Imatinib reduces tumor cell KIT signaling and causes tumor cell apoptosis, which drives TAMs to shift from M1- to M2-like in mouse and human GIST.
Abstract
Tumor-associated macrophages (TAMs) are a major component of the cancer microenvironment. Modulation of TAMs is under intense investigation because they are thought to be nearly always of the M2 subtype, which supports tumor growth. Gastrointestinal stromal tumor (GIST) is the most common human sarcoma and typically results from an activating mutation in the KIT oncogene. Using a spontaneous mouse model of GIST and 57 freshly procured human GISTs, we discovered that TAMs displayed an M1-like phenotype and function at baseline. In both mice and humans, the KIT oncoprotein inhibitor imatinib polarized TAMs to become M2-like, a process which involved TAM interaction with apoptotic tumor cells leading to the induction of CCAAT/enhancer binding protein (C/EBP) transcription factors. In human GISTs that eventually developed resistance to imatinib, TAMs reverted to an M1-like phenotype and had a similar gene expression profile as TAMs from untreated human GISTs. Therefore, TAM polarization depends on tumor cell oncogene activity and has important implications for immunotherapeutic strategies in human cancers.
GIST is the most common sarcoma in humans (Ducimetière et al., 2011). The majority of GISTs are driven by activating mutations in either KIT (Hirota et al., 1998) or PDGFRA (Heinrich et al., 2003). Imatinib mesylate (Gleevec) is a molecular inhibitor of the KIT and PDGFRA oncoproteins and has increased the median survival in advanced GIST from <1 yr (Gold et al., 2007) to 5 yr (Joensuu and DeMatteo, 2012), making it one of the most successful examples of targeted therapy. Unfortunately, imatinib is rarely curative and half of the patients develop resistance by 2 yr (Joensuu and DeMatteo, 2012), most often because of secondary KIT mutations (Antonescu et al., 2005).
Although it has long been recognized that the immune system contributes to tumor development and control of tumor growth (Dunn et al., 2004), there are now considerable data that it plays a major role in the response to cancer therapy (Zitvogel et al., 2008). Recently, we showed in a spontaneous mouse model of GIST (Sommer et al., 2003) that imatinib’s anti-tumor activity depended partially on CD8+ T cells (Balachandran et al., 2011). Imatinib treatment caused a striking reduction in tumor cell production of the immunosuppressive enzyme indoleamine 2,3-dioxygenase, thereby decreasing regulatory T cells (T reg cells) and increasing CD8+ T cells within the tumor. Furthermore, we found that the immune modulating agent α-CTLA-4 was synergistic with imatinib.
TAMs play a central role in cancer biology because they constitute a substantial portion of the tumor mass and interact with numerous effector cells (Biswas and Mantovani, 2010). Although it is an oversimplification of their diverse and intricate biology, macrophages have been categorized as classically (M1) or alternatively (M2) activated (Lewis and Pollard, 2006; Biswas and Mantovani, 2010; Qian and Pollard, 2010; Lawrence and Natoli, 2011; Ruffell et al., 2012; Schmieder et al., 2012). M1 macrophages are induced by LPS or IFN-γ and stimulate a Th1 response, whereas M2 macrophages are polarized by IL-4 or IL-13 and promote a Th2 response. M1 macrophages are anti-tumoral because they secrete inflammatory cytokines (TNF, IL-6, IL-1β, and IL-12), present antigen, and recruit effector T cells. In contrast, M2 macrophages are anti-inflammatory, as they produce IL-10, express scavenger and IL-1 decoy receptors, and recruit T reg cells via CCL22 secretion (Curiel et al., 2004; Biswas and Mantovani, 2010). M2 macrophages also suppress effector T cells via arginase (Schmieder et al., 2012) and support angiogenesis and metastasis through a variety of mechanisms. TAMs are almost always M2 and usually confer worse prognosis in both mice (Qian and Pollard, 2010) and humans (Heusinkveld and van der Burg, 2011). There is scant evidence for M1 TAMs in cancer. In a murine flank tumor model of breast cancer, TAMs had an MHC class IIhi phenotype but actually suppressed T cell proliferation in vitro (Movahedi et al., 2010). In a subcutaneous model of liver cancer, TAMs had an M1 phenotype and did increase T cell proliferation in vitro (Wang et al., 2011). TAMs in human cancer are generally regarded as pro-tumoral, but the data derive almost entirely from limited immunohistochemical analyses, and functional studies are lacking (Heusinkveld and van der Burg, 2011).
Because TAMs are a potential immunotherapeutic target (Beatty et al., 2011; DeNardo et al., 2011; Shiao et al., 2011; Hume and MacDonald, 2012), we investigated their role in GIST. Here, we demonstrate in mouse and human GISTs that tumor cell oncogene activity determined TAM phenotype and function. In mice, established tumors contained M1-like TAMs, which were anti-tumoral, as proven by depletion studies. Imatinib therapy in mouse GIST polarized TAMs to become M2-like through the activation of CCAAT/enhancer binding protein (C/EBP) β. Consistent with our mouse findings, human TAMs were also M1-like at baseline and became M2-like after imatinib therapy. In patients whose tumors developed resistance to imatinib, TAMs reverted to M1-like and had a remarkably similar gene expression profile as M1-like TAMs from untreated patients. Our findings reveal the central importance of tumor cell oncogene activity in TAM polarization.
RESULTS
Mouse GIST TAMs are inflammatory
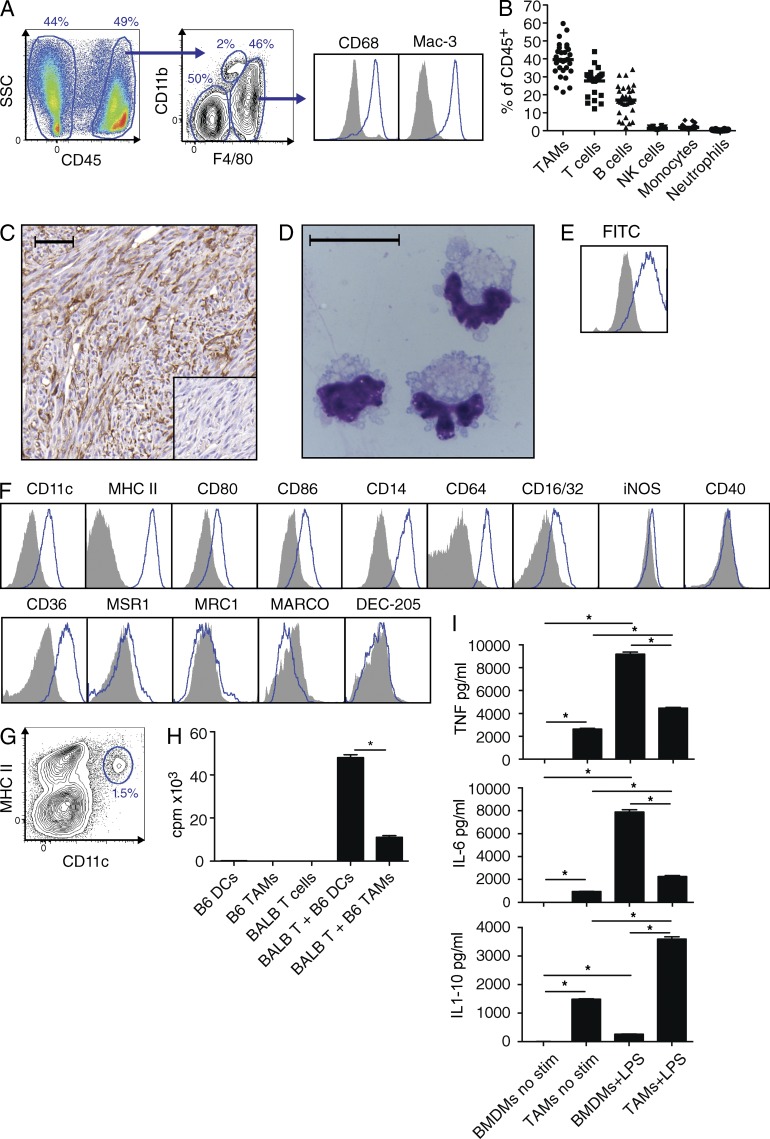
GIST mice spontaneously develop a single intestinal tumor by 3–4 wk of age and die from intestinal obstruction at a median of ∼6 mo (Sommer et al., 2003). After digesting tumors with collagenase, we found that F4/80hiCD11b+/−CD68+Mac-3+ cells (Fig. 1 A) comprised ∼40% of intratumoral CD45+ cells (Fig. 1 B). The F4/80hi cells did not express the monocyte and neutrophil markers Ly6C and Ly6G or the tumor cell markers KIT and CD34 (unpublished data). F4/80+ cells diffusely infiltrated the tumors (Fig. 1 C), were large and contained multiple cytoplasmic granules (Fig. 1 D) after bead isolation to >90% purity (not depicted), and efficiently phagocytosed latex beads (Fig. 1 E). Overall, the data were consistent with these cells being TAMs.
Figure 1.
Mouse GIST TAMs are inflammatory. (A) GIST tumors from untreated mice were processed and stained for CD45, CD11b, and F4/80. Histograms (right) were gated on F4/80hi cells and isotypes are shown as solid gray. (B) Composition of intratumoral leukocytes from 31 untreated GIST mice is shown as a percentage of CD45+ cells using the following definitions: TAMs, F4/80hi; T cells, F4/80−B220−NK1.1−CD3+; B cells, F4/80−CD3−B220+; NK cells, F4/80−B220−CD3−NK1.1+; monocytes, CD11bhiF4/80int; and neutrophils, CD11b+Ly6G+. Medians are indicated (horizontal bars). (C) F4/80 staining of an untreated tumor. Bar, 50 µm. Inset shows isotype control. (D) Giemsa stain of a cytospin of TAMs from an untreated GIST mouse. Bar, 20 µm. (E) TAM phagocytosis of FITC-labeled latex beads was measured by flow cytometry after 24 h compared with cells incubated without beads (shaded histogram). (F) Flow cytometry for various proteins after gating on TAMs. MSR1, macrophage scavenger receptor 1; MRC1, mannose receptor C type 1; MARCO, macrophage receptor with a collagenous structure; and DEC-205, DCs and epithelial cells 205 kD. (G) DCs are shown among CD45+F4/80− cells from an untreated GIST mouse. (H) TAMs and spleen DCs were isolated from untreated GIST mice (B6) and cultured with naive CD4+ T cells from BALB/c spleens in a mixed leukocyte reaction. After 72 h, proliferation was measured by 3H-thymidine incorporation (cpm, counts per minute). (I) 2 × 105 BMDMs (day 7) or freshly isolated GIST TAMs were cultured alone or with LPS 1 µg/ml in a 24-well plate for 12 h. Supernatant cytokines were measured by cytometric bead array. C–E are representative of two to five experiments, whereas H and I were done in triplicate in one experiment. Bar graphs show mean ± SEM. *, P < 0.05.
To evaluate the polarization of TAMs in murine GIST, we performed flow cytometry for M1 and M2 macrophage markers. TAMs expressed high amounts of the inflammatory markers CD11c and MHC class II (Fig. 1 F, top) but were distinct from the small population of CD11chiMHCIIhi conventional DCs contained among F4/80− cells (Fig. 1 G) and had much less ability to present antigen than DCs (Fig. 1 H). TAMs expressed multiple other inflammatory markers, including CD80 and CD86, the LPS coreceptor CD14, and Fc receptors, but lacked the M1 markers iNOS and CD40 (Fig. 1 F, top). Of five scavenger receptors typical of M2 macrophages, staining was only substantial in CD36 (Fig. 1 F, bottom). That TAMs were more M1-like was further supported by a gene expression array performed on sorted TAMs (purity > 90%; not depicted) from untreated animals (Table S1). TAMs expressed IRF5, a transcription factor associated with M1 macrophages (Krausgruber et al., 2011), but not IRF4, which is associated with the M2 program (Satoh et al., 2010). Compared with BM-derived macrophages (BMDMs), TAMs made significantly higher levels of cytokines at baseline. This included high levels of inflammatory cytokines TNF, IL-6, and IL-1β (Fig. 1 I and not depicted), and also higher levels of the anti-inflammatory cytokine IL-10, perhaps as a homeostatic feedback to the high inflammatory cytokines (Fig. 1 I). Neither cell type produced detectable IL-12 (unpublished data). After culture with LPS, which promotes M1 polarization, TAMs made even higher levels of these cytokines. Collectively, our findings demonstrated that TAMs in murine GIST were predominantly M1-like in phenotype and function.
TAMs require CSF1R, but not CCR2 or CX3CR1
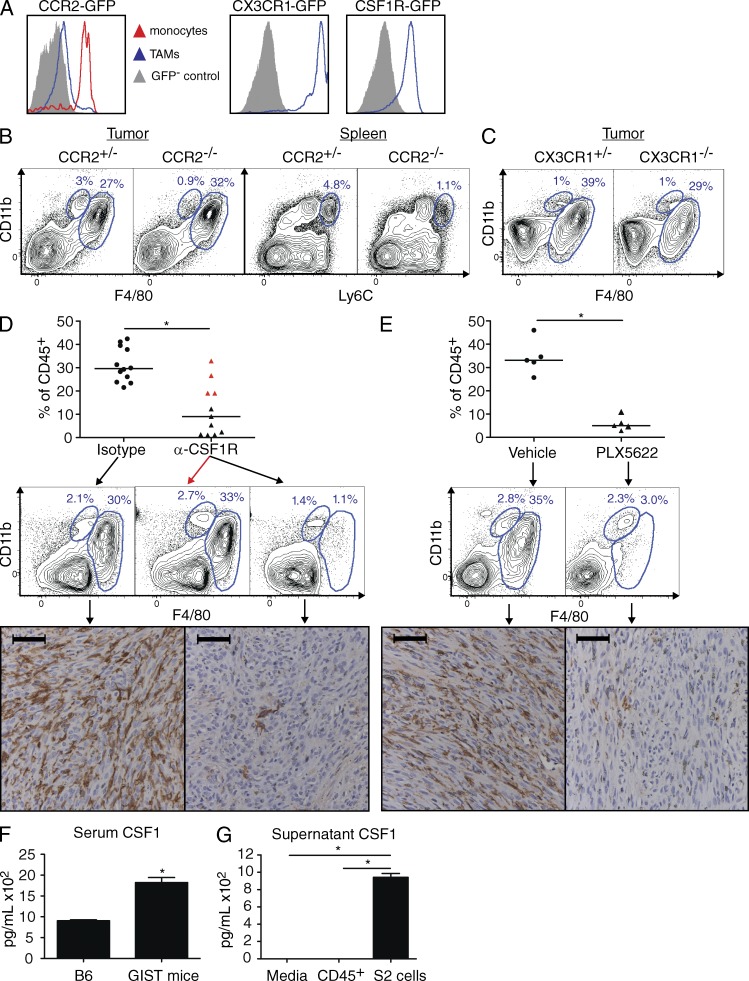
We sought to deplete TAMs to determine their relevance in vivo. We identified several monocyte/macrophage developmental genes from our gene expression array as potential targets, including Ccr2 (Qian et al., 2011), Cx3cr1 (Geissmann et al., 2003; Hart et al., 2009), and Csf1r (Lin et al., 2001; DeNardo et al., 2011; Table S1). TAMs from CCR2-GFP+-GIST mice contained only a small GFP+ (i.e., CCR2+) subset, whereas intratumoral inflammatory monocytes (F4/80intCD11b+), which are thought to give rise to TAMs (Qian et al., 2011), were uniformly CCR2+ (Fig. 2 A, left). We hypothesized that inflammatory monocytes might have down-regulated CCR2 after differentiating into TAMs, but CCR2−/−-GIST mice actually had normal numbers of TAMs, even though inflammatory monocytes were reduced within the tumor and spleen (Fig. 2 B). We next investigated CX3CR1, which is found on tissue-resident monocytes (Geissmann et al., 2003) and some TAMs (Hart et al., 2009). TAMs in CX3CR1-GFP+-GIST mice expressed high levels of CX3CR1 (Fig. 2 A, middle), yet CX3CR1−/−-GIST mice had only a modest reduction in TAMs (Fig. 2 C). Tumor weight was unchanged in CCR2−/−-GIST and CX3CR1−/−-GIST mice (unpublished data). Thus, mouse GIST TAMs did not depend substantially on CCR2 or CX3CR1.
Figure 2.
TAMs require CSF1R, but not CCR2 or CX3CR1. (A) GIST mice were crossed to CCR2-, CX3CR1-, and CSF1R-GFP reporter mice and TAMs were analyzed from 6–8-wk-old animals. (B and C) Tumors and spleens from CCR2−/−-GIST mice (B) and tumors from CX3CR1−/−-GIST mice (C) were analyzed. Monocytes are indicated as CD11bhiF4/80int or CD11bhiLy6Chi. Plots shown for each reporter mouse are representative of two experiments with one to three mice per group. (D and E) After 4 wk of treatment with α-CSF1R (D) or PLX5622 (E), tumors from GIST mice were analyzed and TAMs as a percentage of CD45+ cells were determined. Bars represent medians. (D, top and middle) Composite of two similar experiments (total n = 11–12 mice per group). Red triangles in the α-CSF1R treatment group indicate mice depleted <50% of the median of isotype-treated controls. Representative flow plots show control (left), nondepleted (middle, red arrow), and depleted (right) animals. (E, top and middle) One of two similar experiments (n = 5 mice per group). (D and E, bottom) Representative F4/80 immunohistochemistry is shown below matched groups. Bars, 50 µM. (F) Serum CSF1 in age-matched B6 mice (n = 2) or GIST mice (n = 4) measured by ELISA. (G) 5 × 104 freshly isolated CD45+ leukocytes from a spontaneous tumor in a GIST mouse or S2 cells were cultured for 48 h and supernatant CSF1 was measured by ELISA. Experiments show in F and G were performed once, with individual mice serving as replicates for F and triplicate wells for G. Bar graphs show mean ± SEM. *, P < 0.05.
GIST TAMs expressed high levels of CSF1R (Fig. 2 A, right), whereas other intratumoral leukocytes had low or no expression and tumor cells lacked staining (not depicted). Targeting CSF1R with a blocking antibody (Sudo et al., 1995) markedly reduced the percentage of TAMs in two thirds of mice after 4 wk both by flow cytometry and immunohistochemistry (Fig. 2 D). We confirmed the importance of CSF1R by treating mice with PLX5622 (Coniglio et al., 2012; Hamilton and Achuthan, 2013), an oral inhibitor with nanomolar specificity for CSF1R, which also depleted TAMs (Fig. 2 E). Although TAMs were profoundly sensitive to CSF1R blockade by either treatment, spleen and BM macrophages, monocytes, and neutrophils were less affected (Fig. S1). CSF1 is a ligand for CSF1R and is often elevated in the serum of cancer patients. Accordingly, we found that serum CSF1 was increased in GIST mice compared with age-matched WT controls (Fig. 2 F). Furthermore, a cell line (called S2) derived from a spontaneous GIST mouse tumor secreted high levels of CSF1, unlike freshly isolated CD45+ intratumoral leukocytes from a mouse GIST tumor (Fig. 2 G). Therefore, GIST cells appear to exert paracrine effects on TAMs via CSF1.
M1-like TAMs inhibit tumor growth by multiple mechanisms
Although TAMs generally support tumor growth (Biswas and Mantovani, 2010), we discovered that GIST mice depleted of TAMs with α-CSF1R or PLX5622 for 4 wk had increased tumor weight (Fig. 3, A and B) and more KIT+ tumor cells (Fig. 3 C). These findings were consistent with the TAMs being M1-like and inflammatory (Fig. 1). Notably, mice in which α-CSF1R treatment failed to deplete TAMs at 4 wk (Fig. 2 D, red triangles), perhaps due to the development of anti–rat antibodies, had tumors of comparable size to those from isotype control-treated mice (Fig. 3 A, red triangles).
Figure 3.
M1-like TAMs inhibit tumor growth by multiple mechanisms. (A, left) Tumor weight from GIST mice treated with α-CSF1R for 4 wk. Red triangles represent mice depleted <50% of control group median (horizontal bars; Fig. 2 D). (A, right) Tumor weight excluding mice that were not depleted after α-CSF1R treatment. Shown are composites of two to three experiments per time point (n = 7–17 mice per group). (B) Tumor weight after PLX5622 therapy. Composite of two experiments per time point, 6–10 mice per group. (C) Live CD45−KIT+ cells by trypan blue exclusion after 4 wk of therapy. Only depleted mice are shown for α-CSF1R. Composite of two experiments, 7–12 mice per group. (D) Cell viability at 72 h of co-culture of S2 cells with TAMs from untreated GIST mice. 5 × 103 S2 cells were plated in a flat-bottom 96-well plate either alone or with TAMs in various ratios. After 72 h, wells with S2 cells alone were fully confluent. Cell viability was assessed by optical density at 450 nm (OD450). Shown is a representative of two experiments, with measurements in at least triplicate. (E) Splenic T cells from untreated GIST mice were cultured in α-CD3–coated plates with various ratios of TAMs from GIST mice. Proliferation at 72 h (left) or supernatant IFN-γ at 48 h (right) were measured. Representative of at least three experiments. Cpm, counts per minute. x indicates a single replicate of that dilution. TAMs alone did not produce IFN-γ (not depicted). (F) After 4 wk of treatment with PLX5622, intracellular IFN-γ was measured in tumors and mesenteric lymph nodes. Shown is a representative of two experiments, total 10–11 mice/group. (G) Splenic T cells were cultured in α-CD3–coated plates with various ratios of TAMs from S2 or B16 flank tumors, and proliferation was measured at 72 h. Shown is a representative of two experiments in at least triplicate. (H) S2 cell flank tumor volume after treatment with PLX5622 (n = 5–10 mice/group). Bar and line graphs show mean ± SEM. *, P < 0.05.
We next investigated the mechanism of TAM inhibition of tumor growth in mice with established GISTs and found that TAMs exhibited both direct and indirect effects. TAMs isolated from untreated GIST mice directly inhibited the growth of S2 GIST cells in vitro (Fig. 3 D). TAMs also robustly stimulated CD4+ and CD8+ T cell proliferation and IFN-γ production (Fig. 3 E). Although TAM depletion using either α-CSF1R or PLX5622 did not alter the frequency of intratumoral CD4+, CD8+, or T reg cells (not depicted), intratumoral CD4+ and CD8+ T cells from GIST mice treated with PLX5622 made significantly less intracellular IFN-γ (Fig. 3 F). In contrast to our findings in the native GIST mouse, TAMs isolated from flank S2 GIST or B16 melanoma tumors were M2-like and inhibited T cell proliferation (Fig. 3 G) and IFN-γ production (not depicted). LiHa sarcoma flank tumors also had an M2-like phenotype (unpublished data). As expected, depletion of these M2-like TAMs (control CD11bhiF4/80hi 81.6 ± 5.3% vs. PLX5622 18.8 ± 9.3% at 44 d, P < 0.05, n = 5–10 mice per group) decreased the size of subcutaneous S2 GIST tumors (Fig. 3 H).
TAMs become M2-like during imatinib therapy via C/EBPβ
Because imatinib is the first line of treatment in advanced GIST, we studied how TAMs were affected by imatinib in our spontaneous mouse model. TAMs were depleted, but the effect was delayed compared with tumor shrinkage (Fig. 4 A) and coincided with an almost two-thirds reduction in TAM proliferation after 2 wk of imatinib (Fig. 4 B). To elucidate further the effects of 2 wk of imatinib on TAMs, we analyzed their gene and protein expression. The inflammatory markers CD11c, MHC class II, CD80, and CD86 were reduced in TAMs (Fig. 4 C), but not in matched spleen macrophages (not depicted). Multiple M2 markers (Biswas and Mantovani, 2010) were increased by transcriptome profiling (Fig. 4 D), including Il10, Chi3l3, Pla2g7, CXCR4, and Msr1, which were validated by RT-PCR (Fig. 4 E) or flow cytometry (Fig. 4 F). Notably, there was greater expression of the transcription factor C/EBPβ, a driver of the M2 program and regulator of both Il10 and Msr1 (Ruffell et al., 2009) and tumor-induced immunosuppression (Marigo et al., 2010). C/EBPβ up-regulation was validated by Western blot of isolated TAMs (Fig. 4 G). In vivo, the predominant C/EBPβ isoform was the long, transactivating liver-enriched activator protein (LAP; C/EBPβ), whereas a short repressive isoform liver-enriched inhibitory protein (LIP; C/EBPβ) was expressed only at low levels. In addition, we found that cyclic AMP response element-binding (CREB), a transcription factor which directly regulates C/EBPβ transcription (Ruffell et al., 2009), was also up-regulated and activated, as was the CREB-related protein ATF1 (Fig. 4 G). Finally, in mice treated with imatinib for 2 wk, freshly isolated TAMs were less inflammatory because they produced lower amounts of TNF and IL-6 both with and without LPS stimulus (Fig. 4 H). Collectively, then, imatinib induced TAMs to shift in vivo from M1- to M2-like. After 4 wk of imatinib therapy, we found that intratumoral CD8+ T cells were less activated with lower CD69 staining and less IFN-γ production after in vitro stimulation (unpublished data).
Figure 4.
TAMs become M2-like during imatinib therapy. (A) GIST mice were treated with imatinib or vehicle control and sacrificed at the indicated time points. Means of tumor weight (left) and TAM percentage of CD45+ cells (right) are shown normalized to matched vehicle controls. There were 8–16 mice per time point for tumor weight and at least 3 mice per time point for TAM percentage, but only 1 imatinib-treated mouse and 2 controls existed at 24 wk (marked by an x). (B) GIST mice were treated with vehicle or imatinib for 2 wk and TAM proliferation was assessed by flow cytometry of intracellular Ki67 staining (seven to eight mice/group). (C and D) Mean fluorescence intensity (MFI) of various proteins on TAMs was measured by flow cytometry (C; represents at least two experiments, n = 6–7 per group) or TAMs were sorted from GIST mice (n = 8 pooled per group) and subjected to gene expression array (D). Selected statistically significant genes with a false discovery rate of <0.05 and a fold change greater than two are shown. Color of box depicts fold change (scale on left). On the right, mean value of absolute expression is shown for each group. (E and F) Selected genes were validated by RT-PCR (E; pooled RNA from eight per group) or flow cytometry (F; representative of at least two experiments, n = 6–7 per group). (G) Western blot analysis of C/EBPβ isoform expression (C/EBPβ, LAP, and LIP) on isolated TAMs from vehicle- or imatinib-treated mice (at least five mice per group). MW, molecular weight. (H) GIST mice were treated with imatinib for 2 wk, and then 5 × 104 freshly isolated TAMs were cultured with or without 1 µg/ml LPS overnight in a 96-well plate. Supernatant cytokines were then measured by cytometric bead array. Shown is a representative of two experiments, five to seven mice per group. Bar graphs show mean ± SEM. *, P < 0.05.
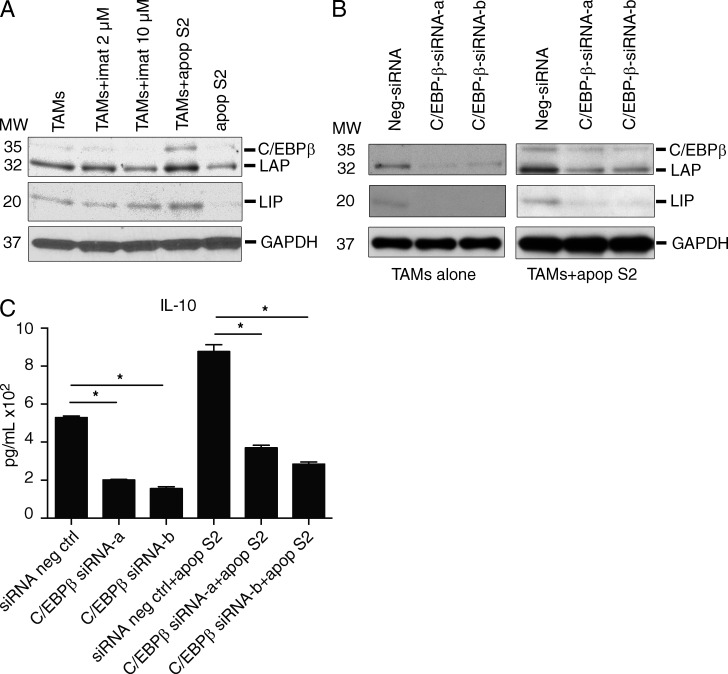
To identify the mechanism of the shift in TAM polarization, we performed in vitro studies. Imatinib did not directly alter the expression of C/EBPβ in TAMs at a concentration of 2 µM, which is similar to the serum level in humans taking imatinib (Demetri et al., 2009) and our treated mice (mean 3.8 ± 0.4 µM, n = 4 mice, 4 wk of treatment), or at a supraphysiological dose of 10 µM (Fig. 5 A). However, TAMs up-regulated C/EBPβ and the LAP isoform when cultured with S2 GIST cells that had been rendered apoptotic by irradiation and pretreatment with imatinib (Fig. 5 A). To evaluate the specific role of C/EBPβ in TAM polarization, we used RNA interference to knock down C/EBPβ in freshly isolated TAMs that were cultured alone or with apoptotic S2 GIST cells. Knockdown of C/EBPβ isoforms was confirmed by Western blotting in both conditions (Fig. 5 B). In TAMs cultured alone, C/EBPβ knockdown caused a decrease in IL-10, indicating a role of C/EBPβ in baseline polarization (Fig. 5 C). In TAMs cultured with apoptotic S2 GIST cells, IL-10 expression was increased, an effect which was abrogated by C/EBPβ knockdown. Thus, C/EBPβ plays a central role in the polarization of GIST TAMs both at baseline and in the polarization shift that occurs during TAM interaction with apoptotic cells during imatinib treatment.
Figure 5.
Apoptotic tumor cells induce an M2-like shift via C/EBPβ. (A) S2 cells were rendered apoptotic by irradiation with 20 Gy and treatment with imatinib for 3 h and then washed to remove imatinib. 106 apoptotic S2 cells (apop S2) were then cultured with 2 × 106 TAMs for 48 h in 6-well plates. Some wells were treated with 2 or 10 µM imatinib (imat). After 48 h, cell lysates were analyzed by Western blot. Shown is a representative of two experiments. MW, molecular weight. (B and C) 106 TAMs were transfected with either control (siRNA neg ctrl) or C/EBPβ-targeted siRNA (constructs a or b) for 24 h and then cultured with or without apoptotic S2 cells (apop S2). After 48 h, cell lysates were analyzed by Western blotting (B) and supernatant IL-10 was measured by cytometric bead array (C). Shown is a representative of two experiments. Bar graphs show mean ± SEM. *, P < 0.05.
Human GIST TAMs become M2-like during therapy
To ascertain the clinical relevance of our findings in the GIST mouse, we analyzed 57 freshly obtained human GIST specimens from 50 patients (Table 1). Untreated GISTs contained a large population of CD45+CD11b+CD14+CD68+ cells that were lineage (CD3, CD19, and CD56) negative (Fig. 6 A). In addition, they expressed CD11c, HLA-DR, CD86, and CD64, but also the M2 marker CD163 (Fig. 6 B). Bead-isolated cells had >90% purity (not depicted) and were large with multiple cytoplasmic granules (Fig. 6 C). In vitro, TAMs from untreated human GISTs secreted high amounts of TNF, IL-1β, and IL-6 (Fig. 6 D). Thus, TAMs from untreated human GISTs were M1-like, as in mouse GIST.
Table 1.
Clinicopathologic characteristics of 57 specimens from 50 patients
| Characteristic | Untreated | Sensitive | Resistant |
| Number of patients | 25 | 18 | 9a |
| Number of specimens | 25 | 23 | 9 |
| Median age (range) | 65 (38-85) | 58 (43-81) | 53 (39-66) |
| Female | 13 | 4 | 3 |
| Male | 12 | 14 | 6 |
| Primary | 25 | 9 | 2 |
| Metastatic | 0 | 14 | 6 |
| Local recurrence | 0 | 0 | 1 |
| Primary location | |||
| Stomach | 23 | 7 | 4 |
| Small intestine | 1 | 12 | 4 |
| Other | 1 | 4 | 1 |
| Tyrosine kinase inhibitor | |||
| Imatinib | Not applicable | 23 | 9 |
| Sunitinib | Not applicable | 0 | 5 |
| Other | Not applicable | 0 | 2 |
| Median treatment duration (months) | Not applicable | 6 (3-84) | 84 (36-132)b |
| Mutational status | |||
| KIT exon 9 | 0 | 2 | 1 |
| KIT exon 11 | 6 | 9c | 8d |
| KIT exon 13 | 0 | 1 | 0 |
| PDGFRA | 2 | 0 | 0 |
| WT | 2 | 0 | 0 |
| Unavailable | 15 | 6 | 0 |
Two patients also had sensitive tumors.
Shows duration of any treatment.
One patient had a secondary KIT exon 17 mutation but was still sensitive by clinical criteria.
Two patients had secondary KIT exon 13 mutations and 1 had a secondary KIT exon 17 mutation.
Figure 6.
Human GIST TAMs become M2-like during therapy. (A) Representative flow plots of a freshly obtained and processed GIST from an untreated patient. Histograms are gated on CD45+Lin−CD11b+CD14+ cells and isotypes are gray. (B) Expression of various proteins on human TAMs from an untreated patient. (C) Cytospin of TAMs with Giemsa stain. Bar, 20 µm. (D) 2.5 × 104 TAMs were isolated from untreated patients and stimulated with 1 µg/ml LPS for 24 h and supernatant cytokines were measured by cytometric bead array. Shown is a representative of seven untreated tumors. (E, left) TAMs as a percentage of CD45+ cells in untreated (n = 25), sensitive (n = 23), and resistant (n = 9) tumors. (E, right) Blood monocytes (CD45+lin−CD11b+CD14+) as a percentage of CD45+ cells in 18 untreated, 13 sensitive, and 5 resistant patients. (F) CD14 in bulk tumor measured by gene expression array of an independent cohort of 37 untreated, 9 sensitive, and 39 resistant patients. Bars in E and F represent medians. (G) CD14 immunohistochemistry showing representative staining from untreated, sensitive, and resistant human GIST specimens. Inset shows isotype control. Bars, 50 µm. (H) Proportion of TAMs expressing HLA-DR by flow cytometry (n = 17 untreated, 15 sensitive, and 8 resistant). (I) Matched peripheral blood CD4+ T cells were cultured in the presence of α-CD3 and various ratios of TAMs and proliferation was measured by 3H-thymidine incorporation after 3–4 d (representative of two untreated, three of six sensitive, and three resistant tumors, x indicates single replicate of that dilution). Bar graphs show mean ± SEM. *, P < 0.05.
Tumors were categorized, as previously (Balachandran et al., 2011), as untreated, sensitive, or resistant based on their response to imatinib at the time of surgery (Table 1). TAMs constituted a lower median percentage of intratumoral leukocytes in sensitive tumors than in untreated tumors (10 versus 25%), but there was no difference between untreated and resistant tumors (Fig. 6 E, left). We validated this finding with gene expression array of CD14 in an independent cohort of 85 human GISTs (Fig. 6 F) and with immunohistochemistry (Fig. 6 G). Meanwhile, the percentage of monocytes in matched peripheral blood was similar among the three patient groups (Fig. 6 E, right).
Given that TAMs were reduced in sensitive human GISTs as we had observed in our mouse model after imatinib therapy, we then ascertained whether they also became M2-like. Sensitive TAMs had less HLA Class II expression (Fig. 6 H). TAMs from three of six sensitive tumors suppressed T cell proliferation, indicating an M2 shift (Fig. 6 I), whereas those from two others had no effect and one caused stimulation. In contrast, TAMs from resistant tumors were M1-like in function as they uniformly (three of three) stimulated T cell proliferation, as did TAMs from untreated (two of two) tumors (Fig. 6 I). Meanwhile, TAMs isolated from two human pancreatic adenocarcinomas failed to induce T cell proliferation (unpublished data).
To further investigate human TAM polarization, we performed gene expression profiling on TAMs that had been freshly bead-isolated from 11 untreated, 5 sensitive, and 4 resistant human tumors. TAMs from sensitive tumors had 689 genes with at least a twofold expression difference compared with TAMs from untreated tumors. Multiple M2 and related genes were up-regulated in TAMs from sensitive GISTs (Fig. 7), consistent with our functional data. Based on global gene expression changes, Ingenuity pathway analysis software identified multiple members of the C/EBP transcription factor family as important in sensitive tumors, including C/EBPα, C/EBPδ, and C/EBPε (Table 2). Strikingly, isolated TAMs from untreated and resistant patients did not have a single gene with an expression difference of at least twofold. Hence, in accordance with our mouse data, tumor cell oncogenic activity, which is fully active in untreated human GISTs and human GISTs that have acquired resistance to imatinib, was associated with TAM polarization. Furthermore, the C/EBP transcription factor family was up-regulated in sensitive human GISTs as it was in mouse GIST after imatinib therapy.
Figure 7.
Gene expression in TAMs of patients with GIST. TAMs isolated from five sensitive human tumors were compared with TAMs from 11 untreated and 4 resistant tumors by gene expression array. RNA was isolated from TAMs that were freshly bead-isolated from human GISTs. Differentially regulated genes were those with a fold change >2 at a false discovery rate of <0.05. For the comparison of untreated to sensitive TAMs, selected genes are shown by category, with the magnitude of fold change depicted on the scale. There were 689 genes differentially expressed between untreated and sensitive TAMs (310 up and 379 down on sensitive compared with untreated), 0 genes differentially expressed between untreated and resistant TAMs, and 44 genes differentially expressed between sensitive and resistant TAMs (24 up and 20 down on sensitive compared with resistant).
Table 2.
C/EBP transcription factors are activated in sensitive human tumors
| Transcription regulator | Regulation z-score | P-value of overlap | Target molecules in dataset |
| CEBPA | 4.0 | 8.3−6 | 36 |
| TP53 | 3.9 | 2.2−22 | 117 |
| NF-κB (complex) | 3.7 | 2.5−5 | 47 |
| SP1 | 3.4 | 1.6−9 | 52 |
| Nfat (family) | 3.3 | 1.2−2 | 9 |
| CDKN2A | 3.3 | 2.6−8 | 28 |
| SMARCB1 | 3.1 | 6.5−3 | 16 |
| PGR | 3.0 | 9.7−2 | 11 |
| RELA | 2.9 | 3.2−2 | 21 |
| HNF4A | 2.8 | 2.0−1 | 82 |
| CEBPE | 2.8 | 1.4−3 | 8 |
| CEBPD | 2.7 | 4.3−3 | 10 |
| TCF3 | 2.6 | 2.6−3 | 20 |
| RB1 | 2.6 | 9.6−13 | 40 |
| TP63 | 2.5 | 7.9−2 | 12 |
| HMGB1 | 2.5 | 2.9−3 | 8 |
| IFI16 | 2.5 | 2.1−1 | 5 |
| EGR1 | 2.3 | 1.1−4 | 18 |
| NFKBIB | 2.3 | 1.1−2 | 7 |
| SMAD3 | 2.3 | 8.1−2 | 13 |
| PDX1 | 2.2 | 4.6−1 | 8 |
| SMARCE1 | 2.2 | 2.0−3 | 5 |
| Rb | 2.2 | 1.2−5 | 12 |
| ETS1 | 2.2 | 3.2−1 | 10 |
| NFATC1 | 2.1 | 1.8−2 | 6 |
| ETV4 | 2.1 | 5.8−2 | 4 |
| JUND | 2.1 | 6.4−2 | 6 |
| Ap1 | 2.0 | 1.2−3 | 15 |
| Hdac | −2.0 | 1.6−2 | 6 |
| MYBL2 | −2.0 | 1.7−3 | 7 |
| MYCN | −2.1 | 3.2−1 | 9 |
| E2f | −2.4 | 3.2−5 | 16 |
| MYOD1 | −2.4 | 2.7−2 | 14 |
| MYC | −2.5 | 1.7−5 | 57 |
| KLF2 | −2.6 | 1.6−2 | 13 |
| FOXO1 | −2.9 | 3.4−5 | 24 |
| MEOX2 | −3.1 | 4.7−4 | 8 |
| FOXM1 | −3.7 | 1.2−11 | 19 |
| TBX2 | −3.9 | 1.1−10 | 20 |
Bolding highlights CEBP family genes among other genes that were found significant in the Ingenuity pathway analysis. A positive regulation z-score indicates activation, whereas a negative score indicates inhibition.
DISCUSSION
Although TAMs are almost always M2-like in mice and humans, we established that mouse and human GISTs contained TAMs that were M1-like in phenotype and function at baseline. The exception to the classical M1 polarity was the expression of some scavenger receptors (CD36 in mouse and CD163 in humans), which are typically associated with the M2 program. Based on CD163 immunohistochemistry, a previous study characterized TAMs in human GIST as M2, despite also finding high HLA-DR expression (van Dongen et al., 2010). Thus, the nearly exclusive use of immunohistochemistry to categorize human TAMs (Heusinkveld and van der Burg, 2011) may underestimate the frequency of M1 TAMs in human cancer. Our findings also underscore the potential limitations of studying heterotopic tumors in mice using cell lines because TAMs from subcutaneous tumors of S2 GIST cells were M2-like, unlike TAMs from the parent tumor.
TAMs from untreated mouse and human GISTs were functionally M1-like, as they secreted inflammatory cytokines and stimulated T cell proliferation and IFN-γ production. Consistent with these findings, TAM depletion increased tumor size in GIST mice with established tumors. In contrast, depletion of M2 TAMs is known to decrease tumor burden in models of breast, colon, ovarian, lung, and prostate cancer and melanoma (Qian and Pollard, 2010). It has been suggested that early carcinogenesis may be dominated by M1-like TAMs, whereas an M2-like phenotype emerges as the tumor microenvironment evolves (Qian and Pollard, 2010; Wang et al., 2011; Schmieder et al., 2012). However, we found that TAMs isolated from 7-, 24-, and 47-wk-old mice had a similar ability to stimulate T cell proliferation (unpublished data). Thus, TAMs in our model do not naturally become M2-like over time. To determine the cause of the M1-like TAM phenotype in GIST, we crossed GIST mice to IFN-γ−/− mice because IFN-γ is a major determinant of M1 polarization (Biswas and Mantovani, 2010). TAMs from GIST-IFN-γ−/− mice showed only minor decreases in CD11c and MHC class II, whereas CD80 and CD86 expression, TNF production, and stimulation of T cell proliferation were unchanged (unpublished data). Future studies will focus on the contributions of other local stimuli in skewing TAMs to be M1-like in GIST.
TAMs are thought to arise from circulating blood monocytes, particularly CCR2+ inflammatory monocytes (Movahedi et al., 2010; Qian et al., 2011), although other studies suggest that TAMs may derive from resident CX3CR1hiCCR2lo monocytes (Lawrence and Natoli, 2011). We proved that genetic deletion of CCR2 did not alter the number of mouse GIST TAMs, whereas deletion of CX3CR1 had only a partial effect. Thus, chemokine receptors on TAMs have redundant functions. In our mouse model, TAMs were profoundly sensitive to CSF1R blockade by an antibody or molecular inhibitor, even though the major myeloid populations in the spleen and BM were only modestly affected. The “addiction” of M1-like TAMs to CSF1R signaling in mouse GIST was unexpected given the general contention that CSF1 is an M2-polarizing cytokine (Biswas and Mantovani, 2010). Indeed, we found that in vitro treatment of TAMs with CSF1 did not up-regulate C/EBPβ (unpublished data).
Imatinib therapy decreased TAMs in our mouse model and in human GISTs. Imatinib does inhibit CSF1R modestly, albeit at a higher IC50 than KIT inhibition (Dewar et al., 2005; Manley et al., 2010). Imatinib depleted TAMs in GIST mice by only 20% at 4 wk, whereas α-CSF1R and PLX5622 achieved 80% depletion after just 1 wk (unpublished data). In addition to CSF1, which was secreted by the S2 GIST cell line, tumor cells produce numerous other growth factors and chemokines that support TAM recruitment and maintenance, including VEGF and CCL2, 5, 7, and 8 (Solinas et al., 2009). TAMs in GIST mice had lower Ki67 expression after imatinib therapy and numerous markers of proliferation and cell cycle progression were down-regulated in human GIST TAMs from sensitive tumors. Overall, it seems that the eventual decline of TAMs with imatinib treatment in mouse and human GIST is due to moderate CSF1R inhibition as well as the reduction of tumor cell–derived growth factors (e.g., CSF1) caused by oncogene inhibition.
Imatinib caused TAMs to become M2-like in phenotype and function in both mouse and human GIST. Thus, oncogene activity might drive TAM polarization (Gabrilovich et al., 2012). There was less Class II, CD80, and CD86 expression and several M2-like markers increased, including phospholipase A2 group VII, chitinase genes, cathepsins, and scavenger receptors. Other increases in M2-like genes did not overlap between mouse and human and were generally more pronounced in humans, perhaps reflecting the longer treatment period (median, 6 mo vs. 2 wk in the mouse). Human TAMs also up-regulated numerous genes for angiogenesis, matrix metallopeptidases, matrix synthesis genes, chemokines/chemokine receptors involved in neutrophil recruitment, and arginase. TAMs isolated from imatinib-treated mice were less inflammatory, as they produced lower amounts of TNF. Similarly, TAMs isolated from imatinib-sensitive human GISTs tended to suppress T cell proliferation, unlike TAMs from untreated or resistant patients, which consistently stimulated T cells. Therefore, the frequency and polarization of TAMs depended on the oncogenic activity of the tumor cells. TAMs were M1-like at baseline, became M2-like with imatinib via indirect induction of C/EBP (through tumor cell apoptosis in mice), and in humans reverted to the M1-like polarity and gene expression profile of untreated tumors upon acquiring resistance to imatinib.
The C/EBP family of transcription factors was implicated in the switch to M2-like TAMs after imatinib therapy of mouse and human GISTs. There is >90% homology among the C/EBP isoforms, although the exact function of each is unknown (Ramji and Foka, 2002). C/EBP binding sites exist in numerous genes that are important for myeloid cells, including CD14, COX-2, IL-1β, IL-6, IL-8, IL-12, CSF1R, iNOS, and TNF (Ramji and Foka, 2002). In particular, C/EBPβ is a central regulator in normal mouse macrophages (Gautier et al., 2012) and regulates the M2 genes MSR1, IL-10, and Arg1 (Csóka et al., 2007; Ruffell et al., 2009; Lawrence and Natoli, 2011; Schmieder et al., 2012). It has been reported that C/EBPβ−/− macrophages failed to produce IL-10 in response to Escherichia coli (Csóka et al., 2007). Given that we found mouse TAMs to have a high level of phagocytic activity, imatinib induces tumor cell apoptosis, and TAM phagocytosis of apoptotic tumor cells has been shown previously to induce M2 polarization (Voll et al., 1997), it is possible that TAM phagocytosis of dying GIST cells promoted M2 polarization in vivo.
TAM depletion in untreated mice increased tumor growth, consistent with their M1-like profile. Surprisingly, though, depletion of the M2-like TAMs by either anti-CSF1R or PLX5622 did not further alter tumor weight during imatinib therapy (unpublished data). It is possible that our mouse model is not aggressive enough to demonstrate an additive benefit of oncogene inhibition combined with depletion of M2-like TAMs. The tumor cells in GIST mice have a single, homogenous mutation and the mice do not develop metastases or imatinib resistance. Alternatively, it is possible that M2-like TAMs are not required for tumor cell maintenance during inhibition of tumor cell oncogene activity.
It is now clear that the immune response is critical in mouse and human GIST. Others have shown in GIST patients that IFN-γ production by blood NK cells and an immunosuppressive NKp30 isoform are associated with survival (Ménard et al., 2009; Delahaye et al., 2011). Our previous work demonstrated the importance of the adaptive immune system in GIST (Balachandran et al., 2011). Although imatinib-sensitive mouse and human GISTs had more CD8+ T cells and fewer T reg cells, here we show that they also have fewer TAMs with a shift from predominantly M1-like to M2-like TAMs, which would be expected to support tumor growth. The role of the adaptive immune system in GIST was validated in another recent study, in which a high density of CD3+ T cells correlated with progression-free survival (Rusakiewicz et al., 2013). Given that targeted molecular agents are almost never curative in solid tumors (including GIST), other approaches, such as modulation of TAMs, are being intensely studied. However, depletion of TAMs in human GIST either in the absence of imatinib therapy or after imatinib resistance has developed may actually be detrimental because the TAMs are M1-like. Whether TAM depletion is beneficial during imatinib response in GIST patients remains to be seen. Overall, our findings are relevant to the clinical application of TAM-directed therapies in GIST and have implications for targeted and immune therapy of other human cancers.
MATERIALS AND METHODS
Mice.
Heterozygous 6–8-wk-old GIST mice (KITΔV558/+; Sommer et al., 2003) on a C57BL/6 (B6) background (The Jackson Laboratory), CCR2−/− (Boring et al., 1997), CCR2-GFP (Serbina et al., 2009), CX3CR1-GFP (Jung et al., 2000), CSF1R-GFP (Burnett et al., 2004; all Jackson), and CX3CR1−/− (Combadière et al., 2003; Taconic) mice were used. Animal procedures were approved by the Sloan-Kettering Institutional Animal Care and Use Committee.
Treatments.
Purified anti-CSF1R (clone AFS98; Sudo et al., 1995) was produced by the Monoclonal Antibody Core Facility, Sloan-Kettering Institute. GIST mice received a loading dose of AFS98 or rat IgG2a (clone 2A3; Bio X Cell) 500 µg i.p. on day 1, then 250 µg on days 3 and 5 and twice weekly thereafter. PLX5622 (Coniglio et al., 2012; Hamilton and Achuthan, 2013) chow (1,200 mg/kg) was provided by Plexxikon and control chow was rodent diet AIN-76A (Plexxikon). Imatinib was obtained from Novartis and LC Laboratories and dissolved in the drinking water at 600 mg/liter.
Implantable flank tumors.
106 S2 cells (see below) in PBS were mixed 1:1 with growth factor reduced Matrigel (BD) and injected subcutaneously in the left flank of B6 mice. 5 × 104 B16 melanoma cells (a gift of A. Houghton, Memorial Sloan-Kettering Cancer Center, New York, NY) in PBS were implanted in other mice. Flank tumor volume was calculated using the ellipse formula (1/2 length × width × height as measured with calipers).
Tissue processing.
Mice were sacrificed with CO2 inhalation and tumors were weighed, minced, and digested in 12 mg/ml collagenase type II (Worthington Biochemical) and 0.5 mg/ml DNase I (Roche) for 30 min in a shaker at 37°C. Tumor suspensions were washed through a 100-µm cell strainer with 1% FCS, then passed through a 40-µm filter, and washed again. Flank tumors were excised and processed as above. After mashing through a 70-µm cell strainer, spleens were centrifuged and red blood cells lysed using ammonium chloride lysis buffer (eBioscience), quenched with 1% FCS, and passed through a 40-µm filter. BM was flushed from a single tibia per mouse, pooled by group, homogenized by repeated aspiration through an 18-gauge needle, and washed in 1% FCS. With patient consent, we obtained fresh human tumors from the operating room under an IRB-approved protocol and processed immediately using the same protocol as mouse tumors, with the only modification being that a red blood cell lysis step was added if the cell pellet contained substantial heme. Matched peripheral blood was collected in heparinized tubes before surgical incision. Peripheral blood mononuclear cells were obtained using density centrifugation over a Ficoll-Paque gradient (GE Healthcare).
Cell isolation.
To obtain murine TAMs, tumor single cell suspensions were incubated with anti–mouse CD117 microbeads (Miltenyi Biotec), washed, filtered through a 40-µm cell strainer, centrifuged, and then run through a single LS column (Miltenyi Biotec) per 108 cells. Unbound KIT− cells were collected from the negative fraction, counted, and incubated with 10 µl biotinylated anti–mouse F4/80 (clone BM8; eBioscience) per 107 cells, washed, and incubated with 20 µl streptavidin microbeads per 107 cells (Miltenyi Biotec). Positive selection was then performed using two sequential LS columns. TAMs were isolated from subcutaneous S2 or B16 tumors using the same protocol, omitting the KIT negative selection. Human TAMs were isolated using a similar protocol, except negative selection used anti–human CD3, CD56, and CD117 microbeads and positive selection used anti–human CD14 microbeads (Miltenyi Biotec). Mouse spleen CD4+ and CD8+ T cells and human peripheral blood CD4+ T cells were isolated using the appropriate kits according to the manufacturer’s protocol (Miltenyi Biotec).
Flow cytometry.
Cellular analysis was performed using a FACSAria (BD) as previously described (Balachandran et al., 2011). Mouse-specific antibodies conjugated to various fluorochromes were purchased from BD (CD45, clone 30-F11; CD3, 145-2C11; CD11b, M1/70; CD11c, HL3; CD34, RAM34; CD40, 3/23; CD80, 16-10A1; CD86, GL1; CD117 [KIT], 2B8; Mac-3, M3/84; B220, RA3-6B2; NK1.1, PK136; Ly-6C, AL-21; and Ly-6G, 1A8), eBioscience (CD8, 53–6.7; CD36, No.72-1; CD45, 30-F11; F4/80, BM8; FoxP3, FJK-16s; Ki67, SolA15; MHC Class II [I-A/I-E], M5/114.15.2; and DEC-205, 205yekta), Invitrogen (F4/80, BM8), BioLegend (CD14, Sa14-2; CD16/32, 93; CD64, X54-5/7.1; CD68, FA-11; and MRC1, MR5D3), R&D Systems (MSR1, 268318), and AbD Serotec (MARCO, ED31). Purified rabbit anti–mouse iNOS (Millipore) was detected using FITC-conjugated goat anti–rabbit IgG (Abcam). Human-specific antibodies were purchased from BD (CD3, SK7; CD11b, D12; CD14, M5E2; CD16, 3G8; CD19, SJ25C1; CD45, 2D1; CD56, B159; CD64, 10.1; CD80, L307.4; CD86, 2331 [FUN-1]; and HLA-DR, L243 [G46-6]), eBioscience (CD163, GHI/61), BioLegend (CD11c, 3.9; and CD68, Y1/82A), or Miltenyi Biotec (CD45, 5B1). Appropriate isotype controls were used where applicable. Histograms are shown with staining intensity on the x axis and percentage of maximum on the y axis.
Histochemistry.
Tumors were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at a thickness of 5 µm. Antigen retrieval was achieved with citrate buffer. Sections were stained with anti–mouse F4/80 (1:100 dilution; clone BM8; BioLegend) or anti–human CD14 (prediluted; clone 7; Thermo Fisher Scientific) as previously described (Zeng et al., 2004). Murine or human TAMs were isolated as above and centrifuged onto a glass slide (Cytospin 4; Thermo Fisher Scientific). Giemsa stains were then performed using standard methods.
Cytokine detection.
Cell culture supernatant or serum cytokines were measured using cytometric bead array according to manufacturer’s instructions (Mouse Inflammation kit, Mouse Th1-Th2-Th17 Cytokine kit, IL-1β and Mouse Flex Set, Human Inflammation kit; BD). Mouse serum and supernatant CSF1 were measured by ELISA as instructed (R&D Systems). For intracellular TNF detection, freshly isolated tumor cell suspensions were stimulated with 1 µg/ml LPS in the presence of a 1:1,000 dilution of Brefeldin A (Golgi plug; BD). After 4 h, cells were washed, surface stained, fixed, and permeabilized (Cytofix/Cytoperm; BD). TNF was then quantified in TAMs on a per-cell basis (clone MP6-XT22; BD). For IFN-γ, single cell suspensions were stimulated with 20 ng/ml PMA and 1 µM ionomycin for 1 h, after which Brefeldin A was added. After 6 h, cells were surface stained and then fixed/permeabilized and stained intracellularly for IFN-γ (clone XMG1.2; BD). IL-10 in FACS-sorted TAMs was quantified by real-time PCR using probes purchased from Applied Biosystems.
Cell assays.
For tumor cell viability assays, 5 × 103 S2 cells were plated in a flat-bottom 96-well plate either alone or with TAMs in various ratios. After 72 h, wells with S2 cells alone were fully confluent. A cell viability assay was then performed according to the manufacturer (Dojindo) and viability was assessed by optical density at 450 nm (OD450). For T cell proliferation assays, mouse or human TAMs were plated in a flat-bottom, anti-CD3–coated, 96-well plate (BD) alone or in various ratios with or without 5 × 104 mouse or human CD4+ or CD8+ T cell responders as previously described (Movahedi et al., 2010). After 2–3 d of culture (when responder cells in control wells were blastic and forming rosettes), 1 µCi 3H-thymidine was added to each well. Cells were harvested (Tomtec automated harvester) the next day and lysed onto filter paper (Perkin Elmer). Beta decay was then measured using a Microbeta Trilux counter (Perkin Elmer; Wallac Microbeta software). For mixed leukocyte reactions, CD4+ T cells were isolated from BALB/c spleens using beads. GIST mouse spleens were incubated with type II collagenase to enhance DC yield and then processed by mashing through a 70-µm filter, washing in 1% FCS, and passing through a 40-µm filter. Spleen DCs were enriched over 2 LS columns using anti–mouse CD11c microbeads (Miltenyi Biotec). 105 responder T cells were plated in a 96-well plate with either 2.5 × 104 TAMs or spleen DCs and cultured for 3 d, with 1 µCi 3H-thymidine added for last 18 h of culture. The plate was then harvested and β counted as above. For the phagocytosis assay, 2.5 × 105 TAMs were incubated in media in a 24-well plate overnight. FITC-labeled latex beads (Miltenyi Biotec) were then added to some wells (Cayman Chemical). After 24 h, TAMs were dissociated, washed, and incorporation of beads was measured by flow cytometry. BMDMs were generated from B6 BM, as previously (Mosser and Zhang, 2008), and were used on day 7. For transient C/EBPβ gene knockdown, 106 TAMs from untreated GIST mice were cultured overnight in a 6-well plate in serum free medium. TAMs were then transfected with 30 nM C/EBPβ ON-target plus SMARTpool siRNA (construct a, b; Thermo Fisher Scientific) or nontargeting negative control siRNA (Thermo Fisher Scientific) using lipofectamine (Invitrogen) according to the manufacturer’s instructions. RNA interference sequences are listed in the manufacturer’s data sheet. After 24 h, wells were changed to full medium and 2 × 105 apoptotic S2 cells (5:1 ratio of TAMs/S2 cells) were added and co-cultured for another 48 h. Supernatant was then harvested to measure IL-10 by cytometric bead array. Cell lysates were used to measure protein by Western blotting.
Generation of a murine GIST cell line (S2).
GIST mouse tumor single cell suspensions were incubated in RPMI 1640 medium containing 10% FCS, glutamine, 2-mercaptoethanol, and antibiotics. Non-adherent cells were removed and media was replaced after 48 h. This was repeated until colonies of tumor cells were visible. These were then dissociated using a trypsin alternative (Tryple Express; Invitrogen) and serially passaged. After 10 passages, the cell line stabilized.
Microarray.
Isolated TAMs were pelleted, snap frozen, and RNA was isolated using the RNeasy Plus Mini kit (QIAGEN). Gene expression microarray was then performed by the Genomics Core Laboratory (Sloan-Kettering Institute) using the Mouse Genome 430A 2.0 and Human Genome U133A 2.0 microarrays according to manufacturer instructions (Affymetrix). Microarray data were analyzed using Partek Genomics Suite version 6.5. After log transformation and quantile normalization, ANOVA was performed to compare multiple groups. Selected statistically significant genes with a False Discovery Rate of <0.05 and a fold change >2 are shown. Heat maps were generated using Matrix2png software (Pavlidis and Noble, 2003). Statistically significant genes were analyzed using Ingenuity pathway analysis software (Ingenuity Systems, Inc.). Whole tumor CD14 expression was analyzed in an independent cohort of patients (Affymetrix Human Genome U133A microarray). Array data have been deposited in NCBI’s GEO database (GSE51697 and GSE51698).
Western blot.
Protein was isolated and Western blotting performed as previously (Balachandran et al., 2011). Anti–mouse C/EBPβ (clone 1H7; BioLegend) was used at a 1:1,000 dilution and detects the three isoforms C/EBPβ, LAP, and LIP. Antibodies for p-CREB (Ser133, clone 87G3, which also reacts with p-ATF1) and GAPDH (clone D16H11; Cell Signaling Technology) were used at a 1:1,000 dilution.
Statistical analysis.
Unpaired two-tailed Student’s t tests and one-way ANOVA comparisons were done where appropriate using Prism 5.0 (GraphPad Software). A p-value <0.05 was considered significant.
Online supplemental material.
Fig. S1 depicts the gating strategy of spleen and BM myeloid populations during CSF1R blockade. Table S1 shows a summary of a gene expression array of murine GIST TAMs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130875/DC1.
Supplementary Material
Acknowledgments
We are grateful to the Tissue Procurement Service for assistance in the acquisition of human tumor specimens; Laboratory of Comparative Pathology; Tanya Henry and Peter Romanienko from the Colony Management Group; Research Animal Resource Center; and the Genomics, Monoclonal Antibody, and Molecular Cytology core facilities. We thank Gideon Bollag and Brian West of Plexxikon for providing PLX5622 and technical suggestions, Miriam Merad for the AFS98 hybridoma, and Eric Pamer for CCR2−/− mice. We thank Carl Nathan and members of the laboratories of Alan Houghton, Jedd Wolchok, and James Allison for helpful discussions and technical support, and Russell Holmes for logistical and administrative support.
This work was supported by CA102613; T32 CA09501; the Geoffrey Beene Cancer Foundation; Mr. JHL Pit, Mrs. Pit-van Karnebeek, and the Dutch GIST Foundation; the GIST Cancer Research Fund; Swim Across America; Stephanie and Fred Shuman through the Windmill Lane Foundation; David and Monica Gorin (R.P. DeMatteo); CA102774 (P. Besmer); CA140146 (P. Besmer and C.R. Antonescu); and CA162721 (T.S. Kim). The Molecular Cytology Core Facility was supported by Cancer Center Support Grant P30 CA008748.
The authors have no conflicting financial interests.
All authors contributed to the experimental design. M.J. Cavnar, S. Zeng, T.S. Kim, and C.R. Antonescu performed the experiments. All authors participated in data analysis. M.J. Cavnar and R.P. DeMatteo wrote the manuscript with critical comments from all authors.
Footnotes
Abbreviations used:
- BMDM
- BM-derived macrophage
- C/EBP
- CCAAT/enhancer binding protein
- CREB
- cyclic AMP response element-binding
- GIST
- gastrointestinal stromal tumor
- LAP
- liver-enriched activator protein
- LIP
- liver-enriched inhibitory protein
- TAM
- tumor-associated macrophage
References
- Antonescu C.R., Besmer P., Guo T., Arkun K., Hom G., Koryotowski B., Leversha M.A., Jeffrey P.D., Desantis D., Singer S., et al. 2005. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin. Cancer Res. 11:4182–4190 10.1158/1078-0432.CCR-04-2245 [DOI] [PubMed] [Google Scholar]
- Balachandran V.P., Cavnar M.J., Zeng S., Bamboat Z.M., Ocuin L.M., Obaid H., Sorenson E.C., Popow R., Ariyan C., Rossi F., et al. 2011. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat. Med. 17:1094–1100 10.1038/nm.2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty G.L., Chiorean E.G., Fishman M.P., Saboury B., Teitelbaum U.R., Sun W., Huhn R.D., Song W., Li D., Sharp L.L., et al. 2011. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 331:1612–1616 10.1126/science.1198443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.K., Mantovani A. 2010. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11:889–896 10.1038/ni.1937 [DOI] [PubMed] [Google Scholar]
- Boring L., Gosling J., Chensue S.W., Kunkel S.L., Farese R.V., Jr, Broxmeyer H.E., Charo I.F. 1997. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J. Clin. Invest. 100:2552–2561 10.1172/JCI119798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett S.H., Kershen E.J., Zhang J., Zeng L., Straley S.C., Kaplan A.M., Cohen D.A. 2004. Conditional macrophage ablation in transgenic mice expressing a Fas-based suicide gene. J. Leukoc. Biol. 75:612–623 10.1189/jlb.0903442 [DOI] [PubMed] [Google Scholar]
- Combadière C., Potteaux S., Gao J.L., Esposito B., Casanova S., Lee E.J., Debré P., Tedgui A., Murphy P.M., Mallat Z. 2003. Decreased atherosclerotic lesion formation in CX3CR1/apolipoprotein E double knockout mice. Circulation. 107:1009–1016 10.1161/01.CIR.0000057548.68243.42 [DOI] [PubMed] [Google Scholar]
- Coniglio S.J., Eugenin E., Dobrenis K., Stanley E.R., West B.L., Symons M.H., Segall J.E. 2012. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol. Med. 18:519–527 10.2119/molmed.2011.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csóka B., Németh Z.H., Virág L., Gergely P., Leibovich S.J., Pacher P., Sun C.X., Blackburn M.R., Vizi E.S., Deitch E.A., Haskó G. 2007. A2A adenosine receptors and C/EBPbeta are crucially required for IL-10 production by macrophages exposed to Escherichia coli. Blood. 110:2685–2695 10.1182/blood-2007-01-065870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel T.J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J.R., Zhang L., Burow M., et al. 2004. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10:942–949 10.1038/nm1093 [DOI] [PubMed] [Google Scholar]
- Delahaye N.F., Rusakiewicz S., Martins I., Ménard C., Roux S., Lyonnet L., Paul P., Sarabi M., Chaput N., Semeraro M., et al. 2011. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat. Med. 17:700–707 10.1038/nm.2366 [DOI] [PubMed] [Google Scholar]
- Demetri G.D., Wang Y., Wehrle E., Racine A., Nikolova Z., Blanke C.D., Joensuu H., von Mehren M. 2009. Imatinib plasma levels are correlated with clinical benefit in patients with unresectable/metastatic gastrointestinal stromal tumors. J. Clin. Oncol. 27:3141–3147 10.1200/JCO.2008.20.4818 [DOI] [PubMed] [Google Scholar]
- DeNardo D.G., Brennan D.J., Rexhepaj E., Ruffell B., Shiao S.L., Madden S.F., Gallagher W.M., Wadhwani N., Keil S.D., Junaid S.A., et al. 2011. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1:54–67 10.1158/2159-8274.CD-10-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar A.L., Zannettino A.C., Hughes T.P., Lyons A.B. 2005. Inhibition of c-fms by imatinib: expanding the spectrum of treatment. Cell Cycle. 4:851–853 10.4161/cc.4.7.1788 [DOI] [PubMed] [Google Scholar]
- Ducimetière F., Lurkin A., Ranchère-Vince D., Decouvelaere A.V., Péoc’h M., Istier L., Chalabreysse P., Muller C., Alberti L., Bringuier P.P., et al. 2011. Incidence of sarcoma histotypes and molecular subtypes in a prospective epidemiological study with central pathology review and molecular testing. PLoS ONE. 6:e20294 10.1371/journal.pone.0020294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G.P., Old L.J., Schreiber R.D. 2004. The three Es of cancer immunoediting. Annu. Rev. Immunol. 22:329–360 10.1146/annurev.immunol.22.012703.104803 [DOI] [PubMed] [Google Scholar]
- Gabrilovich D.I., Ostrand-Rosenberg S., Bronte V. 2012. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 12:253–268 10.1038/nri3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. ; Immunological Genome Consortium 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Jung S., Littman D.R. 2003. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 19:71–82 10.1016/S1074-7613(03)00174-2 [DOI] [PubMed] [Google Scholar]
- Gold J.S., van der Zwan S.M., Gönen M., Maki R.G., Singer S., Brennan M.F., Antonescu C.R., De Matteo R.P. 2007. Outcome of metastatic GIST in the era before tyrosine kinase inhibitors. Ann. Surg. Oncol. 14:134–142 10.1245/s10434-006-9177-7 [DOI] [PubMed] [Google Scholar]
- Hamilton J.A., Achuthan A. 2013. Colony stimulating factors and myeloid cell biology in health and disease. Trends Immunol. 34:81–89 10.1016/j.it.2012.08.006 [DOI] [PubMed] [Google Scholar]
- Hart K.M., Bak S.P., Alonso A., Berwin B. 2009. Phenotypic and functional delineation of murine CX(3)CR1 monocyte-derived cells in ovarian cancer. Neoplasia. 11:564–573: 1: 573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M.C., Corless C.L., Duensing A., McGreevey L., Chen C.J., Joseph N., Singer S., Griffith D.J., Haley A., Town A., et al. 2003. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 299:708–710 10.1126/science.1079666 [DOI] [PubMed] [Google Scholar]
- Heusinkveld M., van der Burg S.H. 2011. Identification and manipulation of tumor associated macrophages in human cancers. J. Transl. Med. 9:216 10.1186/1479-5876-9-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota S., Isozaki K., Moriyama Y., Hashimoto K., Nishida T., Ishiguro S., Kawano K., Hanada M., Kurata A., Takeda M., et al. 1998. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 279:577–580 10.1126/science.279.5350.577 [DOI] [PubMed] [Google Scholar]
- Hume D.A., MacDonald K.P. 2012. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood. 119:1810–1820 10.1182/blood-2011-09-379214 [DOI] [PubMed] [Google Scholar]
- Joensuu H., DeMatteo R.P. 2012. The management of gastrointestinal stromal tumors: a model for targeted and multidisciplinary therapy of malignancy. Annu. Rev. Med. 63:247–258 10.1146/annurev-med-043010-091813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., Littman D.R. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausgruber T., Blazek K., Smallie T., Alzabin S., Lockstone H., Sahgal N., Hussell T., Feldmann M., Udalova I.A. 2011. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 12:231–238 10.1038/ni.1990 [DOI] [PubMed] [Google Scholar]
- Lawrence T., Natoli G. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11:750–761 10.1038/nri3088 [DOI] [PubMed] [Google Scholar]
- Lewis C.E., Pollard J.W. 2006. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 66:605–612 10.1158/0008-5472.CAN-05-4005 [DOI] [PubMed] [Google Scholar]
- Lin E.Y., Nguyen A.V., Russell R.G., Pollard J.W. 2001. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 193:727–740 10.1084/jem.193.6.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley P.W., Stiefl N., Cowan-Jacob S.W., Kaufman S., Mestan J., Wartmann M., Wiesmann M., Woodman R., Gallagher N. 2010. Structural resemblances and comparisons of the relative pharmacological properties of imatinib and nilotinib. Bioorg. Med. Chem. 18:6977–6986 10.1016/j.bmc.2010.08.026 [DOI] [PubMed] [Google Scholar]
- Marigo I., Bosio E., Solito S., Mesa C., Fernandez A., Dolcetti L., Ugel S., Sonda N., Bicciato S., Falisi E., et al. 2010. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 32:790–802 10.1016/j.immuni.2010.05.010 [DOI] [PubMed] [Google Scholar]
- Ménard C., Blay J.Y., Borg C., Michiels S., Ghiringhelli F., Robert C., Nonn C., Chaput N., Taïeb J., Delahaye N.F., et al. 2009. Natural killer cell IFN-gamma levels predict long-term survival with imatinib mesylate therapy in gastrointestinal stromal tumor-bearing patients. Cancer Res. 69:3563–3569 10.1158/0008-5472.CAN-08-3807 [DOI] [PubMed] [Google Scholar]
- Mosser D.M., Zhang X. 2008. Activation of murine macrophages. Curr. Protoc. Immunol. Chapter 14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi K., Laoui D., Gysemans C., Baeten M., Stangé G., Van den Bossche J., Mack M., Pipeleers D., In’t Veld P., De Baetselier P., Van Ginderachter J.A. 2010. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 70:5728–5739 10.1158/0008-5472.CAN-09-4672 [DOI] [PubMed] [Google Scholar]
- Pavlidis P., Noble W.S. 2003. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 19:295–296 10.1093/bioinformatics/19.2.295 [DOI] [PubMed] [Google Scholar]
- Qian B.Z., Pollard J.W. 2010. Macrophage diversity enhances tumor progression and metastasis. Cell. 141:39–51 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R., Kaiser E.A., Snyder L.A., Pollard J.W. 2011. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 475:222–225 10.1038/nature10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramji D.P., Foka P. 2002. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365:561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell D., Mourkioti F., Gambardella A., Kirstetter P., Lopez R.G., Rosenthal N., Nerlov C. 2009. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA. 106:17475–17480 10.1073/pnas.0908641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell B., Affara N.I., Coussens L.M. 2012. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 33:119–126 10.1016/j.it.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusakiewicz S., Semeraro M., Sarabi M., Desbois M., Locher C., Mendez R., Vimond N., Concha A., Garrido F., Isambert N., et al. 2013. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res. 73:3499–3510 10.1158/0008-5472.CAN-13-0371 [DOI] [PubMed] [Google Scholar]
- Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., et al. 2010. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11:936–944 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- Schmieder A., Michel J., Schönhaar K., Goerdt S., Schledzewski K. 2012. Differentiation and gene expression profile of tumor-associated macrophages. Semin. Cancer Biol. 22:289–297 10.1016/j.semcancer.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Hohl T.M., Cherny M., Pamer E.G. 2009. Selective expansion of the monocytic lineage directed by bacterial infection. J. Immunol. 183:1900–1910 10.4049/jimmunol.0900612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao S.L., Ganesan A.P., Rugo H.S., Coussens L.M. 2011. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 25:2559–2572 10.1101/gad.169029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas G., Germano G., Mantovani A., Allavena P. 2009. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J. Leukoc. Biol. 86:1065–1073 10.1189/jlb.0609385 [DOI] [PubMed] [Google Scholar]
- Sommer G., Agosti V., Ehlers I., Rossi F., Corbacioglu S., Farkas J., Moore M., Manova K., Antonescu C.R., Besmer P. 2003. Gastrointestinal stromal tumors in a mouse model by targeted mutation of the Kit receptor tyrosine kinase. Proc. Natl. Acad. Sci. USA. 100:6706–6711 10.1073/pnas.1037763100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo T., Nishikawa S., Ogawa M., Kataoka H., Ohno N., Izawa A., Hayashi S., Nishikawa S. 1995. Functional hierarchy of c-kit and c-fms in intramarrow production of CFU-M. Oncogene. 11:2469–2476 [PubMed] [Google Scholar]
- van Dongen M., Savage N.D., Jordanova E.S., Briaire-de Bruijn I.H., Walburg K.V., Ottenhoff T.H., Hogendoorn P.C., van der Burg S.H., Gelderblom H., van Hall T. 2010. Anti-inflammatory M2 type macrophages characterize metastasized and tyrosine kinase inhibitor-treated gastrointestinal stromal tumors. Int. J. Cancer. 127:899–909 [DOI] [PubMed] [Google Scholar]
- Voll R.E., Herrmann M., Roth E.A., Stach C., Kalden J.R., Girkontaite I. 1997. Immunosuppressive effects of apoptotic cells. Nature. 390:350–351 10.1038/37022 [DOI] [PubMed] [Google Scholar]
- Wang B., Li Q., Qin L., Zhao S., Wang J., Chen X. 2011. Transition of tumor-associated macrophages from MHC class II(hi) to MHC class II(low) mediates tumor progression in mice. BMC Immunol. 12:43 10.1186/1471-2172-12-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S., Feirt N., Goldstein M., Guarrera J., Ippagunta N., Ekong U., Dun H., Lu Y., Qu W., Schmidt A.M., Emond J.C. 2004. Blockade of receptor for advanced glycation end product (RAGE) attenuates ischemia and reperfusion injury to the liver in mice. Hepatology. 39:422–432 10.1002/hep.20045 [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Apetoh L., Ghiringhelli F., André F., Tesniere A., Kroemer G. 2008. The anticancer immune response: indispensable for therapeutic success? J. Clin. Invest. 118:1991–2001 10.1172/JCI35180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.