Abstract
The 1100delC mutation in the CHEK2 gene has a carrier frequency of up to 1.5% in individuals from North-West Europe. Women heterozygous for 1100delC have an increased breast cancer risk (odds ratio 2.7). To explore the prevalence and clinical consequences of 1100delC homozygosity in the Netherlands, we genotyped a sporadic breast cancer hospital-based cohort, a group of non-BRCA1/2 breast cancer families, and breast tumors from a tumor tissue bank. Three 1100delC homozygous patients were found in the cohort of 1434 sporadic breast cancer patients, suggesting an increased breast cancer risk for 1100delC homozygotes (odds ratio 3.4, 95% confidence interval 0.4–32.6, P=0.3). Another 1100delC homozygote was found in 592 individuals from 108 non-BRCA1/2 breast cancer families, and two more were found after testing 1706 breast tumors and confirming homozygosity on their wild-type DNA. Follow-up data was available for five homozygous patients, and remarkably, three of them had developed contralateral breast cancer. A possible relationship between 1100delC and lung cancer risk was investigated in 457 unrelated lung cancer patients but could not be confirmed. Due to the small number of 1100delC homozygotes identified, the breast cancer risk estimate associated with this genotype had limited accuracy but is probably higher than the risk in heterozygous females. Screening for CHEK2 1100delC could be beneficial in countries with a relatively high allele frequency.
Keywords: CHEK2, 1100delC homozygotes, genotype risk, bilateral breast cancer, lung cancer
Introduction
The CHEK2 gene on chromosome 22 is a tumor-suppressor gene encoding the protein kinase CHEK2, which is involved in cell-cycle control and DNA repair in response to DNA double-strand breaks.1, 2, 3 A deletion of a cytosine at position 1100 (1100delC) in CHEK2 can be found in individuals originating from North-West Europe, with a heterozygous carrier frequency of up to 1.5% in some populations. In other geographical areas the mutation is much rarer or even absent.3, 4, 5 The 1100delC frameshift mutation causes a premature stop codon, which triggers nonsense-mediated decay, resulting in a lower expression of CHEK2 mRNA in heterozygous carriers.6, 7, 8 Although a low level of mutant mRNA remains detectable, the presence of the mutated protein could not be demonstrated in lymphoblastoid cell lines from humans heterozygous for the 1100delC mutation.9
Heterozygous female carriers of the 1100delC mutation have an increased breast cancer risk, with an odds ratio (OR) of 2.7 (95% confidence interval (95% CI): 2.1–3.4) in sporadic breast cancer cases and an OR of 4.8 (95% CI: 3.3–7.2) in familial breast cancer cases.3, 10, 11, 12, 13 Breast cancer patients heterozygous for 1100delC also have an increased risk of developing contralateral breast cancer when compared with wild-type breast cancer patients.5, 13, 14, 15, 16, 17, 18, 19, 20 The contralateral breast cancer risk may be even higher when radiotherapy has been given to treat the first tumor.15, 19 It must be noted that in women who also carry a pathogenic BRCA1/2 mutation the 1100delC allele does not seem to modify breast cancer risk.3, 14, 21
An association of the 1100delC allele with increased colon cancer and prostate cancer risk has been described, while an association with melanoma could not be found.22, 23, 24, 25 Other variants in CHEK2 have been associated with lung cancer, but a possible association of 1100delC with lung cancer has never been investigated.26, 27, 28, 29, 30, 31
Homozygosity for 1100delC is expected to be rare, and until recently, there had only been two reports on homozygous carriers, a male who developed colon cancer at age 52 years32 and a female who developed bilateral breast cancer at ages 47 and 61 years and uterine sarcoma at age 58 years.33 Recently, Adank et al.34 reported 12 homozygous 1100delC mutation carriers from 8 breast cancer families from the Netherlands. All 10 female 1100delC homozygotes had developed breast cancer. All these cases were derived from selected patient cohorts, mostly on the basis of family history, and hence do not allow conclusions to be drawn about breast cancer risk for 1100delC homozygous women in the general population.
To further explore the prevalence and clinical consequences of 1100delC homozygosity in humans in a geographical area with a high allele frequency, we genotyped a hospital-based sporadic breast cancer cohort from the South-West region of the Netherlands and breast tumors from the Rotterdam tumor tissue bank. Genotyping was extended in a previously reported cohort of non-BRCA1/2 breast cancer families.11 The phenotypes of homozygous 1100delC carriers were studied.
Materials and methods
Cohorts
The ORIGO cohort is a Dutch hospital-based cohort of 1434 breast cancer patients diagnosed with a first primary breast cancer between 1996 and 2005 in two academic hospitals in the South-West area of the Netherlands.17, 35 Patients were included regardless of family history of breast cancer. Average age at diagnosis was 53.4 years (SD 11.2 years). A subset of the ORIGO cohort had been genotyped for the 1100delC mutation before17 but with a genotyping technique that precluded the identification of homozygous 1100delC individuals.
Female family members (n=592) from 108 non-BRCA1/2 breast cancer families (range 1–24 individuals per family) were ascertained through the Departments of Clinical Genetics in Leiden, Rotterdam and Nijmegen, as well as through the Netherlands Foundation for the Detection of Hereditary Tumors. Families were included if there were at least three cases of breast cancer diagnosed before the age of 60 years from whom genotypes could be determined or could be inferred by genotyping close relatives. Mutations in BRCA1 or BRCA2 were excluded in these families as described previously.11 A total of 325 affected and 267 unaffected females were genotyped for the 1100delC mutation.
The Rotterdam Medical Oncology Tumorbank (RMOT) has been described previously.36, 37 DNA samples from 1706 breast tumor specimens from the three study cohorts were available for analysis. The first series of 503 samples was drawn from a consecutive series of unselected breast cancer cases diagnosed in the year 1990, median age at diagnosis of these cases was 62.1 years (range 22.7–89.6 years). The second series (n=269) comprises cases who were diagnosed before 40 years of age, with exclusion of the early-onset cases from the first cohort. Median age at diagnosis was 36.2 years (range 22–40 years). The remaining cases (n=934) were drawn from ongoing studies on prognostic and predictive markers of breast cancer, median age at diagnosis was 59.3 years (range 27.4–85.8 years). Cases originated from several hospitals in the South-West area of the Netherlands and were recruited between 1978 and 1992, the year 1992 being chosen to allow for sufficient follow-up.36, 37
The lung cancer cohort of 457 lung cancer patients was collected as an extension of the POLYGENE project.38 Patients with lung cancer were identified through the population-based cancer registry of the Comprehensive Cancer Center IKO, Nijmegen, the Netherlands. Patients diagnosed in one of the three hospitals from the east of the Netherlands (Radboud University Medical Center and Canisius Wilhelmina Hospital in Nijmegen and Rijnstate Hospital in Arnhem) who were still alive on 15 April 2008 were recruited for a study on gene–environment interactions in lung cancer. Year of diagnosis was between 1989 and 2008, average age at diagnosis was 60.5 years (SD 9.1 years), and 278 patients (60.1%) were male.
Three control groups from the Southwestern part of the Netherlands were used. The first control group consists of 783 females from cystic fibrosis (CF) families who were either spouses of CF family members or CF family members tested negative for a mutation in the CFTR gene.39 Average age at blood draw was 43.3 years (SD 12.6 years). The second control population of 183 females had been referred for mutation testing for hereditary diseases not related to cancer syndromes, including Duchenne Muscular Dystrophy, Huntington's Disease and Hemophilia type A and had been tested negative. Average age at blood draw was 41.9 years (SD 16.0 years). The third group consists of 612 females between the ages of 35 and 70 years who donated blood at the blood bank in April and May of 2007. People with a diagnosis of invasive cancer are not allowed to donate blood in the Netherlands. Of these anonymous blood donors, only the age at blood collection is known, average age at blood draw was 54.7 years (SD 6.9 years).
All breast cancer patients from the ORIGO cohort, all included family members from non-BRCA1/2 families and all lung cancer patients gave informed consent to the use of their DNA for cancer research, and the ethical committee of the hospitals involved approved of the studies. Separate consent was not required for the retrospective RMOT study, which was approved by the medical ethical committee of the Erasmus Medical Center Rotterdam and which is in accordance with the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands. All controls were irreversibly anonymized.
DNA extraction to genotype RMOT cases
Genotyping of the RMOT cohort was first performed on DNA isolated from fresh-frozen tumor tissue, as described previously.37 If patients were found to be homozygous for the 1100delC allele in their tumor, genomic DNA was extracted from normal formalin-fixed paraffin-embedded tissue and was used for genotyping to study genomic 1100delC homozygosity.
DNA was extracted from normal formalin-fixed paraffin-embedded tissue using xylene and ethanol to deparaffinize the samples and proteinase K to extract the DNA.40, 41
Genotyping
Screening for 1100delC was performed in 384-well plates. In each 384-well plate, six different samples with known genotypes were present at random positions, two samples for each genotype. The results for these samples were always consistent.
To avoid genotyping errors caused by the presence of highly homologous CHEK2 pseudogenes in the genome, a nested PCR was performed. First a 537-bp fragment of CHEK2 was amplified using 20 ng of genomic DNA and primers specific for the CHEK2 gene (see Supplementary Table S1 for primers and Supplementary Table S2 for PCR conditions).42 Subsequently, 2.5 μl of the resulting PCR product was genotyped in an 8 μl reaction volume using a custom Taqman SNP genotyping assay according to the manufacturer's recommendations (Supplementary Table S1 and Applied Biosystems, Carlsbad, CA, USA).
For 12 individuals from the ORIGO cohort, 4 from the lung cancer cohort and 3 from the blood bank controls, genotyping failed repeatedly (PCR and Taqman analyses were repeated at least twice). These samples were then excluded from further analyses.
Whereas our previous genotyping technique had only allowed the identification of 1100delC carriers, the method used here allowed the distinction between heterozygous and homozygous carriers. To confirm the specificity of this procedure and of the Taqman probes, all 1100delC homozygous samples were sequenced as well as several heterozygous and wild-type samples, using the specific primers (Supplementary Table S1), 35 cycles of PCR (Supplementary Table S2) and Sanger sequencing. Sequencing always confirmed the genotypes as determined by the Taqman assay.
Statistics
PASW Statistics 17.0 (IBM Corporation, Armonk, NY, USA) was used for statistical calculations with the following exceptions.
For efficient estimation of breast cancer risk for heterozygous and homozygous carriers of the 1100delC allele, a combined method using all data simultaneously was applied.43 In short, the 1100delC genotype distribution was modeled assuming Hardy–Weinberg equilibrium and using genotype data from the cases, blood bank controls and other controls (with unknown disease status). A disease prevalence of 5% (for females with an average age of 50 years) was used to weigh the genotype distribution of cases and of controls. Repeating the analysis with other prevalence values between 1 and 10% showed that results were robust to prevalence misspecification. A logistic regression model based on breast cancer cases and blood bank controls (excluding the controls with unknown disease status) was used to estimate breast cancer risk for 1100delC carriers. In order to be able to compute the risk for 1100delC homozygous individuals, the number of homozygotes in the controls was modeled using the allele frequency. Calculations were made using R version 2.14.1. Further details on this combined approach are described in a separate document (see Supplementary Methods).
In the non-BRCA1/2 cohort, the presence of family relationships was taken into account by applying a generalized linear model (GLM) to estimate allele frequencies and ORs, using the geeglm function from the R package geepack.44 By using the family indicator as cluster (id) variable, SEs are valid in the case of within family correlations.
For power calculations regarding the CHEK2 1100delC allele frequencies in the lung cancer cohort and the control cohort, an online Statistical Power Calculator for two sample tests using percentage values was used (DSS Research, Fort Worth, TX, USA; http://www.dssresearch.com/toolkit/spcalc/power_p2.asp).
Results
CHEK2 1100delC homozygotes in sporadic breast cancer patients
In the cohort of 1422 sporadic breast cancer patients (ORIGO), 42 heterozygotes for the 1100delC variant were found, versus 19 heterozygotes among 1575 controls (Table 1). Thus, for heterozygotes the OR to develop breast cancer was 2.3 (joint likelihood model, 95% CI: 2.1–2.5), which is in concordance with previously reported breast cancer risk estimates for 1100delC heterozygous women.
Table 1. Overview of 1100delC genotypes in all the cohortsa.
| CHEK2 1100delC genotype (number of individuals) | ||||
|---|---|---|---|---|
|
Cohort |
Homozygous wild type |
Heterozygous |
Homozygous 1100delC |
Total |
| ORIGO cohort (sporadic breast cancer) | 1377 | 42 | 3 | 1422 |
| Non-BRCA1/2 breast cancer families | 562 from 108 families | 29 from 14 families | 1 | 592 from 108 families |
| Lung cancer cohort | 449 | 4 | 0 | 453 |
| Controls: from blood bank | 601 | 8 | 0 | 609 |
| Controls: individuals from CF families, unaffected by CF | 774 | 9 | 0 | 783 |
| Controls: individuals from families with hereditary diseases unrelated to cancer, not affected by the disease running in their family | 180 | 2 | 1 | 183 |
| Controls: total | 1555 | 19 | 1 | 1575 |
Abbreviation: CF, cystic fibrosis.
The RMOT (Rotterdam Medical Oncology Tumorbank) cohort is not mentioned in this table. It consists of several subgroups with different selection criteria.
Among the 1422 sporadic breast cancer patients in the ORIGO cohort, 3 1100delC homozygotes were observed, while 1 1100delC homozygote was identified among 1575 controls. This control was a woman who had been referred for mutation testing for hereditary diseases not related to cancer syndromes. She was 60 years old at blood draw. Information on her medical history was not accessible, thus her breast cancer status is unknown. Based on all controls, the OR to develop breast cancer for CHEK2 1100delC homozygotes was 3.4 (binary logistic regression, 95% CI: 0.4–32.6, P=0.3).
However, if only controls are included of whom we are certain that they have not had a diagnosis of breast cancer, only the 609 blood bank donors remain and there are no 1100delC homozygotes present in the control group. Applying a combined approach, which simultaneously estimates the allele frequency and genotypic ORs, the 1100delC allele frequency was estimated to be 0.007 (95% CI: 6.9 × 10−3–7.0 × 10−3), and the expected number of homozygotes in the 609 controls was thus 0.03. The OR for homozygotes was then estimated to be 44.3 (joint likelihood model, 95% CI: 11.3–174.2).
CHEK2 1100delC homozygotes in non-BRCA1/2 families
A subset of 71 of our non-BRCA1/2 breast cancer families had been genotyped for 1100delC before,11 using a technique which could not discriminate between heterozygous and homozygous carriers of 1100delC. Therefore, we now re-examined and expanded the previously reported genotyping results in a cohort of 592 women from 108 non-BRCA1/2 breast cancer families. The current analysis confirmed all previously detected carriers of the 1100delC mutation as heterozygotes, except for 1 carrier now being identified as homozygous 1100delC. This individual was a breast cancer patient from a family with four other breast cancer patients (one of whom was heterozygous for 1100delC, the others were wild type). Using a GLM to adjust for the family relationships present in the cohort, the frequency of homozygotes in unrelated individuals from non-BRCA1/2 breast cancer families was estimated to be 0.2% (geeglm, 95% CI: 0%–0.5%). This genotype frequency is comparable with the frequency in the sporadic breast cancer cohort (3 of 1422 patients).
The phenotype of homozygous 1100delC carriers
The three 1100delC homozygous patients from the sporadic breast cancer cohort (ORIGO) were 40, 52 and 55 years at breast cancer diagnosis, while the average age at breast cancer diagnosis was 56.1 years for the wild-type patients from the same cohort. The homozygous female in the non-BRCA1/2 cohort was 56 years at diagnosis. Two more 1100delC homozygotes were identified in the RMOT cohort, diagnosed with breast cancer at ages 60 and 62 years, respectively.
Histology of the primary breast cancer was invasive ductal carcinoma in five patients, while one patient had an atypical medullary carcinoma. The average size of the tumor was 2.3 cm (range 1.7–3.0 cm). Lymph node status was known for five patients and was positive in four and negative in one patient. For three patients, the estrogen receptor (ER) status was known, all being ER positive. The ER status of the atypical medullary carcinoma was not determined.
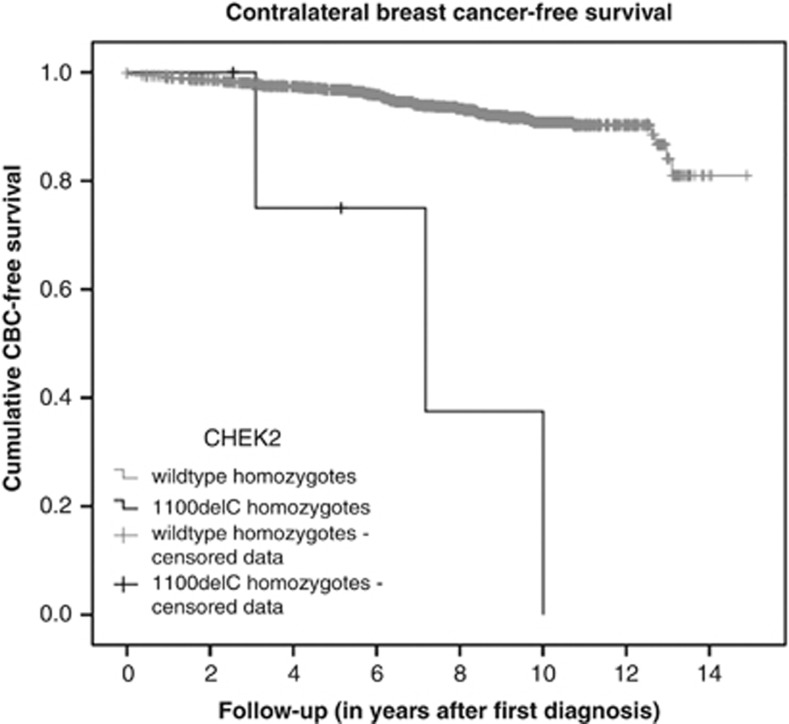
Follow-up information was available for five out of six patients, with an average follow-up of 8.5 years. Three of these five patients developed an invasive ductal carcinoma in the contralateral breast 3, 7 and 10 years after the first breast cancer diagnosis, while a fourth patient developed a melanoma located on the skin of the ipsilateral breast 5 years after the breast cancer diagnosis. The fifth patient died of metastatic disease 2.5 years after the initial breast cancer diagnosis. A Kaplan–Meier analysis showed a statistically significant increase in contralateral breast cancer incidence when compared with the homozygous wild-type breast cancer patients in the ORIGO cohort (Mantel–Cox log rank 31.6, P-value <0.001, Figure 1).
Figure 1.
Comparison of the contralateral breast cancer (CBC)-free survival in breast cancer patients being homozygous wild type (n=1334) versus being homozygous mutant (n=5) for the CHEK2 1100delC mutation. Homozygous wild-type patients are all homozygous wild-type patients from the ORIGO cohort for whom follow-up was available.
The phenotype of the three homozygous 1100delC patients with bilateral breast cancer is similar to that of the wild-type bilateral cancer patients in the sporadic breast cancer cohort, with the exception that for the three patients both tumors were of the invasive ductal subtype (Table 2).
Table 2. Comparison of the clinical parameters of bilateral breast cancer patients being homozygous wild type versus being homozygous mutant for the CHEK2 1100delC mutation.
| Characteristic | Homozygous wild-type genotypea (n=110) | Homozygous 1100delC genotype (n=3) |
|---|---|---|
| Age at first breast cancer (±SD) (years) | 54 (±12.2) | 40, 55 and 56 |
| Average follow-up (±SD) (years) | 8.6 (±2.8) | 11, 10 and 14 |
| Interval between first and contralateral breast cancer (±SD) (years) | 5.1 (±3.3) | 3, 7 and 10 |
| First breast cancer | ||
| Size (±SD) (cm) | 2.1 (±1.4) | 1.7, 1.7 and 2 |
| Nodal status | 63% N0 | N0, N1 and unknown |
| Invasive ductal carcinoma (%) | 70 | 100 (n=3) |
| ER status | 73% positive | 2 positive, 1 unknown |
| Contralateral breast cancer | ||
| Invasive ductal carcinoma (%) | 59 | 100 (n=3) |
Abbreviations: ER, estrogen receptor; N0, negative nodal status, N1, positive nodal status.
All homozygous wild-type patients were from the sporadic breast cancer cohort (ORIGO).
Genotyping of CHEK2 1100delC in lung cancer cohort
Adank et al.34 report two possible and two confirmed lung cancer diagnoses in relatives of 1100delC homozygotes. All three of our 1100delC homozygous breast cancer patients from the ORIGO cohort had one family member with lung cancer. Other variants in CHEK2 have been associated with lung cancer risk, but a possible association of 1100delC with lung cancer risk had never been investigated before.26, 27, 28, 29, 30, 31
Thus, we examined whether the 1100delC variant is associated with lung cancer risk by investigating a prevalent cohort of 457 lung cancer patients. Of these, 449 were homozygous wild type and 4 were heterozygous for the 1100delC mutation; no 1100delC homozygous patients were found (genotyping failed repeatedly for the remaining four patients). There was no significant difference in the frequency of 1100delC heterozygous individuals when comparing the lung cancer cohort and the control cohort (0.9 versus 1.2%, Pearson's Chi-Square, P=0.8).
Discussion
We present the first estimate of the frequency of CHEK2 1100delC homozygotes among sporadic breast cancer patients in a geographical locale with a high 1100delC allele frequency. Approximately 1 in 500 sporadic breast cancer cases was found to be homozygous. Depending on which control cohorts were included in the analysis (respectively, without and with certainty of the absence of breast cancer in these controls), the OR for developing breast cancer was either 3.4 (95% CI 0.4–32.6) or 44 (95% CI 11–174). Adank et al.34 report a genotype relative risk of 101.34 for CHEK2 1100delC homozygous females and a breast cancer risk more than twice the risk of heterozygous females within a cohort of breast cancer families. Thus, breast cancer risk in 1100delC homozygous women could be as high as the risks conferred by pathogenic BRCA1/2 mutations, but due to the small number of 1100delC homozygotes the 95% CIs of the risk estimate remain very wide.
In the non-BRCA1/2 families, one homozygous 1100delC carrier was found, yielding a genotype frequency comparable with that in the sporadic breast cancer cohort. Despite the small numbers, this indicates that the chance of being homozygous for 1100delC in the Netherlands is similar for a patient with a positive family history of breast cancer as for a sporadic breast cancer patient. Similar findings were made in a recent study on the prevalence of 1100delC homozygous females in non-BRCA1/2 families.34 More controls and breast cancer cases should be genotyped to improve the accuracy of the estimates of breast cancer risk for 1100delC homozygotes.
We investigated the association between 1100delC and lung cancer risk after observing a high incidence of lung cancer in family members of 1100delC homozygotes, as reported in this article and by Adank et al.34 Reported associations of other variants in CHEK2 and lung cancer risk further validated this investigation. Our prevalent cohort of 457 lung cancer patients had sufficient power (0.86) at a significance level of 0.05 to find a three times higher 1100delC allele frequency in cases than in controls. However, the allele frequency of 1100delC turned out to be about equal in our cases and controls. Some of these lung cancer cases survived up to 19 years before being included in this prevalent cohort. If the 1100delC mutation would increase lung cancer risk but also reduce lung cancer survival, it could be that an existing association was missed due to the construction of our lung cancer cohort. More lung cancer patients should be genotyped to investigate a smaller effect of 1100delC on lung cancer risk.
The phenotype of a total of six 1100delC homozygous females who developed breast cancer was studied. Their average age at first breast cancer diagnosis was not significantly younger than the average age at first diagnosis in the sporadic breast cancer cohort (54 versus 56.1 years). However, three out of five patients of whom follow-up was available developed a second primary breast tumor within 10 years of the first diagnosis. Adank et al.34 report bilateral breast cancer in 4 out of 10 1100delC homozygous females, with the second diagnosis being made within 4 years of the first. It has been reported that CHEK2 1100delC heterozygotes have an OR of about 6 for developing a second primary breast cancer when compared with unselected patients with a first primary breast cancer.15, 18, 20 In 1100delC homozygotes, this risk seems comparable or even higher, with potential consequences for clinical management.
With the current developments in DNA sequencing techniques, it is expected that in the near future the CHEK2 1100delC mutation will either be detected automatically during whole-exome sequencing or will be included in a large panel of breast cancer-associated genetic variants that are screened routinely. Then, it is important that health-care providers know whether this finding has implications for the prognosis or preferred treatment of the breast cancer patient or healthy woman under their care.
The observed increased risk to develop a second primary breast cancer for a breast cancer patient heterozygous for 1100delC could justify intensified follow-up, as has been suggested.5, 13, 14, 15, 16, 17, 18, 19 For homozygous carriers, our observations indicate that (contralateral) breast cancer risk may be as high as to justify preventive mastectomy (or at least intensified screening), but longitudinal studies on more cases are needed to arrive at more accurate risk estimates.
As CHEK2 is involved in the response pathway to DNA damage, breast tumors from CHEK2 1100delC heterozygotes and homozygotes may respond differently to some common treatment options such as radiotherapy, chemotherapy and PARP inhibitors. One study has reported an increased contralateral breast cancer risk when heterozygous patients had received radiotherapy as part of the treatment for their first breast tumor; another reported a similar but not statistically significant tendency.15, 19 Genotyping of larger cohorts is needed to better understand the effect of different treatments on breast cancer patients carrying the 1100delC allele. In the future, this can lead to personalized treatment strategies for 1100delC heterozygous and homozygous patients.
In conclusion, an increasing body of evidence indicates that screening for CHEK2 1100delC could be clinically meaningful and even cost-effective in countries with a relatively high allele frequency. Larger numbers of homozygotes will need to be studied to arrive at more accurate estimates of the associated cancer risks.
Acknowledgments
We thank the women who took part in this research, and all the support staff who made this study possible. Marion Meijer-van Gelder is acknowledged for data collection of RMOT. The ORIGO study was supported by a grant from the Dutch Cancer Society (UL 1997-1505). This study was, in part, financed by the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (JWMM's and EMJJB's contribution).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy —two sides of the same coin. Nat Rev Cancer. 2007;7:925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- Bahassi elM, Penner CG, Robbins SB, et al. The breast cancer susceptibility allele CHEK2*1100delC promotes genomic instability in a knock-in mouse model. Mutat Res. 2007;616:201–209. doi: 10.1016/j.mrfmmm.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- Hollestelle A, Wasielewski M, Martens JW, Schutte M. Discovering moderate-risk breast cancer susceptibility genes. Curr Opin Genet Dev. 2010;20:268–276. doi: 10.1016/j.gde.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahassi elM., Robbins SB, Yin M, et al. Mice with the CHEK2*1100delC SNP are predisposed to cancer with a strong gender bias. Proc Natl Acad Sci USA. 2009;106:17111–17116. doi: 10.1073/pnas.0909237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Dumont T, Jordheim LP, Michelon J, et al. Detecting differential allelic expression using high-resolution melting curve analysis: validation on the breast cancer susceptibility gene CHEK2. BMC Med Genomics. 2011;4:39–39. doi: 10.1186/1755-8794-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel JH, Peeters JK, Smid M, et al. Gene expression profiling assigns CHEK2 1100delC breast cancers to the luminal intrinsic subtypes. Breast Cancer Res Treat. 2012;132:439–448. doi: 10.1007/s10549-011-1588-x. [DOI] [PubMed] [Google Scholar]
- Anczukow O, Ware MD, Buisson M, et al. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins. Hum Mutat. 2008;29:65–73. doi: 10.1002/humu.20590. [DOI] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- Oldenburg RA, Kroeze-Jansema K, Kraan J, et al. The CHEK2*1100delC variant acts as a breast cancer risk modifier in non-BRCA1/BRCA2 multiple-case families. Cancer Res. 2003;63:8153–8157. [PubMed] [Google Scholar]
- Thompson D, Easton D. The genetic epidemiology of breast cancer genes. J Mammary Gland Biol Neoplasia. 2004;9:221–236. doi: 10.1023/B:JOMG.0000048770.90334.3b. [DOI] [PubMed] [Google Scholar]
- Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008;26:542–548. doi: 10.1200/JCO.2007.12.5922. [DOI] [PubMed] [Google Scholar]
- Bell DW, Kim SH, Godwin AK, et al. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer. 2007;121:2661–2667. doi: 10.1002/ijc.23026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks A, de, Witte L, Nooijen A, et al. Excess risk for contralateral breast cancer in CHEK2*1100delC germline mutation carriers. Breast Cancer Res Treat. 2004;83:91–93. doi: 10.1023/B:BREA.0000010697.49896.03. [DOI] [PubMed] [Google Scholar]
- Broeks A, Braaf LM, Huseinovic A, et al. Identification of women with an increased risk of developing radiation-induced breast cancer: a case only study. Breast Cancer Res. 2007;9:R26–R26. doi: 10.1186/bcr1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bock GH, Schutte M, Krol-Warmerdam EM, et al. Tumour characteristics and prognosis of breast cancer patients carrying the germline CHEK2*1100delC variant. J Med Genet. 2004;41:731–735. doi: 10.1136/jmg.2004.019737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher O, Johnson N, Dos, Santos Silva I, et al. Family history, genetic testing, and clinical risk prediction: pooled analysis of CHEK2 1100delC in 1,828 bilateral breast cancers and 7,030 controls. Cancer Epidemiol Biomarkers Prev. 2009;18:230–234. doi: 10.1158/1055-9965.EPI-08-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellemkjaer L, Dahl C, Olsen JH, et al. Risk for contralateral breast cancer among carriers of the CHEK2*1100delC mutation in the WECARE Study. Br J Cancer. 2008;98:728–733. doi: 10.1038/sj.bjc.6604228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuligina E, Reiner A, Imyanitov EN, Begg CB. Evaluating cancer epidemiologic risk factors using multiple primary malignancies. Epidemiology. 2010;21:366–372. doi: 10.1097/EDE.0b013e3181cc8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Seal S, Renwick A, et al. Gene-gene interactions in breast cancer susceptibility. Hum Mol Genet. 2012;21:958–962. doi: 10.1093/hmg/ddr525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debniak T, Scott RJ, Gorski B, et al. Common variants of DNA repair genes and malignant melanoma. Eur J Cancer. 2008;44:110–114. doi: 10.1016/j.ejca.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Meijers-Heijboer H, Wijnen J, Vasen H, et al. The CHEK2 1100delC mutation identifies families with a hereditary breast and colorectal cancer phenotype. Am J Hum Genet. 2003;72:1308–1314. doi: 10.1086/375121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Seal S, Schutte M, et al. A multicenter study of cancer incidence in CHEK2 1100delC mutation carriers. Cancer Epidemiol Biomarkers Prev. 2006;15:2542–2545. doi: 10.1158/1055-9965.EPI-06-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang HP, Geng XP, Ge WW, Li H. Meta-analysis of CHEK2 1100delC variant and colorectal cancer susceptibility. Eur J Cancer. 2011;47:2546–2551. doi: 10.1016/j.ejca.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12:399–408. doi: 10.1016/S1470-2045(10)70126-1. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lu J, Zhao X, et al. A variant in the CHEK2 promoter at a methylation site relieves transcriptional repression and confers reduced risk of lung cancer. Carcinogenesis. 2010;31:1251–1258. doi: 10.1093/carcin/bgq089. [DOI] [PubMed] [Google Scholar]
- Hall J, Marcel V, Bolin C, et al. The associations of sequence variants in DNA-repair and cell-cycle genes with cancer risk: genotype-phenotype correlations. Biochem Soc Trans. 2009;37:527–533. doi: 10.1042/BST0370527. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Masojc B, Oszutowska D, et al. Constitutional CHEK2 mutations are associated with a decreased risk of lung and laryngeal cancers. Carcinogenesis. 2008;29:762–765. doi: 10.1093/carcin/bgn044. [DOI] [PubMed] [Google Scholar]
- Hung RJ, Baragatti M, Thomas D, et al. Inherited predisposition of lung cancer: a hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiol Biomarkers Prev. 2007;16:2736–2744. doi: 10.1158/1055-9965.EPI-07-0494. [DOI] [PubMed] [Google Scholar]
- Brennan P, McKay J, Moore L, et al. Uncommon CHEK2 mis-sense variant and reduced risk of tobacco-related cancers: case control study. Hum Mol Genet. 2007;16:1794–1801. doi: 10.1093/hmg/ddm127. [DOI] [PubMed] [Google Scholar]
- van Puijenbroek M, van Asperen CJ, van Mil A, Devilee P, van Wezel T, Morreau H. Homozygosity for a CHEK2*1100delC mutation identified in familial colorectal cancer does not lead to a severe clinical phenotype. J Pathol. 2005;206:198–204. doi: 10.1002/path.1764. [DOI] [PubMed] [Google Scholar]
- Zhang S, Phelan CM, Zhang P, et al. Frequency of the CHEK2 1100delC mutation among women with breast cancer: an international study. Cancer Res. 2008;68:2154–2157. doi: 10.1158/0008-5472.CAN-07-5187. [DOI] [PubMed] [Google Scholar]
- Adank MA, Jonker MA, Kluijt I, et al. CHEK2*1100delC homozygosity is associated with a high breast cancer risk in women. J Med Genet. 2011;48:860–863. doi: 10.1136/jmedgenet-2011-100380. [DOI] [PubMed] [Google Scholar]
- Huijts PE, Vreeswijk MP, Kroeze-Jansema KH, et al. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9:R78–R78. doi: 10.1186/bcr1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEK2 Breast Cancer Case-Control Consortium: CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies Am J Hum Genet 2004741175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh SM, Look MP, Sieuwerts AM, Foekens JA, Hunter KW. Distinct inherited metastasis susceptibility exists for different breast cancer subtypes: a prognosis study. Breast Cancer Res. 2009;11:R75–R75. doi: 10.1186/bcr2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollestelle A, Pelletier C, Hooning M, et al. Prevalence of the variant allele rs61764370 T>G in the 3'UTR of KRAS among Dutch BRCA1, BRCA2 and non-BRCA1/BRCA2 breast cancer families. Breast Cancer Res Treat. 2011;128:79–84. doi: 10.1007/s10549-010-1080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs NJ, Gough AC, Primrose JN. Optimisation of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res. 1999;27:e12–e12. doi: 10.1093/nar/27.16.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kumar P, Schwarz M, Malone G, Haworth A, Kumar S. PCR amplification of 40-year-old paraffin-embedded tumor-tissues - comparison of 4 different DNA extraction and purification methods. Int J Oncol. 1994;5:453–457. doi: 10.3892/ijo.5.3.453. [DOI] [PubMed] [Google Scholar]
- Jekimovs CR, Chen X, Arnold J, et al. Low frequency of CHEK2 1100delC allele in Australian multiple-case breast cancer families: functional analysis in heterozygous individuals. Br J Cancer. 2005;92:784–790. doi: 10.1038/sj.bjc.6602381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balliu B, Tsonaka R, van der Woude D, Boehringer S, Houwing-Duistermaat JJ. Combining family and twin data in association studies to estimate the noninherited maternal antigens effect. Genet Epidemiol. 2012;36:811–819. doi: 10.1002/gepi.21667. [DOI] [PubMed] [Google Scholar]
- Højsgaard S, Halekoh U, Yan J. The R Package geepack for generalized estimating equations. J Stat Software. 2006;15:1–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.