Abstract
Rapidly accumulating data from genome-wide association studies (GWASs) and other large-scale studies are most useful when synthesized with existing databases. To address this opportunity, we developed the Phenotype–Genotype Integrator (PheGenI), a user-friendly web interface that integrates various National Center for Biotechnology Information (NCBI) genomic databases with association data from the National Human Genome Research Institute GWAS Catalog and supports downloads of search results. Here, we describe the rationale for and development of this resource. Integrating over 66 000 association records with extensive single nucleotide polymorphism (SNP), gene, and expression quantitative trait loci data already available from the NCBI, PheGenI enables deeper investigation and interrogation of SNPs associated with a wide range of traits, facilitating the examination of the relationships between genetic variation and human diseases.
Keywords: database, data integration, genome sequence, genome-wide association study, phenotype, single nucleotide polymorphism
Introduction
The genome-wide association study (GWAS) design has identified over 8900 genetic variants associated with over 250 human traits and diseases.1 Rarely are the functional consequences of these variants understood. Thus, replication, functional, and follow-up studies are the crucial next steps. Integration of GWAS results with existing complementary databases can facilitate prioritization of variants for the follow-up, study design considerations, and generation of biological hypotheses.
A number of existing genomic resources are housed at the National Center for Biotechnology Information (NCBI), including dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP), NCBI Gene (http://www.ncbi.nlm.nih.gov/gene), and the Genotype-Tissue Expression (GTEx) eQTL (expression quantitative trait loci) browser (http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi/). GWAS data and results are now readily available through two other NIH resources, the database of genotypes and phenotypes (dbGaP, http://www.ncbi.nlm.nih.gov/gap)2 and the National Human Genome Research Institute (NHGRI) GWAS catalog (http://www.genome.gov/GWAStudies/).1 Although comprehensive information is available at each of these online resources, the ability to navigate easily between them is limited. We sought to develop a user-friendly online resource that incorporates this layer of genotype–phenotype association data with existing databases, targeting genetics, epidemiologists, and clinical researchers who use or produce GWAS data. With the intent to design a simple, intuitive interface synthesizing data from multiple NIH databases and allowing users to download search results, we developed the Phenotype–Genotype Integrator (PheGenI, http://www.ncbi.nlm.nih.gov/gap/PheGenI).
Implementation
The PheGenI resource integrates content from several NIH resources: dbGaP, which archives and distributes the primary data of studies investigating associations between genotypes and phenotypes as well as their results; the NHGRI GWAS catalog, which curates published GWAS papers for genotype–phenotype associations from the scientific literature; dbSNP, which includes data on single nucleotide polymorphisms (SNPs) and their frequencies and genotypes; NCBI Gene, which includes gene-specific data, such as nomenclature, chromosomal localization, gene products, phenotypes, and links to related resources; and eQTL data from the GTEx program, which archives and displays associations between genetic variation and high-throughput molecular-level phenotypes.
The search queries were organized into two types: phenotype-oriented and genotype-oriented (Figure 1). Phenotype searches are linked to the association results from dbGaP and GWAS catalog, which are assigned to phenotype categories by NCBI curators using Medical Subject Headings (MeSH) concepts.3 Currently, phenotypes are matched to exact MeSH terms; parent, child, and synonyms are not indexed. The dbSNP rs numbers and genes mapped to those rs numbers are subsequently used to query dbSNP, NCBI Gene, and GTEx in a series of parallel searches. Similar searches can be performed for chromosomal location, gene, or SNP, and results are filtered accordingly (Supplementary Figure 1). Additional filters based on P-value of association and SNP functional class are also available. Future updates will incorporate NCBI Entrez Programming Utilities to programmatically retrieve data. The documentation is available at http://www.ncbi.nlm.nih.gov/books/NBK25501/.
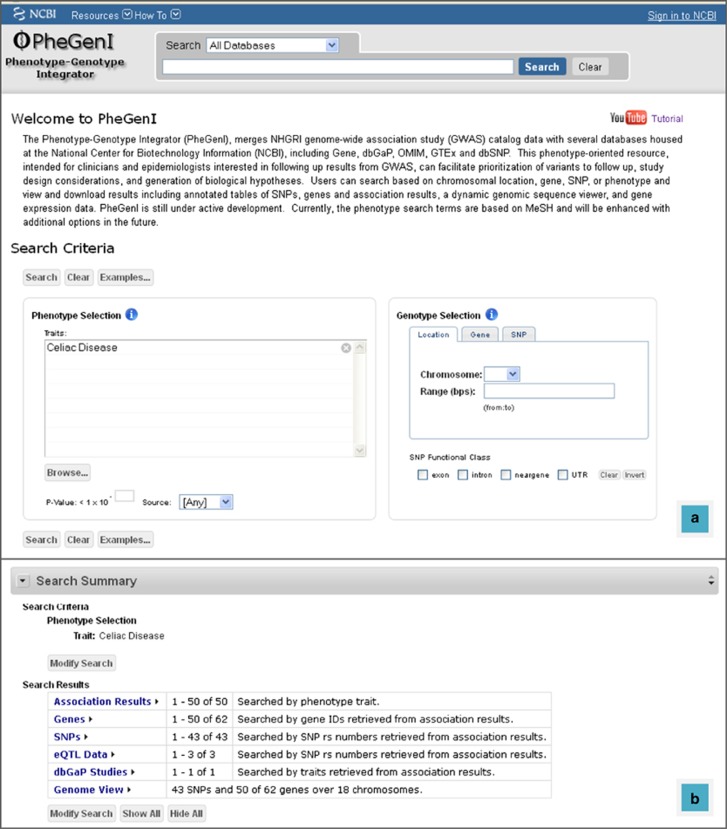
Figure 1.
PheGenI search interface and results summary. (a) The PheGenI search interface. In this example, a search was performed for the celiac disease trait. (b) The summary table of results returned for the celiac disease query.
Features
Search results are displayed in individual sections comprising: (1) a search summary; (2) association results; (3) interactive genome view/ideogram; (4) gene results; (5) SNP results; (6) eQTL results; and (7) a summary of relevant dbGaP studies that contain individual-level genotype and phenotype data available for authorized access. Results are annotated and hyperlinked using related information from their respective databases. For example, the association table includes the rs number (linked to dbSNP record), functional context of the SNP, gene (linked to Entrez Gene record), genomic location (linked to genomic sequence viewer), P-value of the association (linked to the dbGaP association browser), source record (linked to NHGRI GWAS catalog or dbGaP), and study ID or PubMed ID (linked to dbGaP or PubMed). PheGenI also provides structured URLs for stable links to records based on chromosomal location, gene, SNP, or phenotype (Supplementary Text).
The relative location of each section within the PheGenI display can be user-customized, and information links provide documentation for each section. Following a search, users may download data tables including annotated tables of SNPs, genes, association results, and gene expression data (Supplementary Figure 2). Associated loci are displayed on a chromosomal ideogram with customizable display features, which can be downloaded as a high-resolution image in multiple formats. Individual loci can be explored further using an interactive sequence viewer, which displays the genomic context of each SNP using customizable tracks (Figure 2).
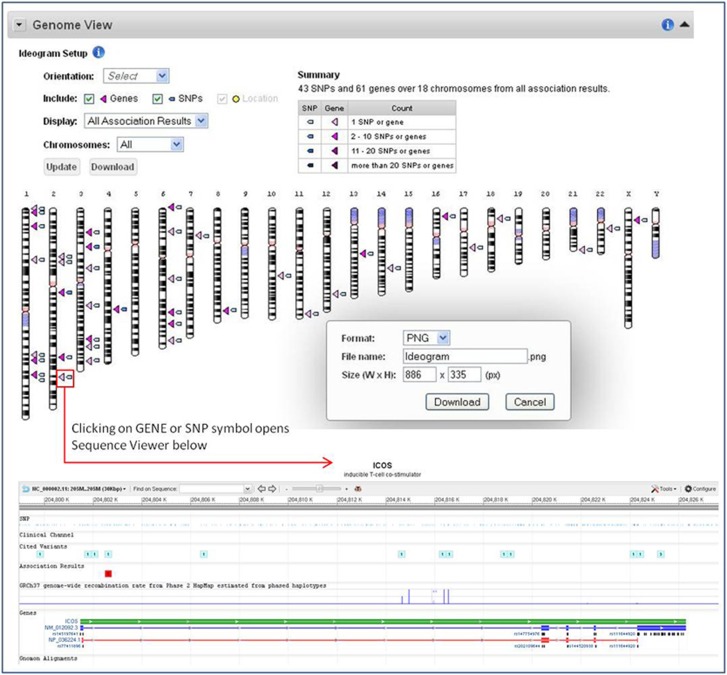
Figure 2.
PheGenI genome view and sequence viewer. PheGenI association results displayed on a customizable and downloadable ideogram. In this example, 43 SNPs (blue triangles) and 61 genes (pink triangles) across 18 chromosomes were identified as associated with celiac disease (P-value threshold set at 1 × 10−8). Clicking on the SNP or GENE triangle opens up a customizable sequence viewer to further explore a particular genomic region.
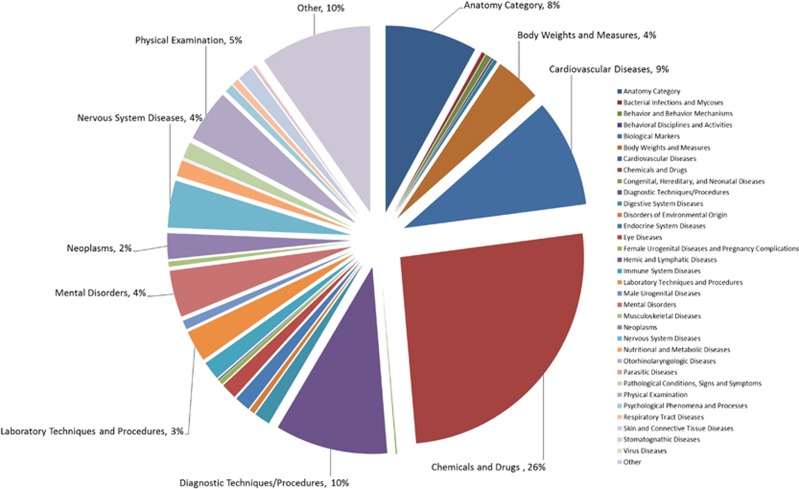
As of 1 March 2013, 54 282 association records from dbGaP and 11 781 from the NHGRI GWAS catalog (66 063 total) are available, corresponding to 30 885 unique rs numbers. These association records are integrated with ∼54 million records from dbSNP, 40 000 records from the NCBI Gene, and 61 000 eQTL records. After accounting for replicate SNP–trait associations from multiple publications and associations of the same SNP with multiple traits spanning multiple broad phenotype categories, 70% of the variants are distributed among a few categories: anatomy, body weights and measures, cardiovascular diseases, chemicals and drugs, diagnostic techniques and procedures, mental disorders, nervous system diseases, and physical examination (Figure 3).
Figure 3.
Distribution of PheGenI SNPs with association data across broad phenotype categories. The majority of SNP–trait associations can be assigned to a few broad MeSH categories.
Discussion
With the development of this integrated PheGenI resource, GWAS results can be further explored in the genomic context, and linked to attributes of SNP, gene, and eQTL data. By building in capabilities to download data tables, customize the view, and interactively browse features of the genomic sequence, the data are readily available in a user-friendly format tailored toward population scientists and clinical researchers who wish to follow up genetic association results in more detail. The component databases are regularly updated and maintained, reflecting the current state of the field and providing stable links to external resources. Documentation and user support are provided in the form of information links, a YouTube video (http://www.youtube.com/watch?v=v_yEy--HcKc) and a link to submit questions directly to the NCBI help desk.
Several improvements are targeted for the near future, including broadening the phenotype search to include synonyms, adding additional data sources, including those focused on functional elements, and annotating supporting results to provide added confidence in reported association results. PheGenI complements several existing resources that also provide information about genetic associations in a genomic and/or phenotypic context, including the CDC's HuGE Navigator (http://www.hugenavigator.org/HuGENavigator/home.do), the Ensembl Genome Browser (http://useast.ensembl.org/index.html), UCSC's Genome Browser (http://genome.ucsc.edu/), the EU-GEN2PHEN-funded GWAS Central resource (http://www.gwascentral.org), and other efforts.4, 5 However, to evaluate the potential for genetic knowledge to be relevant to clinical care and public health, additional evaluation related to clinical relevance and clinical utility are necessary. Common standards for annotating genetic variants in this way will be needed, as well as a comprehensive database of relevant genetic variants that spans a range of phenotypes. PheGenI is one component of this evolving knowledge base, and this regularly updated resource will provide much-needed genomic-level and phenotypic-level annotation of GWAS results to enable future studies.
Acknowledgments
We thank M Kimura, J Paschall, and T Manolio for thoughtful input throughout the development of PheGenI. This research was supported, in part, by the Intramural Research Program of the US National Institutes of Health, National Library of Medicine.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGap database of genotypes and phenotypes. Nat Genet. 2007;2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage A. Changes in MeSH data Structure. NLM Tech Bull. 2000;313:e2. [Google Scholar]
- Johnson AD, O'Donnell CJ. An open access database of genome-wide association results. BMC Med Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schully SD, Yu W, McCallum V, et al. Cancer GAMAdb: database of cancer genetic associations from meta-analyses and genome-wide association studies. Eur J Hum Genet. 2011;19:928–930. doi: 10.1038/ejhg.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.