Of the 3240 samples referred for the CMA-COMP array, 162 (5%) had at least one region of contiguous AOH larger than 10 Mb. A total of 59 consecutive cases with one or more AOH regions measuring >10 Mb in size as identified by the CMA-COMP array were also tested on the Illumina SNP array for quality control. Fifteen of these cases had interstitial, terminal, or centromeric AOH in single chromosomes (Table 1). The AOH regions were confirmed by the Illumina array in all 15 cases; however, the AOH calls in two cases were found to represent smaller-sized AOH regions by the higher-resolution Illumina SNP array. Twenty seven cases had AOH regions totaling 100 Mb or higher, which is suggestive of consanguinity. Such high levels of AOH were confirmed by the Illumina array for all these cases (Table 2). Therefore, the CMA-COMP platform, even though it has a lower density of SNP probes than the Illumina array, was able to reliably detect AOH events >10 Mb.
Table 1. Comparison of the results of CMA-COMP and Illumina array for AOH regions greater than 10 Mb in size and limited to single chromosomes.
| CMA-COMP array | Illumina SNP array | |||||
|---|---|---|---|---|---|---|
| Chromosome | Genomic interval | Size in Mb | Genomic interval | Size in Mb | Location | UPD confirmed |
| 1 | 166 983 908–183 983 607 | 17.0 | 167 228 031–183 982 100 | 16.7 | Interstitial | — |
| 3 | 38 493 212–52 622 086 | 14.1 | 45 790 685–52 193 685 | 6.4 | Interstitial | — |
| 3 | 45 708 410–59 299 266 | 13.6 | 47 077 306–53 281 955a | 6.2 | Interstitial | — |
| 5 | 97 151 690–123 037 288 | 25.9 | 106 422 150–123 740 802 | 17.3 | Interstitial | — |
| 5 | 163 685 643–178 577 218 | 14.9 | 163 735 878–176 745 707 | 13.0 | Interstitial | — |
| 5 | 125 870 653–146 253 199 | 20.4 | 127 465 300–146 414 993 | 18.9 | Interstitial | — |
| 7 | 107 850 309–120 523 873 | 12.7 | 109 017 420–120 492 482 | 11.5 | Interstitial | — |
| 8 | 35 551 788–61 354 167 | 25.8 | 37 132 731–63 207 089 | 26.1 | Centromeric | — |
| 9 | 4 175 231–14 383 183 | 10.2 | 1 486 230–14 377 817 | 12.9 | Interstitial | UPD |
| 88 061 835–128 350 612 | 40.3 | 66 587 429–126 250 362 | 59.7 | Interstitial | — | |
| 12 | 1–22 919 015 | 22.9 | 1 607 841–22 154 162 | 20.5 | Terminal | — |
| 14 | 35 601 054–50 516 087 | 14.9 | 35 958 318–50 563 154 | 14.6 | Interstitial | No UPD |
| 14 | 33 803 584–70 306 504 | 36.5 | 33 742 889–70 200 059 | 36.4 | Interstitial | No UPD |
| 14 | 32 945 633–94 103 812 | 61.2 | 32 899 071–86 182 374 | 53.3 | Interstitial | — |
| 15 | 21 317 493–68 226 073 | 46.9 | 26 998 850–68 211 801 | 41.2 | Interstitial | UPD |
| X | 95 140 723–119 474 584 | 24.3 | 95 721 890–119378541 | 23.6 | Interstitial | — |
Small non-contiguous segments of AOH were seen within the same AOH interval identified by the CMA-COMP.
Table 2. Comparison of the results of CMA-COMP and Illumina array for detection of AOH regions totaling 100 Mb or higher in size.
| CMA-COMP array | Illumina array | |||
|---|---|---|---|---|
| Patients | AOH level (Mb) | Number of chromosomes involved | Number of blocks | AOH level (Mb) |
| Case 1 | 871 | 15 | 18 | 851 |
| Case 2 | 863 | 20 | 25 | 787 |
| Case 3 | 707 | 16 | 23 | 702 |
| Case 4 | 678 | 18 | 20 | 681 |
| Case 5 | 654 | 14 | 16 | 609 |
| Case 6 | 514 | 10 | 14 | 529 |
| Case 7 | 501 | 10 | 14 | 506 |
| Case 8 | 352 | 8 | 8 | 354 |
| Case 9 | 345 | 8 | 11 | 367 |
| Case 10 | 332 | 7 | 9 | 310 |
| Case 11 | 289 | 9 | 10 | 247 |
| Case 12 | 276 | 8 | 8 | 244 |
| Case 13 | 270 | 8 | 9 | 240 |
| Case 14 | 252 | 8 | 9 | 245 |
| Case 15 | 234 | 5 | 6 | 147 |
| Case 16 | 223 | 8 | 8 | 228 |
| Case 17 | 194 | 5 | 5 | 127 |
| Case 18 | 187 | 7 | 7 | 150 |
| Case 19 | 177 | 6 | 7 | 158 |
| Case 20 | 158 | 6 | 7 | 152 |
| Case 21 | 141 | 6 | 6 | 138 |
| Case 22 | 138 | 6 | 7 | 130 |
| Case 23 | 129 | 5 | 7 | 117 |
| Case 24 | 122 | 3 | 4 | 90 |
| Case 25 | 118 | 4 | 4 | 107 |
| Case 26 | 110 | 3 | 3 | 108 |
| Case 27 | 107 | 5 | 5 | 96 |
Uniparental disomy
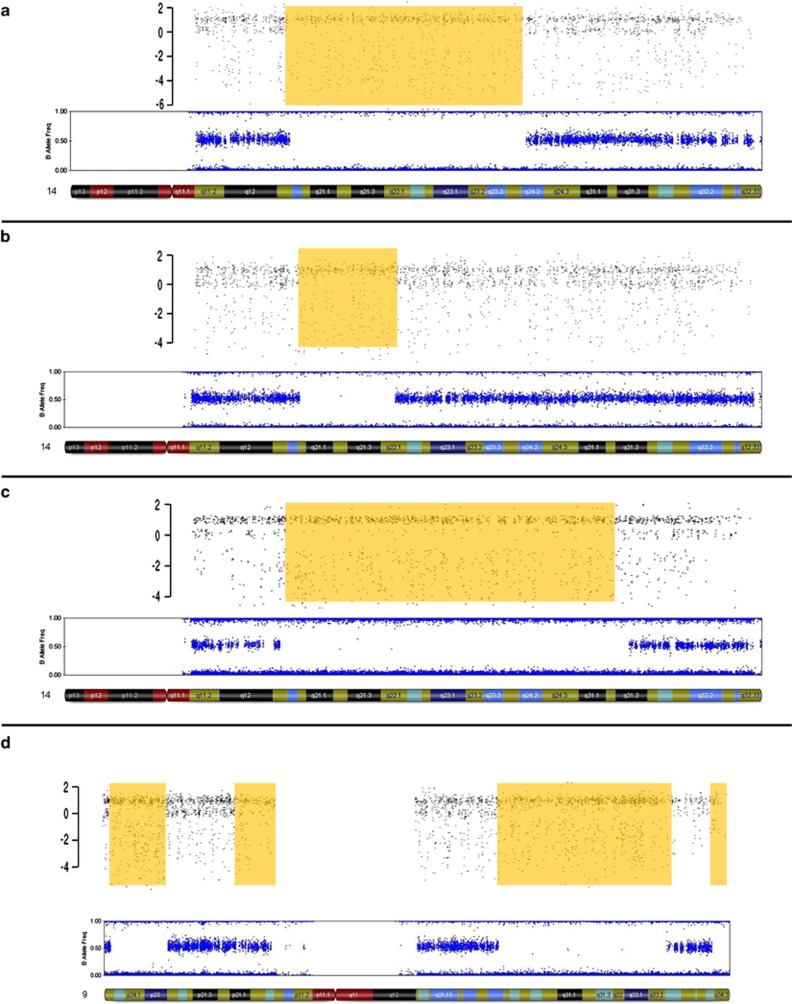
Two cases were identified by CMA-COMP with AOH in a single chromosome within which all interrogated SNPs were homozygous, indicating the presence of uniparental isodisomy. One case showed isodisomy 1 and the other isodisomy 16. Maternal UPD16 has been associated with intrauterine growth restriction and fetal malformations.31 Additionally, 45 cases analyzed by CMA-COMP showed AOH regions exceeding 10 Mb involving one chromosome only, indicating possible uniparental heterodisomy for that particular chromosome. Twelve of these cases involved chromosomes that contain imprinted regions associated with a clinical phenotype [chromosomes 7 (three cases), 14 (seven cases), and 15 (two cases)]. Methylation-specific PCR was performed on two (Figures 1a and b) of the seven cases with AOH limited to chromosome 14; in both cases, there was no evidence of UPD. The B allele frequency plot for a case with AOH of ∼61 Mb on chromosome 14 is shown in Figure 1c. Unfortunately, this case was lost to follow-up, so UPD could not be confirmed. For one case involving AOH on chromosome 15, an interstitial duplication of 15q11.2q13.1 and maternal uniparental trisomy was detected, genomic findings which are all consistent with a diagnosis of Prader–Willi syndrome. A detailed description of this clinical case is described elsewhere.32 All other remaining cases with AOH on imprinted chromosomes were lost to follow-up.
Figure 1.
Large regions of AOH limited to single chromosomes suggestive of heterodisomy. Top panels – CMA-COMP SNP data. Bottom panels – corresponding Illumina B allele frequency data. (a–c) Chromosome 14. Follow-up studies by methylation specific PCR showed presence of both maternal and paternal bands, indicating biparental chromosome 14 inheritance for cases shown in panels (a) and (b). Methylation studies were not available for the case shown in panel (c). (d) Chromosome 9. Follow-up parental SNP array analyses confirmed presence of maternal heterodisomy in this patient.
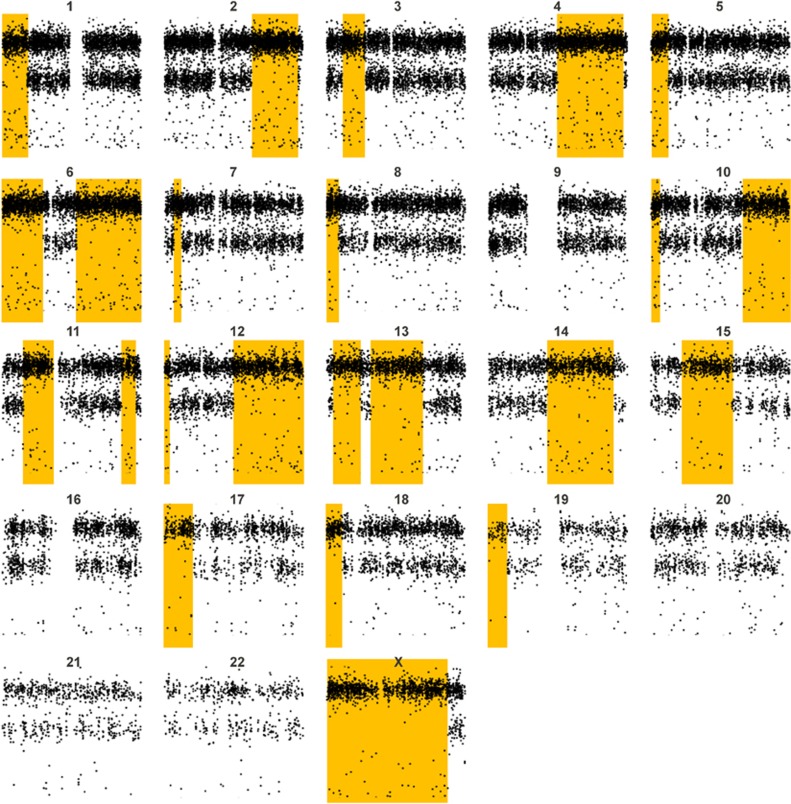
Of interest, one case had multiple AOH regions limited to chromosome 9 (Figure 2), which was confirmed by the Illumina SNP array. Analysis of the proband and parental genotypes indicated the presence of maternal heterodisomy 9.
Figure 2.
CMA-COMP array showing a patient with consanguinity as demonstrated by the multiple blocks of AOH (shaded) on numerous chromosomes.
Consanguinity
Multiple AOH regions >10 Mb involving two or more chromosomes were identified in 115 cases by CMA-COMP, of which 44 cases were also performed on the Illumina SNP array. As shown in Table 2, the AOH calls between the two arrays are comparable in the 27 cases with AOH regions totaling ≥100 Mb. For cases in which the degree of parental relatedness was provided by the referring center (Table 3), we compared the total length of AOH regions to that which is expected from the coefficient of inbreeding (F). Patients whose parents are first cousins (third degree of relationship) are expected to have 1/16 of their genome homozygous or ∼179 Mb of total AOH regions. The total length of AOH regions observed was 225 Mb as shown in case 2. Consistent data were also observed in cases of fourth and sixth degree of parental relationship [expected 90 Mb and observed 89 and 82 for fourth degree (cases 3 and 4), and expected 22.5 Mb and observed 58 for sixth degree (case 5)].
Table 3. Extent of AOH regions detected in patients from known consanguineous families.
| CMA-COMP array | |||||
|---|---|---|---|---|---|
| Patients | AOH level (Mb) | Number of chromosomes involved | Number of blocks | Parental relationship | Expected AOH |
| Case 1a | 707 | 16 | 23 | First-degree relatives | 716 Mb (F=1/4) |
| Case 2 | 225 | 9 | 10 | First cousins | 179 Mb (F=1/16) |
| Case 3 | 89 | 4 | 4 | First cousins once removed | 90 Mb (F=1/32) |
| Case 4 | 82 | 3 | 3 | First cousins once removed | 90 Mb (F=1/32) |
| Case 5 | 58 | 3 | 3 | Second cousins once removed | 22.5 Mb (F=1/128) |
Same as case 3 in Table 2.
Notably, our analyses identified 10 cases in which the observed total length of AOH regions was consistent with that expected for a first degree relationship (F=1/4 or ∼716 Mb). In those cases, the total length of AOH ranged from ∼506 Mb to 851 Mb, with a mean of 677 Mb. Amongst this group of subjects, consanguinity was confirmed in two patients, one of which was known to be the product of a sister–brother mating (Table 3, case 1).
Autosomal recessive disorders and AOH
We evaluated regions of AOH for gene content when the indication was a specific disease. In four patients with an autosomal recessive disorder, the associated gene localized within one of the AOH regions. Sequencing analysis performed elsewhere confirmed the presence of a homozygous mutation in two patients (Table 4).
Table 4. Correlation of AOH regions with autosomal recessive disorders.
| AR disorder | Genes related to diagnosis | Total AOH >10 Mb | Gene within AOH block | AOH size (Mb) | Comment |
|---|---|---|---|---|---|
| Cohen syndrome | VSP13B | 29 | VSP13B | 11 | Homozygous exonic deletion detected |
| HHH syndrome | SLC25A15 | 270 | SLC25A15 | 40 | Homozygous mutation in SLC25A15 detected |
| SCID | 18 genes | 332 | CD3D | 8 | Homozygous mutation in CD3D detected |
| Usher syndrome | Dozens of genes | 214 | USH2A | 18 |
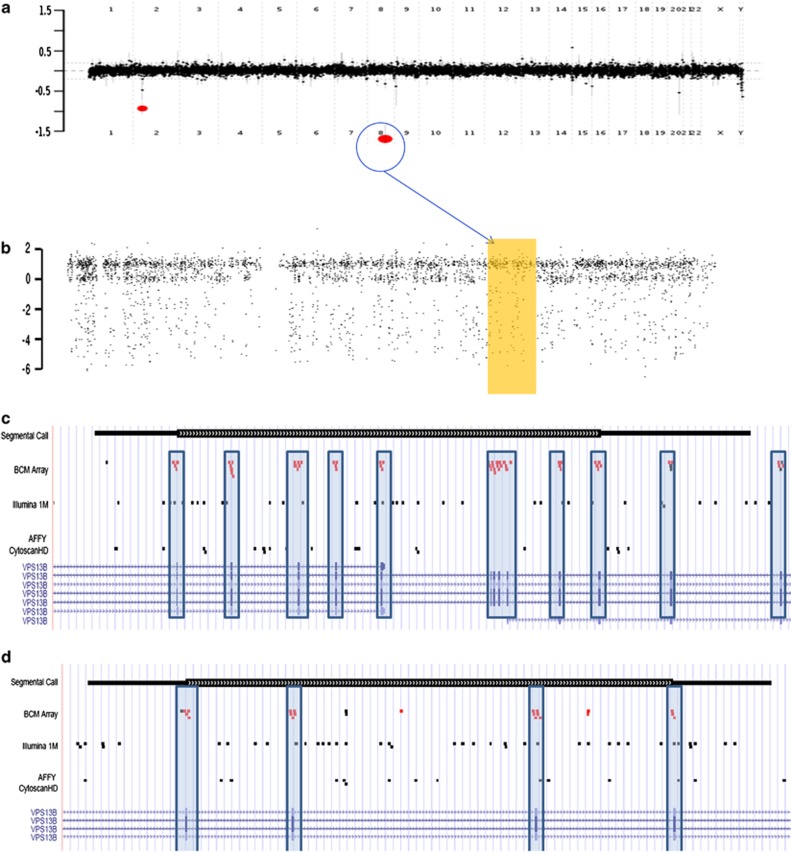
One patient, the product of a consanguineous mating (second cousins once removed), presented with DD/ID and microcephaly, and MRI revealing a mild diffuse reduction in the volume of the cerebral hemispheric white matter with a borderline to increased size of the corpus callosum. CMA-COMP analysis showed AOH events larger than 10 Mb on chromosomes 8 and 14. The AOH event on chromosome 8 also harbored a homozygous deletion of ∼52 kb involving exons 4–14 of the VPS13B (vacuolar protein sorting 13 homolog B) gene (Figures 3a and b), which is associated with Cohen syndrome (OMIM #216550) and consistent with the clinical phenotype of this patient.
Figure 3.
(a) A ∼52-kb homozygous deletion involving exons 4–14 of the VSP13B gene in a patient with Cohen syndrome detected by CMA-COMP array. (b) shows the SNP data plot with an AOH region on chromosome 8, wherein the VSP13B gene is located. (c) and (d) Comparison of the probe distribution between the exon-targeted CMA-COMP array and two other commercial SNP arrays for the portion of the VSP13B gene deleted in cases 6 and 7 (Table 4) (c) Case 6 (same case as in (a)) with a deletion of exons 4–14 (red dots represent the deleted oligos in the exons). Note that the locations of the SNPs (black dots) are outside of almost all of the exons, and therefore single exon deletions would not be detectable by these SNP arrays. (d) Case 7 in Table 4 – red dots represent the deleted oligonucleotides corresponding to exons 22–25 (represented by the hatch marks at the bottom of the figure) as detected by the CMA-COMP array. The black dots represent the SNP distribution within the same region. Again, note that most SNP probes are located within introns.
Intragenic CNVs
An advantage of the CMA-COMP array platform is its ability to detect very small, single-exon copy number changes. Pathogenic genomic deletions and/or duplications were detected in 14% (445/3240) of the cases. CNVs of unclear clinical significance were detected in an additional 13% (421/3240). Intragenic pathogenic CNVs were detected in 21 cases (Table 5) with an average of 21 probes (range 4–90). The smallest CNV was confirmed by sequencing to be 500 bp. A comparison of the distribution of the probes of the CMA-COMP to two commercial SNP arrays for the intragenic deletions involving exons of the VPS13B gene (Table 5, cases 6 and 7) is provided in Figures 3c and d. In addition to the VPS13B gene, intragenic deletions of both NRXN1 and DMD were detected in multiple patients as well as two cases involving genes that predispose to cancer.
Table 5. Pathogenic exonic CNVs detected by CMA-COMP array.
| Case | Gene | Disease association | Exon(s) | Copy number change | Chromosome Location | Inheritance | Gender |
|---|---|---|---|---|---|---|---|
| 1 | NRXN1 | Autism and developmental disorders | 23 | Loss | 2p16.3 | Unknown | F |
| 2 | NRXN1 | Autism and developmental disorders | 3–5 | Loss | 2p16.3 | Paternal | M |
| 3 | NRXN1 | Autism and developmental disorders | 4–5 | Loss | 2p16.3 | Unknown | M |
| 4 | MSH6 | Hereditary nonpolyposis colorectal cancer | 5–6 | Loss | 2p16.3 | Unknown | F |
| 5 | TGFB1 | Corneal dystrophy | 8–10 | Loss | 5q31.1 | Unknown | F |
| 6 | VPS13B | Cohen syndrome | 4–14 | Homozygous loss | 8q22.2 | Unknown | F |
| 7 | VPS13B | Cohen syndrome | 22–25 | Loss | 8q22.2 | Unknown | M |
| 8 | BRCA2 | Increased risk of cancer | 13–14 | Loss | 13q13.1 | Unknown | M |
| 9 | UBE3A | Angelman syndrome | 7–14 | Loss | 15q11.2 | Maternal | F |
| 10 | CHRNA7 | Intellectual disability and seizures | Entire gene | Loss | 15q13.3 | Unknown | F |
| 11 | ANKRD11 | KBG syndrome | 4–13 | Loss | 16q24.3 | Unknown | F |
| 12 | ANKRD11 | KBG syndrome | 2–3 | Loss | 16q24.3 | Unknown | F |
| 13 | HNF1B | Polycystic kidney disease, genital malformations, pancreatic atrophy and maturity-onset diabetes of the young type 5 | 7–8 | Loss | 17q12 | Unknown | M |
| 14 | DMD | Duchenne muscular dystrophy | 15 | Gain | Xp21.1 | Unknown | M |
| 15 | DMD | Duchenne muscular dystrophy | 40–50 | Loss | Xp21.1 | Unknown | M |
| 16 | DMD | Duchenne muscular dystrophy | 63–64 | Gain | Xp21.1 | Unknown | M |
| 17 | DMD | Duchenne muscular dystrophy | 49–51 | Loss | Xp21.1 | Maternal | M |
| 18 | MED12 | FG syndrome and Lujan–Fryns syndrome | 25–28 | Gain | Xq13.1 | De novo | M |
| 19 | GPC3 | Simpson–Golabi–Behmel syndrome | 7 | Gain | Xq26.2 | Unknown | M |
| 20 | FMR1 | X-linked intellectual disability | Entire gene | Gain | Xq27.3 | Unknown | M |
| 21 | L1CAM | X-linked hydrocephalus | 1–2 | Loss | Xq28 | Unknown | M |
Discussion
With the addition of SNP probes to our exon coverage array, we provide a comprehensive approach for the identification of clinically relevant copy number neutral changes in addition to medically actionable CNVs in a single assay.
The principles used to guide this unique array design were to: (1) empirically select the best performing probes to maximize detection and signal to noise ratio, (2) detect CNVs known to be associated with diseases and target coverage of the exons of these known disease genes, (3) maintain 30 kb backbone coverage, and (4) exclude most of the known LCR regions. The rationale for our array design to maximize detection of clinically relevant genomic changes has been independently validated by the observations of Haraksingh et al.33 They compared the CNVs observed in the 1000 Genomes Project with all the currently commercially available high-resolution array platforms and concluded that the sensitivity, total number, size range, and breakpoint resolution of CNV calls were highest for CNV focused arrays that did not compromise the backbone tiling density of the rest of the genome. They also found that probe distribution greatly affected the performance of a platform. A disadvantage for SNP-only arrays is that the probe distribution is restricted by the non-uniform availability of informative SNPs throughout the genome. Additionally, probes that are optimized will provide maximum information for specific loci.
In this study, the CMA-COMP array, with only 60 000 SNP probes, was concordant with a high-density SNP array in detecting AOH events and is thus an excellent tool for genome-wide screening for AOH in order to identify UPD, consanguinity, and map genomic intervals containing potential recessive loci. The percent of the genome manifesting AOH for each case was calculated for both platforms (data not shown) and there was complete concordance between the two platforms within 1–2%. Because of the lower density of SNP probes in the CMA-COMP array, there may be an overestimation of the genomic size of an AOH event, which occurred in two cases in Table 1. In one of these cases, the Illumina array revealed a smaller AOH (6 Mb) event, and in the other case, a series of smaller discrete AOH events interrupted by small regions of heterodisomy were seen in the Illumina array. The CMA-COMP did not resolve the discrete interval events; instead it showed a contiguous AOH event for this region. As both of these cases involved a non-imprinted chromosome, further studies were not recommended.
Added value to the genomic analysis provided by SNP probes is the detection of uniparental isodisomy without requiring analysis of parental samples, as it is with other conventional assays that utilize STR markers. In addition, UPD of virtually any chromosome can be detected in one assay. While SNP arrays will detect virtually 100% of isodisomies,24 not all cases of uniparental heterodisomy may be detected by SNP arrays unless a heterodisomic chromosome underwent a recombination event during meiosis, resulting in isodisomy interrupted by regions of heterozygosity, reflecting the presence of heterodisomy (hetero-isoUPD). In such cases, heterodisomy appears as a large block of AOH (isodisomic segment) confined to a single chromosome. Recently, Papenhausen et al22 found that their cohort of nine confirmed UPD cases all showed an AOH block within the affected chromosome. Similar findings were also observed in five confirmed UPD cases and an additional three were consistent with the phenotype by Bruno et al.34 However, not all AOH regions confined to a single chromosome indicate the presence of heterodisomy, as shown in our patients with AOH on chromosome 14. Similar observations were reported by Papenhausen et al,22 who showed that only 29 out of 46 cases with a single segmental AOH event were confirmed to be UPD. Therefore, a single region of AOH could be a random finding representing a region of identity by descent or linkage disequilibrium (LD) because of a low recombination rate.
Detection of isodisomy has obvious clinical relevance for cases in which chromosomes bearing imprinted genomic regions are involved, but it is also relevant in the context of autosomal recessive (AR) disorders. Most of the non-imprinted UPD cases described so far were ascertained by detection of a homozygous mutation in autosomal recessive genes that did not follow the expected Mendelian allele transmission pattern.26 From a genetic counseling perspective, detection of isodisomy or hetero-isoUPD modifies recurrence risk for AR disease from 25% to <1%. The importance of parental follow-up studies to confirm carrier status in cases in which a homozygous mutation was detected in the child cannot be underemphasized.
The identification of AOH has been previously employed for the determination of the genetic defects underlying genetically heterogeneous recessive disorders by a homozygosity mapping approach in a research setting. AOH regions determined by genome-wide SNPs analysis allows delineation of the critical disease-associated genomic interval and the number of potential candidate genes. Analysis of SNP data obtained for both affected and unaffected individuals enables further refinement of AOH regions and subsequent identification of candidate genes. Implementation of SNP arrays in clinical diagnostics allows the clinician to take advantage of this approach in their daily practice, especially because these data could be combined with clinically available whole-exome sequencing approaches. Clinicians can also utilize SNP genotyping information to devise a cost-effective strategy for determining the molecular etiology of genetically heterogeneous autosomal recessive diseases (e.g., deafness, retinitis pigmentosa, etc) by targeting sequence analysis for a suspected disease gene if it is located in an AOH block. Such was the case for the patients identified with SCID and Usher syndrome (Table 4).
The CMA-COMP array also has the advantage of detecting intragenic copy number changes. While intragenic deletions and duplications are known to account for a significant portion of disease-causing mutations for genes such as DMD, only more recently with the implementation of array CGH are intragenic deletions and duplications being detected for many other genes. Examples of haploinsufficient, intragenic deletions and the molecular mechanism and possible consequences of these intragenic CNVs have been previously reported.4, 12, 13, 16, 35, 36 Detection of intragenic copy number changes also has a role in the diagnosis of autosomal recessive disease, as demonstrated by the case with an intragenic heterozygous deletion of the VPS13B gene (case 7 in Table 4), which is associated with Cohen syndrome. Consistently, deletions have been reported as an important cause of Cohen syndrome.37
Interestingly, genotype–phenotype correlations are being described for deletions occurring at different locations within a gene. For example, the more C-terminal deletions, such as case 1 (Table 5), including those affecting the beta isoform of Neurexin-1, present with an increased head size and a high frequency of epilepsy (88%) when compared with more N-terminal deletions of NRXN1, as seen in cases 2 and 3 (Table 5). Therefore, increasingly specific genotype–phenotype predictions are becoming available to assist with the genetic counseling of the copy number changes identified by array studies.28, 35
However, genetic counseling can also become complicated by incidental findings, such as the intragenic deletions within MSH6 in case 4 and BRCA2 in case 8 (Table 5), which confers an increased life-time susceptibility to cancer in a 13-year-old female and a 10-year-old male, respectively. These findings require careful counseling to address prognosis and surveillance. These findings also have obvious implications for other family members that may also have inherited the CNV. While unexpected, these findings can have a profound impact on optimizing medical management and prevention for the whole family. As intragenic deletions of these genes have been observed,36, 38 it is expected that incidental findings such as these will occur as arrays with targeted exonic coverage are increasingly used in clinical practice.
In conclusion, combining both array CGH and SNP genotyping in a single platform optimizes the clinical diagnostic capability by offering the simultaneous detection of copy number neutral changes and small intragenic copy number changes.
Drs Wiszniewska, Bi, Shaw, Stankiewicz, Lalani, Probst, Scaglia, Beaudet, Lupski, Eng, Cheung, Bacino, Patel are the faculty of the Department of Molecular and Human Genetics at Baylor College of Medicine, which derives revenue from molecular genetic testing offered in the Medical Genetics Laboratory. Dr Lupski is a paid consultant for Athena Diagnostics, holds stock ownership in 23andMe, Inc. and Ion Torrent Systems, Inc., and is a co-inventor on multiple United States and European patents related to molecular diagnostics. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Boone PM, Liu P, Zhang F, et al. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet Med. 2011;13:582–592. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudet AL, Belmont JW. Array-based DNA diagnostics: let the revolution begin. Annu Rev Med. 2008;59:113–129. doi: 10.1146/annurev.med.59.012907.101800. [DOI] [PubMed] [Google Scholar]
- Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39 ((7 Suppl)::S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- McCarroll SA. Copy number variation and human genome maps. Nat Genet. 2010;42:365–366. doi: 10.1038/ng0510-365. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev. 2007;17:182–192. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Brain copy number variants and neuropsychiatric traits. Biol Psychiatr. 2012;72:617–619. doi: 10.1016/j.biopsych.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Bacino CA, Shaw CA, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AC, Dossett CJ, Walton CS, et al. Exon deletions of the EP300 and CREBBP genes in two children with Rubinstein-Taybi syndrome detected by aCGH. Eur J Hum Genet. 2011;19:43–49. doi: 10.1038/ejhg.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19:1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw N, Hehir-Kwa JY, Simons A, et al. SNP array analysis in constitutional and cancer genome diagnostics—copy number variants, genotyping and quality control. Cytogenet Genome Res. 2011;135:212–221. doi: 10.1159/000331273. [DOI] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger AL, Abdel-Rahman R, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altug-Teber O, Dufke A, Poths S, et al. A rapid microarray based whole genome analysis for detection of uniparental disomy. Hum Mutat. 2005;26:153–159. doi: 10.1002/humu.20198. [DOI] [PubMed] [Google Scholar]
- Papenhausen P, Schwartz S, Risheg H, et al. UPD detection using homozygosity profiling with a SNP genotyping microarray. Am J Med Genet A. 2011;155A:757–768. doi: 10.1002/ajmg.a.33939. [DOI] [PubMed] [Google Scholar]
- Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6:137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- Spence JE, Perciaccante RG, Greig GM, et al. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet. 1988;42:217–226. [PMC free article] [PubMed] [Google Scholar]
- Robinson WP, Christian SL, Kuchinka BD, et al. Somatic segregation errors predominantly contribute to the gain or loss of a paternal chromosome leading to uniparental disomy for chromosome 15. Clin Genet. 2000;57:349–358. doi: 10.1034/j.1399-0004.2000.570505.x. [DOI] [PubMed] [Google Scholar]
- Kotzot D. Complex and segmental uniparental disomy updated. J Med Genet. 2008;45:545–556. doi: 10.1136/jmg.2008.058016. [DOI] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, et al. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Soens ZT, Campbell IM, et al. Incidental copy-number variants identified by routine genome testing in a clinical population. Genet Med. 2012;15:45–54. doi: 10.1038/gim.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, et al. Diagnostic interpretation of array data using public databases and internet sources. Hum Mutat. 2012;33:930–940. doi: 10.1002/humu.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Amero SN, Ali Z, Abu-Amero KK, Stanier P, Moore GE. An analysis of common isodisomic regions in five mUPD 16 probands. J Med Genet. 1999;36:204–207. [PMC free article] [PubMed] [Google Scholar]
- Burrage LC, Person RE, Flores A, et al. De novo interstitial duplication of 15q11.2-q13.1 with complex maternal uniparental trisomy for the 15q11-q13 region in a patient with Prader-Willi syndrome. Am J Med Genet A. 2012;158A:2557–2563. doi: 10.1002/ajmg.a.35549. [DOI] [PubMed] [Google Scholar]
- Haraksingh RR, Abyzov A, Gerstein M, Urban AE, Snyder M. Genome-wide mapping of copy number variation in humans: comparative analysis of high resolution array platforms. PLoS One. 2011;6:e27859. doi: 10.1371/journal.pone.0027859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno DL, White SM, Ganesamoorthy D, et al. Pathogenic aberrations revealed exclusively by single nucleotide polymorphism (SNP) genotyping data in 5000 samples tested by molecular karyotyping. J Med Genet. 2011;48:831–839. doi: 10.1136/jmedgenet-2011-100372. [DOI] [PubMed] [Google Scholar]
- Schaaf CP, Boone PM, Sampath S, et al. Phenotypic spectrum and genotype-phenotype correlations of NRXN1 exon deletions. Eur J Hum Genet. 2012;20:1240–1247. doi: 10.1038/ejhg.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klift H, Wijnen J, Wagner A, et al. Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2, MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC) Genes Chromosomes Cancer. 2005;44:123–138. doi: 10.1002/gcc.20219. [DOI] [PubMed] [Google Scholar]
- Balikova I, Lehesjoki AE, de Ravel TJ, et al. Deletions in the VPS13B (COH1) gene as a cause of Cohen syndrome. Hum Mutat. 2009;30:E845–E854. doi: 10.1002/humu.21065. [DOI] [PubMed] [Google Scholar]
- Judkins T, Rosenthal E, Arnell C, et al. Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer. 2012;118:5210–5216. doi: 10.1002/cncr.27556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.