Abstract
J-wave syndromes have been associated with increased risk of ventricular fibrillation and sudden cardiac death. Previous studies have identified the KCNJ8-S422L variant in heterozygous form in individuals with J-wave syndromes. Its absence in over 1500 controls, coupled with in vitro analysis, have led to the conclusion that S422L is pathogenic. We previously performed whole-genome sequencing in a family quartet of Ashkenazi Jewish decent with no history of J-wave syndrome. Re-examination of these data reveals that both parents are heterozygous for the S422L variant, while the 12-year old son carries two copies – thus representing the first reported case of a S422L homozygote. In order to examine whether the S422L mutation might segregate at appreciable frequencies in specific populations, we genotyped the variant in a panel consisting of 722 individuals from 22 European, Middle Eastern non-Jewish, Ashkenazi Jewish, and non-Ashkenazi Jewish populations. We found that the S422L allele was at a significantly higher frequency in Ashkenazi Jews (∼4%) compared with other populations in our survey, which have frequencies <0.25%. We also performed ECGs in both male members of the family quartet. The homozygous boy demonstrated no clinically significant ECG abnormalities, while the heterozygous father presented with a subtle J-wave point elevation. Our results suggest that either (a) previous studies implicating S422L as pathogenic for J-wave syndromes failed to appropriately account for European population structure and the variant is likely benign, or (b) Ashkenazi Jews may be at significantly increased risk of J-wave syndromes and ultimately sudden cardiac death.
Keywords: J-wave syndromes, sudden cardiac arrest, idiopathic ventricular fibrillation, early repolarization syndromes, Brugada syndrome
INTRODUCTION
The elevation in the QRS-ST junction of an electrocardiogram (ECG) that signifies early repolarization (ER) is commonly referred to as a J-wave or Osborn wave. While once thought to be a benign abnormality found in healthy young men and athletes, individuals with J-waves have now been shown in a number of studies to be at increased risk of idiopathic ventricular fibrillation (VF) and sudden cardiac death.1, 2 Depending on the spatial localization and magnitude of the J-wave, individuals may be placed into a series of J-wave syndromes such as one of three early repolarization syndromes (ERS) and Brugada syndrome (BRS).3 A number of genes have now been associated with J-wave syndromes, with the vast majority being genes that encode for ion channels such as SCN5A and CACNA1C.4
Three separate studies have now implicated the gene KCNJ8 in individuals with J-wave syndromes.5, 6, 7 Remarkably, they all reported finding a single mutation at position c.C1265T (chr 12:g.21918667, rs72554071) that leads to a serine to leucine substitution at amino acid 422 (p.S422L). Among two cohorts of predominantly recent European ancestry with J-wave syndromes (N=305), the S422L mutation was found in six patients – all in heterozygous form (four with BRS, two with ERS) (2%). In contrast, the S422L variant was absent in 815 controls (again of predominantly European ancestry).6, 7 KCNJ8 encodes the ATP-sensitive inwardly rectifying potassium channel Kir6.18 that is expressed at high levels in cardiomyocytes.9, 10, 11 In vitro functional analysis of the S422L variant suggests it causes an increased KATP current compared with the wild-type Kir6.1 channel, possibly because of incomplete channel closing. These results have implicated S422L, through a gain-of-function action, as pathogenic in J-wave syndromes and the KCNJ8 gene as a susceptibility factor for VF.6, 7 More recently, the S422L mutation was also found in two cases from a cohort of 311 individuals with atrial fibrillation (AF) (both patients also had evidence of ER) and was again absent amongst 368 controls.12
In this study, we report the first case of a homozygous S422L variant, which we found through whole-genome sequencing (WGS) a family quartet of Ashkenazi Jewish background. We go on to demonstrate that the S422L is at substantially higher frequencies in Ashkenazi Jews in general, which has important ramifications for the mutation's status as a pathogenic variant for J-wave syndromes.
METHODS
Subjects
We examined WGS data generated by Complete Genomics Inc from a quartet previously described in Veeramah et al,13 where the initial proband presented with a severe epileptic encephalopathy (EE) and sudden unexplained death in epilepsy (SUDEP). There was no family history of cardiac abnormalities. Informed consent was obtained from the family quartet, and the study was approved by the University of Arizona institutional review board. The 12-year-old second-born male sibling did not show evidence of epilepsy nor any major cognitive or motor deficits. An ECG was performed on the male sibling in 2011 and on the father in 2012. Standard 12-lead ECGs were obtained in the resting, supine position. Recordings were performed using standard voltage at a paper speed of 25 mm/s using commercially available equipment (Marquette Electronics, Milwaukee, WI, USA; Cardiac Sciences Corporation, Bothell, WA, USA). Additional ECG recordings were obtained with the V1 and V2 leads placed more superiorly at the second intercostal space.14 Repeat ECGs were performed in 2013 on the male sibling, including one following a Valsalva maneuver.
In addition, we examined 722 samples from 22 populations (Table 1) for the presence of the S422L allele. All samples were anonymous, unrelated and gave appropriate informed consent.
Table 1. Genotype counts and allele frequencies for the S422L variant in European, Middle Eastern, Jewish non-Ashkenazi and Ashkenazi Jewish populations.
| KCNJ8 genotype | |||||
|---|---|---|---|---|---|
| Country | N | CC | CT | TT | T allele frequency (%) |
| Europe | |||||
| France | 66 | 66 | 0 | 0 | 0.00 |
| Germany | 10 | 10 | 0 | 0 | 0.00 |
| Netherland | 68 | 68 | 0 | 0 | 0.00 |
| Poland | 27 | 27 | 0 | 0 | 0.00 |
| Romania | 27 | 27 | 0 | 0 | 0.00 |
| Russia | 24 | 22 | 0 | 0 | 0.00 |
| Ukraine | 40 | 40 | 0 | 0 | 0.00 |
| Total | 262 | 262 | 0 | 0 | 0.00 |
| Middle East | |||||
| Iran | 42 | 42 | 0 | 0 | 0.00 |
| Turkey | 8 | 8 | 0 | 0 | 0.00 |
| Total | 50 | 50 | 0 | 0 | 0.00 |
| Jewish non-Ashkenazi | |||||
| Iran | 20 | 20 | 0 | 0 | 0.00 |
| Iraq | 26 | 26 | 0 | 0 | 0.00 |
| Turkey | 25 | 25 | 0 | 0 | 0.00 |
| Italy (Rome) | 48 | 45 | 3 | 0 | 3.13 |
| Total | 119 | 116 | 3 | 0 | 1.26 |
| Ashkenazi Jewish | |||||
| France | 35 | 29 | 6 | 0 | 8.57 |
| Germany | 9 | 8 | 1 | 0 | 5.56 |
| Hungary | 8 | 8 | 0 | 0 | 0.00 |
| Netherlands | 16 | 15 | 1 | 0 | 3.13 |
| Poland | 70 | 67 | 3 | 0 | 2.14 |
| Romania | 19 | 19 | 0 | 0 | 0.00 |
| Russia | 8 | 8 | 0 | 0 | 0.00 |
| Ukraine | 25 | 22 | 3 | 0 | 6.00 |
| USA | 101 | 92 | 9 | 0 | 4.46 |
| Total | 291 | 268 | 23 | 0 | 3.95 |
S422L genotyping
The presence of the S422L (rs72554071) variant in the quartet was confirmed by Sanger sequencing (Supplementary Figure 1). Additional genotyping of the larger sample was assessed using an allele-specific PCR protocol. The following primers were used to amplify the 302-base pair allele-specific band and the 442-bp control band: KCNJ8-U: 5′-TTGAGAGAAACGAGCATACC-3′, KCNJ8-AL: 5′-CAGAAGGAAATCAAAACACATT-3′, KCNJ8-L: 5′-CCTCCAAAAGAGTGAACTGTC-3′. Five nanogram of genomic DNA was amplified in 15 μl final volume containing 0.2 mM of each dNTP, 1 μM, 0.6 μM, 1.4 μM of KCNJ8-U, KCNJ8-L and KCNJ8-AL primers, respectively, 0.5 U of Taq DNA polymerase (Invitrogen), 1.5 mM MgCl2, 75 mM KCl, and 10 mM Tris-HCl (pH 8.3). The cycling conditions were 94 °C for 3 min, followed by 35 cycles at 94 °C for 30 s, 59 °C for 30 s, 72 °C for 45 s, with final extension step at 72 °C for 3 min The genotypes were checked by running PCR products on a 2% agarose gel in TBE buffer (Supplementary Figure 2). All samples showing an allele-specific band (ie, potentially heterozygote for the S422L allele) were verified by sequencing.
Statistical analysis
Whole-genome sequencing variant analysis was performed as previously described in Veeramah et al.13 Briefly, after read mapping and local de novo assembly, a nucleotide position is determined based on a previously calibrated minimum confidence score (20 dB for homozygotes and 40 dB for heterozygotes). Variant call quality was also increased using a hidden Markov model that uses Mendelian inheritance rules.15 Annotation was performed using ANNOVAR16 based on Hg19, and custom tools were used to extract particular inheritance patterns within the quartet (eg, potentially recessive variants). Mutational impact was assessed using Polyphen 2.17 Identity-By-Descent (IBD) Pi_Hat was estimated using PLINK.18 All other statistical analysis was performed using the ‘R' statistical package.
RESULTS
Identification of a homozygous S422L individual
We previously performed WGS on a family quartet and identified a pathogenic mutation in the gene SCN8A in a patient with severe EE who passed away from SUDEP. Further examination of the WGS revealed that both parents and the female proband were heterozygotes for the KCNJ8-S422L variant, while the healthy 12-year-old younger male sibling was a homozygote for this variant. These genotypes were successfully validated using an allele-specific assay, direct Sanger sequences, and whole-exome sequencing (WES) using two different capture technologies (Agilent and Illumina). No other variants from the human reference genome (Hg19) were found in the exons or splice sites of KCNJ8 amongst the family.
Both parents were of self-identified Ashkenazi Jewish origin, but otherwise unrelated, with an estimated pairwise IBD Pi_Hat value of 0.023 compared with an average of 0.025 for any two random Ashkenazi Jews chosen from Behar et al.19
S422L population screening
Given the presence of S422L in a family of Ashkenazi Jewish ethnicity, including the first ever identification of a homozygous individual, we genotyped the variant in a panel of individuals from Ashkenazi Jewish, other Jewish, European and Middle Eastern populations (Table 1). S422L was absent in almost all populations except Ashkenazi Jews with 23 out of 291 (7.9%) heterozygous individuals, no homozygous individuals and an overall allele frequency of 4.0%. The only exception to this was the presence of 3 S422L heterozygotes amongst 48 Roman Jews. Further subdivision of Ashkenazi Jews by geographic location demonstrated that the S422L allele was present across Europe, with the first and second highest allele frequencies found in France (8.6%) and the Ukraine (6.0%) respectively, while also being prevalent in Ashkenazi Jews from the USA (4.5%). Interestingly, the first study to identify the mutation was conducted in France and the authors failed to identify any S422L alleles in 764 additional controls.5
The allele frequency in Ashkenazi Jews was significantly higher than that of other non-Jewish populations examined here (P<0.037 to <<0.05, as assessed using a Fisher's Exact Test), as well as in previously examined controls (P<<0.001). The variant was also identified in heterozygous form in 16/3510 European Americans and 1/1869 African American individuals (allele frequency=0.23 % and 0.03%, respectively), for whom WES was performed by the Exome Sequencing Project (ESP).20 The project consists of individuals from multiple sets of cohorts that may include individuals with heart, lung and blood disorders, though the exact composition of the data set was not available to us. Again the frequency in Ashkenazi Jews was significantly higher (P<<0.001) than the ESP data sets.
ECG results
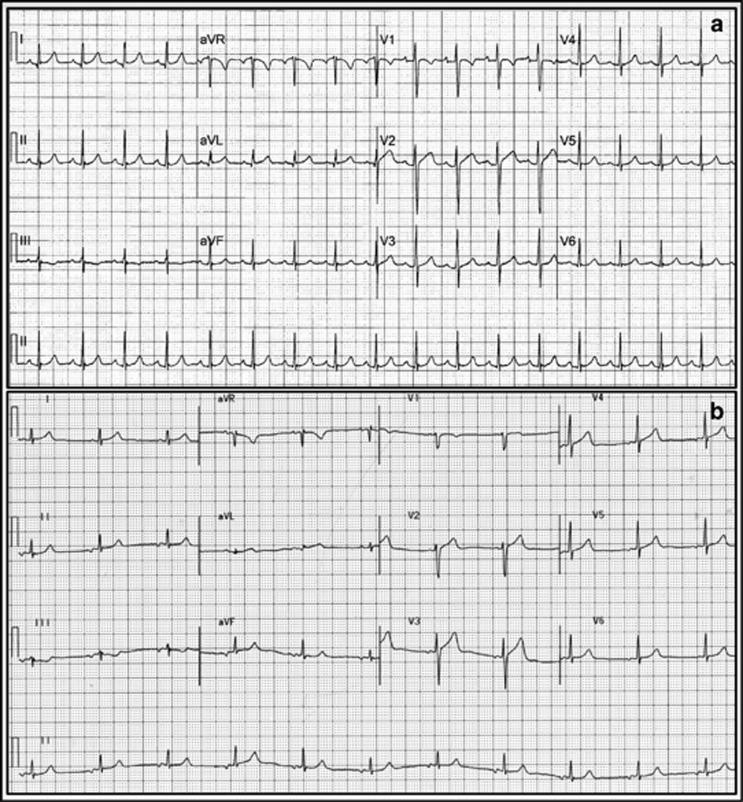
ECGs were performed on the S422L homozygous male sibling and his father. An initial ECG on the homozygous individual (Figure 1a) demonstrated normal sinus rhythm at a rate of 111 beats per minute (bpm) with a normal PR interval of 142 ms, a normal QRS duration of 74 ms and a QT interval of 307 ms (QTc=406, by Bazett's formula). There were no definite abnormalities of the ST–T wave segments. There was a slight fractionation of the terminal QRS in lead II.
Figure 1.
Initial ECG of (a) S422L homozygous sibling and (b) heterozygous father respectively. Twelve-lead ECGs obtained with precordial leads in the standard position. Notch filter set at 60 Hz.
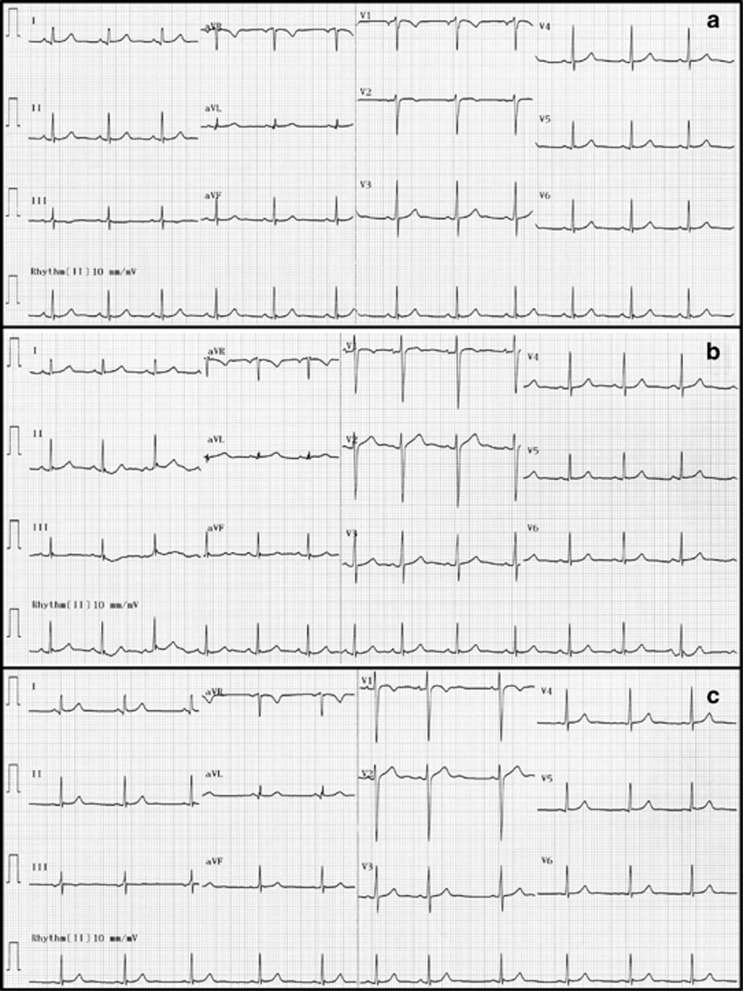
A set of three ECGs were then repeated on this individual at a later date. A 12-lead ECG timed at 0747 hours (with V1 and V2 leads placed at the left and right second intercostal spaces, respectively) showed normal sinus rhythm at a rate of 73 bpm (Figure 2a). The PR interval was normal, measuring 146 ms, the QRS duration was normal at 76 ms, as was the QT interval of 376 ms (QTc=401). There were no definite abnormalities of the ST–T wave segments. There was subtle fractionation of the terminal QRS in leads II, III and avF. A 12-lead ECG of the homozygous individual timed at 0748 hours (with V1 and V2 leads in the standard position) showed a normal sinus rhythm at a rate of 81 bpm (Figure 2b). Baseline artifact secondary to motion was apparent in the first four beats of the recording. The PR, QRS and QTc intervals were normal (140, 82 and 409 ms, respectively). Once again, no definite abnormalities of the ST–T wave segments were noted. Subtle fractionation of the terminal QRS in leads II, III and aVF was again seen. In comparison with the previous ECG (with leads V1, V2 at the left and right second intercostal spaces), there was a suspicion of early repolarization in the V1 and V2.
Figure 2.
Repeat ECGs for the S422L homozygous sibling. (a) Twelve-lead ECG with V1 and V2 leads placed at the left and right second intercostal spaces, respectively. (b) 12-lead ECG with V1 and V2 leads in the standard position. (c) 12-lead ECG obtained following Valsalva maneuver.
A repeat ECG timed at 0754 hours was obtained following a Valsalva maneuver in order to slow the heart rate and potentially unmask further repolarization abnormalities (Figure 2c). This showed sinus arrhythmia at an overall rate of 62 bpm. The PR, QRS and QTc intervals were normal (120, 84 and 389 ms, respectively). Compared with the baseline ECG, subtle fractionation of the terminal QRS in leads II, III and aVF was again apparent but not different from baseline. Of note, there was a very slight (<1 mm) J-point elevation seen in V1 and V2. However, overall there was no evidence of clinically significant abnormalities for this S422L homozygous individual.
The ECG of the father (Figure 1b) demonstrated a low right atrial rhythm, rate 64 bpm with a normal PR interval 0f 158 ms, normal QRS duration of 82 ms and normal QT interval of 358 ms (QTc=375). On careful inspection, there was subtle early repolarization with ∼1–2 mm J-point elevation in leads V2 and V3. There were no differences between the standard ECG and those with leads V1 and V2 placed superiorly at the second intercostal space.
DISCUSSION
Given the finding of a significantly increased KCNJ8-S422L frequency in Ashkenazi Jews, it will be important to establish whether or not previous results reporting the frequency distribution of the S422L mutation that are the basis for the suggestion that it is a risk variant for J-wave syndromes (and ultimately sudden cardiac death) are simply an artifact of cryptic population stratification. Population genetic studies have demonstrated that it is important to take into account Ashkenazi Jewish genetic ancestry for disease association studies involving European populations.21 That no S422L mutations have been identified thus far in control populations may represent a lack of representation of individuals of Ashkenazi Jewish descent compared with the J-wave and AF cohorts, with the current description of control cohorts not going beyond ‘European Caucasian'.6 It is noteworthy that Polyphen 2 analysis results in a benign score of 0.013 for the S422L variant with multiple alignments showing amino-acid differences in non-human species such as hedgehog, lizard, and zebrafish.
On the other hand, in vitro analysis has presented compelling evidence that the mutation does affect channel function.6, 7 Thus, an alternative explanation to cryptic population structure would be that the S422L mutation is pathogenic and that therefore Ashkenazi Jews may have increased risk of J-wave syndromes. Though no data are currently available that would allow us to assess this hypothesis, an increased risk is certainly not unlikely given the generally higher rates of certain diseases in this population because of historical founder effects.22 While Barajas-Martinez et al7 have suggested that S422L is a hotspot for mutation, no case of a de novo S422L mutation has actually been explicitly observed, and simple proliferation of the risk allele from Ashkenazi Jews to other populations through gene flow would likely be able to explain the current distribution. Interestingly, the presence of the allele in Roman Jews, one of the longest surviving Jewish communities in Europe, but not Middle Eastern Jews suggests an origin of the S422L allele sometime in Europe ∼2000 years ago.
The results from our quartet, while useful, do not currently help distinguish between pathogenicity and non-pathogenicity of S422L. The male sibling is the first reported example of a homozygous S422L mutation and thus may be expected to demonstrate a severe J-wave phenotype if the mutation is gain-of-function causal; yet a suite of ECGs showed no obvious abnormalities. However, low penetrance and epistatic interactions may be acting and it is important to appreciate the effect, given his age, that puberty and testosterone are likely to play in J-wave syndromes.23 Furthermore, the heterozygous father demonstrates a slight but inconclusive J-point abnormality. Therefore, it would be of interest to explore the J-wave phenotype in the extended family of this quartet as well as additional samples of Ashkenazim, both with and without J-wave phenotypes, in the future.
Acknowledgments
We thank Dr Charles Antzelevitch for helping with ECG interpretation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Nam GB, Kim YH, Antzelevitch C. Augmentation of J waves and electrical storms in patients with early repolarization. N Engl J Med. 2008;358:2078–2079. doi: 10.1056/NEJMc0708182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haïssaguerre M, Derval N, Sacher F, et al. Sudden cardiac arrest associated with early repolarization. N Engl J Med. 2008;358:2016–2023. doi: 10.1056/NEJMoa071968. [DOI] [PubMed] [Google Scholar]
- Antzelevitch C, Yan GX. J wave syndromes. Heart Rhythm. 2010;7:549–558. doi: 10.1016/j.hrthm.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzelevitch C. Genetic, molecular and cellular mechanisms underlying the J wave syndromes. Circ J. 2012;76:1054–1065. doi: 10.1253/circj.cj-12-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haïssaguerre M, Chatel S, Sacher F, et al. Ventricular fibrillation with prominent early repolarization associated with a rare variant of KCNJ8/K ATP channel. J Cardiovasc Electrophysiol. 2009;20:93–98. doi: 10.1111/j.1540-8167.2008.01326.x. [DOI] [PubMed] [Google Scholar]
- Medeiros-Domingo A, Tan BH, Crotti L, et al. Gain-of-function mutation S422L in the KCNJ8-encoded cardiac KATP channel Kir6.1 as a pathogenic substrate for J-wave syndromes. Heart Rhythm. 2010;7:1466–1471. doi: 10.1016/j.hrthm.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-Martinez H, Hu D, Ferrer T, et al. Molecular genetic and functional association of Brugada and early repolarization syndromes with S422L missense mutation in KCNJ8. Heart Rhythm. 2012;9:548–555. doi: 10.1016/j.hrthm.2011.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Inazawa J, Seino S. cDNA sequence, gene structure, and chromosomal localization of the human ATP-sensitive potassium channel, uKATP-1, gene (KCNJ8) Genomics. 1995;30:102–104. doi: 10.1006/geno.1995.0018. [DOI] [PubMed] [Google Scholar]
- Wu SN, Wu AZ, Sung RJ. Identification of two types of ATP-sensitive K+ channels in rat ventricular myocytes. Life Sci. 2007;80:378–387. doi: 10.1016/j.lfs.2006.09.042. [DOI] [PubMed] [Google Scholar]
- Morrissey A, Rosner E, Lanning J, et al. Immunolocalization of KATP channel subunits in mouse and rat cardiac myocytes and the coronary vasculature. BMC Physiol. 2005;5:1. doi: 10.1186/1472-6793-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Suzuki M, Shibasaki T, et al. Mouse model of Prinzmetal angina by disruption of the inward rectifier Kir6.1. Nat Med. 2002;8:466–472. doi: 10.1038/nm0502-466. [DOI] [PubMed] [Google Scholar]
- Delaney JT, Muhammad R, Blair MA, et al. A KCNJ8 mutation associated with early repolarization and atrial fibrillation. Europace. 2012;14:1428–1432. doi: 10.1093/europace/eus150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah KR, O'Brien JE, Meisler MH, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Yokokawa M, Tanaka K, et al. Diagnostic and prognostic value of a type 1 Brugada electrocardiogram at higher (third or second) v1 to v2 recording in men with Brugada syndrome. Am J Cardiol. 2007;99:53–57. doi: 10.1016/j.amjcard.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Smit AFA, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Yunusbayev B, Metspalu M, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- Exome Variant Server. NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA. Available at: http://evs.gs.washington.edu/EVS/ .
- Price AL, Butler J, Patterson N, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Tang H, Katzenstein H, Ekstein J. Geographic distribution of disease mutations in the Ashkenazi Jewish population supports genetic drift over selection. Am J Hum Genet. 2003;72:812–822. doi: 10.1086/373882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki K, Nakagawa M, Taniguchi Y, et al. Gender differences in the ST segment: effect of androgen-deprivation therapy and possible role of testosterone. Circ J. 2010;74:2448–2454. doi: 10.1253/circj.cj-10-0221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.