Inflammatory cytokines drive NK cell expansion in the absence of the transcription factor Nfil3, and Nfil3 is dispensable for the maintenance and function of mature NK cells.
Abstract
Development of the natural killer (NK) cell lineage is dependent on the transcription factor Nfil3 (or E4BP4), which is thought to act downstream of IL-15 signaling. Nfil3-deficient mice lack NK cells, whereas other lymphocyte lineages (B, T, and NKT cells) remain largely intact. We report the appearance of Ly49H-expressing NK cells in Nfil3−/− mice infected with mouse cytomegalovirus (MCMV) or recombinant viruses expressing the viral m157 glycoprotein. Nfil3−/− NK cells at the peak of antigen-driven expansion were functionally similar to NK cells from infected wild-type mice with respect to IFN-γ production and cytotoxicity, and could comparably produce long-lived memory NK cells that persisted in lymphoid and nonlymphoid tissues for >60 d. We demonstrate that generation and maintenance of NK cell memory is an Nfil3-independent but IL-15–dependent process. Furthermore, specific ablation of Nfil3 in either immature NK cells in the bone marrow or mature peripheral NK cells had no observable effect on NK cell lineage maintenance or homeostasis. Thus, expression of Nfil3 is crucial only early in the development of NK cells, and signals through activating receptors and proinflammatory cytokines during viral infection can bypass the requirement for Nfil3, promoting the proliferation and long-term survival of virus-specific NK cells.
NK cells have historically been considered components of the innate immune system, recognizing virally infected and tumor cells through germline-encoded receptors, and rapidly eliminating these targets through the secretion of lytic granules. However, recent studies using mouse models have shown that NK cells can exhibit features of adaptive immune responses, including antigen-specific and -dependent clonal expansion and the ability to differentiate into long-lived memory cells that display anamnestic responses to secondary antigen exposure (Daniels et al., 2001; Dokun et al., 2001; O’Leary et al., 2006; Cooper et al., 2009; Sun et al., 2009a; Paust et al., 2010). Several groups have demonstrated that analogous antigen-specific effector and memory NK cell populations can also arise in humans during viral infection (Björkström et al., 2011; Lopez-Vergès et al., 2011; Della Chiesa et al., 2012; Foley et al., 2012).
The NK cell response against mouse cytomegalovirus (MCMV) infection has been historically well characterized. In MCMV-resistant WT mouse strains (e.g., C57BL/6), the activating NK cell receptor Ly49H has been shown to specifically recognize the MCMV-encoded glycoprotein m157, which is expressed on infected cells (Arase et al., 2002; Smith et al., 2002). Receptor-ligand engagement triggers the rapid proliferation of Ly49H+ NK cells, generating large numbers of antigen-specific effector cells (representing >90% of the total NK cell population) by day 7 post infection (PI) (Daniels et al., 2001; Dokun et al., 2001; Sun et al., 2009a). After viral clearance, a population of long-lived memory NK cells remain in both lymphoid and nonlymphoid tissues and display enhanced effector functions upon secondary MCMV exposure (Sun et al., 2009a).
The development of NK cells from common lymphoid progenitor (CLP) cells in the bone marrow is critically dependent on IL-15, and mice unable to produce or respond to IL-15 (Il15−/−, Il15ra−/−, and Rag2−/− × Il2rg−/− mice) lack mature NK cells (Lodolce et al., 1998; Kennedy et al., 2000; Cooper et al., 2002). Developmental and survival signals downstream of the IL-15 receptor in NK cells are thought to be mediated by the basic leucine zipper transcription factor Nfil3 (nuclear factor interleukin-3 regulated; also known as E4BP4; Gascoyne et al., 2009; Kamizono et al., 2009; Kashiwada et al., 2010). Similar to Rag2−/− × Il2rγ−/−, Il15−/−, and Il15rα−/− mice, Nfil3−/− mice typically contain <0.1% NK cells in most organs (compared with 2–5% in WT mice).
In addition to its role in NK cell development, Nfil3 has been shown to regulate a wide range of cellular processes in other lymphocyte subsets, including the survival of pro–B cells (Ikushima et al., 1997), IgE class-switching in B cells (Kashiwada et al., 2010), IL-3 transcription in T cells (Zhang et al., 1995), development of CD8α+ dendritic cells (Kashiwada et al., 2011), and modulation of TH2 responses (Kashiwada et al., 2010; Kobayashi et al., 2011; Motomura et al., 2011). Given the breadth of these roles, we considered that Nfil3 may regulate post-development processes such as homeostasis and antiviral responses in NK cells, a hypothesis supported by gene array studies demonstrating continued expression of Nfil3 transcript in mature resting and activated NK cells (Sun et al., 2011). In addition to using Nfil3-deficient mice, we developed and used mice in which the Nfil3 gene could be conditionally deleted to investigate the role of Nfil3 in NK cell homeostasis, activation, clonal expansion, and memory cell generation.
RESULTS
Viral infection drives expansion of NK cells in an Nfil3-independent manner
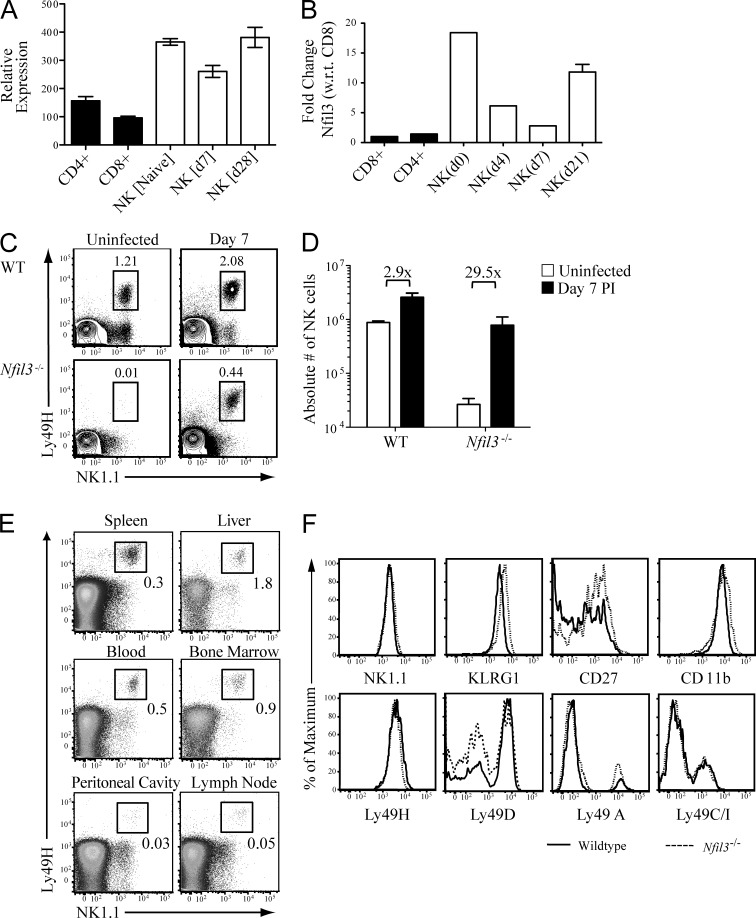
Nfil3 expression levels were evaluated by the ImmGen consortium microarray and confirmed by quantitative RT-PCR in sorted NK cell populations. At rest, NK cells express higher levels of Nfil3 mRNA than CD8+ and CD4+ T cells (Fig. 1, A and B). Nfil3 expression in NK cells modulates immediately after MCMV infection, suggesting activation-induced regulation of Nfil3 expression, but resting and memory NK cells exhibited comparable levels of Nfil3 (Fig. 1, A and B). We investigated whether elevated Nfil3 transcript (relative to T cell controls) correlated with survival and function in NK cells at various stages before and after viral infection.
Figure 1.
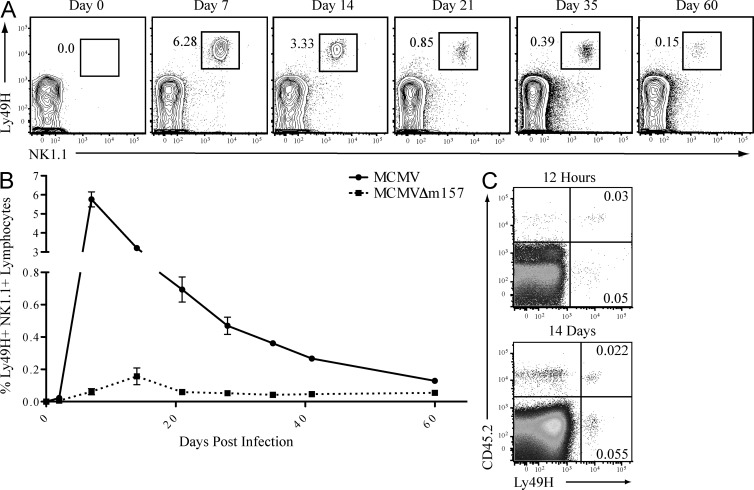
MCMV-induced expansion of an NK1.1+ TCR-β− Ly49H+ NK cell population in Nfil3−/− mice. WT C57BL/6 mice were infected with 6 × 103 pfu MCMV (i.p.), and CD8+ T cells, CD4+ T cells, and NK cells were sorted from spleen at the indicated days after infection. Relative levels of Nfil3 mRNA were determined by the ImmGen microarray (A) or by quantitative RT-PCR (B). (C) Uninfected and MCMV-infected WT and Nfil3−/− mice were analyzed for percentage (indicated on plots) of Ly49H-expressing NK cells in spleens on day 7 PI. (D) Absolute numbers of NK1.1+ Ly49H+ NK cells are shown for uninfected and MCMV-infected WT and Nfil3−/− mice at day 7 PI. Fold expansion is indicated on graph. (E) Percentage of Ly49H+ NK cells in lymphoid and nonlymphoid tissues was determined in Nfil3−/− mice on day 7 PI. (F) NK cells were isolated from MCMV-infected WT and Nfil3−/− mice at day 7 PI and expression of the indicated marker was determined by flow cytometry. Error bars for all graphs show SEM and all data are representative of three independent experiments with n = 3–4 mice per group.
Although NK cells are nearly undetectable in Nfil3−/− mice at steady-state, MCMV infection elicited a profound increase in the overall percentage NK cells, of which the majority (>90%) expressed the activating receptor Ly49H (Fig. 1 C). The expansion of Nfil3−/− NK cells amounted to an ∼30-fold increase in total cell numbers, in contrast to a roughly threefold increase in WT NK cells (Fig. 1 D). The MCMV-expanded Nfil3−/− Ly49H+ NK cells were detected in both lymphoid and nonlymphoid tissues at day 7 PI (Fig. 1 E). Surface expression of Ly49 family activating receptors, as well as markers of activation and maturation, by WT and Nfil3−/− NK cells at day 7 PI, indicated that MCMV-expanded Nfil3−/− NK cells were phenotypically similar to WT NK cells (Fig. 1 F). NK cells from Nfil3−/− mice expressed comparable levels of activating and inhibitory Ly49 receptors to WT mice, with slightly increased expression of activation markers CD11b and KLRG1, likely a reflection of the greater fold expansion of Nfil3−/− NK cells compared with WT NK cells (Fig. 1 D). Thus, although Nfil3 is required for NK cell development, the virus-specific NK cells in Nfil3−/− mice can be driven to expand after MCMV infection.
MCMV-expanded Nfil3−/− NK cells are functionally competent
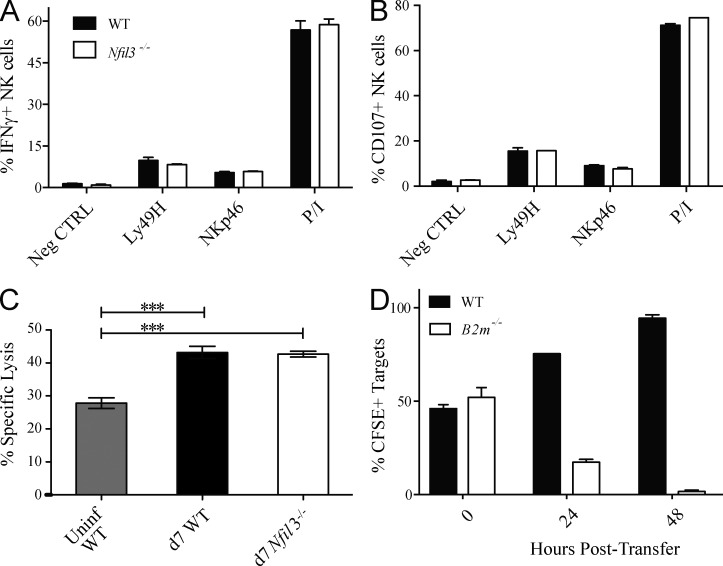
To address functionality, NK cells from MCMV-infected Nfil3−/− and WT mice were assessed by ex vivo stimulation using plate-bound antibodies specific for activating NK cell receptors and in vitro killing assays. Ly49H+ NK cells from Nfil3−/− and WT mice produced comparable amounts of IFN-γ, and degranulated similarly (measured by CD107a expression) in response to plate-bound antibodies, or PMA and ionomycin (Fig. 2, A and B). Nfil3−/− NK cells were capable of mediating in vitro killing of YAC-1 targets similar to WT NK cells (Fig. 2 C). Additionally, MCMV-infected Nfil3−/− mice were able to reject “missing-self” B2m−/− splenocytes at day 7 PI (Fig. 2 D), demonstrating the presence of functional Nfil3−/− NK cells in vivo. Altogether, these findings indicate that NK cells that proliferate in Nfil3−/− mice in response to MCMV infection represent fully functional effector cells.
Figure 2.
Comparable function of WT and Nfil3−/− NK cells after MCMV infection. Splenic WT and Nfil3−/− NK cells were stimulated for 5 h with anti-Ly49H, anti-NKp46, or PMA + Ionomycin (P/I) and evaluated for IFN-γ production (A) and degranulation (B; as determined by CD107a expression) by flow cytometry. (C) 51Cr-labeled YAC-I target cells were incubated with splenic WT or Nfil3−/− NK cells isolated at day 7 PI. Specific lysis was determined 4 h later. (D) CFSE-labeled WT and B2m−/− targets were injected (i.v.) into MCMV-infected Nfil3−/− mice at day 7 PI, and specific killing (percentage of CFSE+ cells remaining) was evaluated at the indicated time points after transfer. Error bars for all graphs show SEM and data are representative of three independent experiments with n = 3 mice per group.
Cognate viral ligand required for NK cell expansion in Nfil3−/− mice
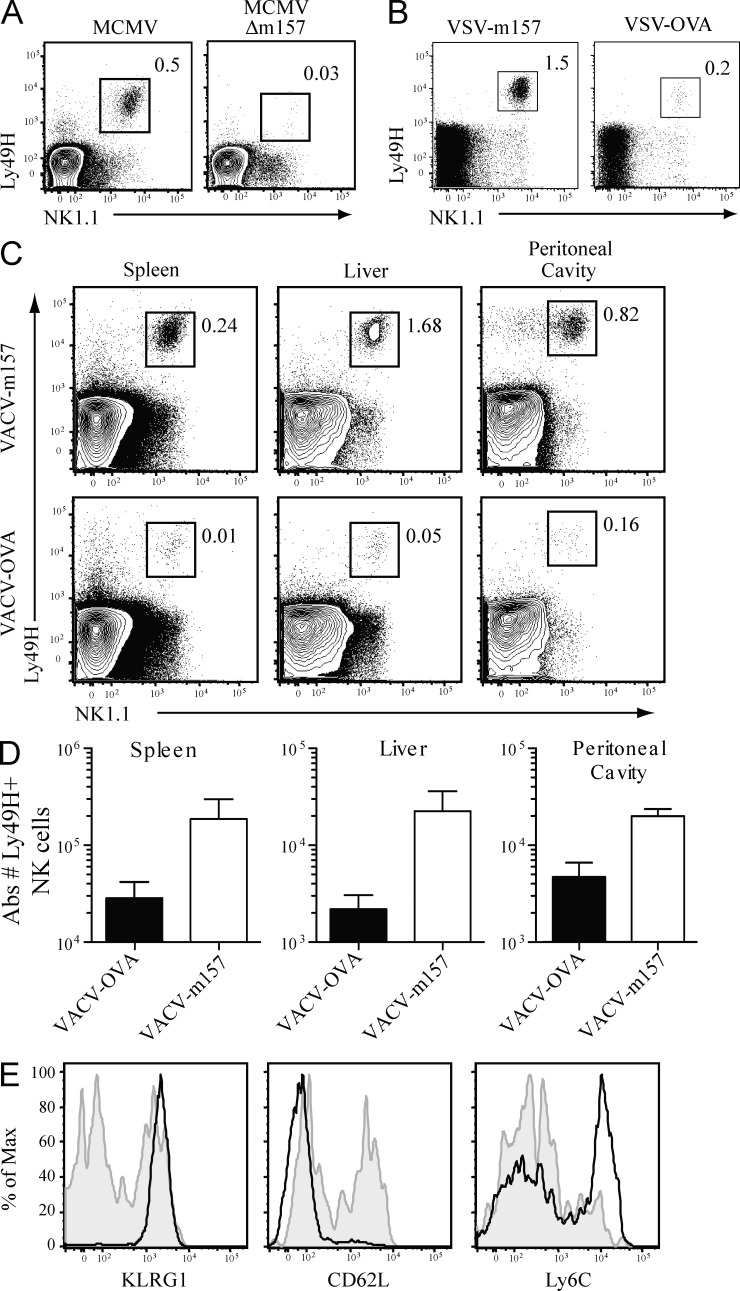
Given the selective expansion of Ly49H+ NK cells in Nfil3−/− mice, we investigated whether the MCMV glycoprotein m157 was required for the expansion of Nfil3−/− NK cells. To test this, we first infected mice with a strain of MCMV in which the m157 gene had been deleted (MCMVΔm157). Nfil3−/− mice did not demonstrate an increase in the percentage of Ly49H+ NK cells after infection with MCMVΔm157, in contrast to infection with parental MCMV, suggesting that expansion of Nfil3−/− Ly49H+ NK cells during MCMV infection depended on recognition of the m157 antigen (Fig. 3 A). To confirm this finding, we engineered a recombinant vesicular stomatitis virus expressing m157 (VSV-m157) and a vaccinia virus expressing m157 (VACV-m157). Nfil3−/− mice infected with either of these recombinant viruses demonstrated an expansion of the Ly49H+ NK cell population that was not observed during infection with control recombinant viruses expressing OVA (Fig. 3, B and C). In all experiments, significant expansion of Ly49H+ NK cells was observed in lymphoid and nonlymphoid organs in Nfil3−/− mice infected with m157-expressing viruses (Fig. 3 D and not depicted). Furthermore, only infection with m157-expressing viruses, but not with OVA-expressing viruses, induced a full maturation phenotype (KLRG1hi CD62Llo Ly6Chi) of Nfil3−/− NK cells (Fig. 3 E and not depicted). Thus, the presence of a cognate viral ligand is required to promote a robust proliferative program in NK cells lacking Nfil3.
Figure 3.
Proliferation of Nfil3−/− Ly49H+ NK cells requires cognate viral ligand m157. (A) Nfil3−/− mice were infected with MCMV (6 × 102 pfu) and MCMV-Δm157 (4.5 × 104 pfu). Percentage of Ly49H+ NK cells within total splenocyte populations were determined on day 7 PI. (B) Nfil3−/− mice were infected with recombinant VSV expressing m157 or OVA (107 pfu), and the percentage of Ly49H+ NK cells within the total splenocyte population was determined at day 7 PI. (C) Nfil3−/− mice were infected with recombinant VACV expressing m157 or OVA as in B, and percentage of Ly49H+ NK cells was determined in the indicated organs at day 7 PI. (D) Absolute numbers of Ly49H+ NK cells in Nfil3−/− mice infected with VACV-m157 or VACV-OVA are shown for the indicated tissues. (E) Nfil3−/− mice were infected with VACV-m157 (solid black lines) or VACV-OVA (shaded gray) and expression of KLRG1, CD62L, and Ly6C on splenic NK1.1+ Ly49H+ NK cells was determined at day 7 PI. Error bars for all graphs show SEM and data are representative of n = 3 mice per group, and experiments were performed twice.
Proinflammatory cytokines drive NK cell expansion in Nfil3−/− mice
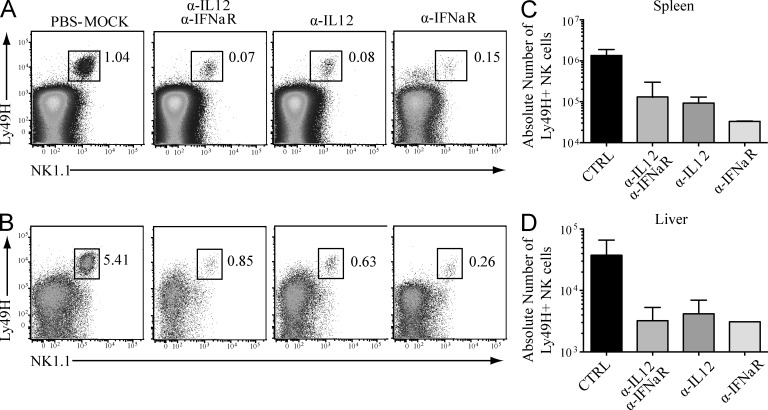
In addition to antigen receptor triggering in T and NK cells, signaling via proinflammatory cytokines such as IL-12 and type I IFNs, generated in response to viral infection, have been shown to play an important role in lymphocyte proliferation and effector function (Sun and Lanier, 2011). To examine the influence of these cytokines during the expansion of Ly49H+ NK cells in Nfil3−/− mice infected with MCMV, Nfil3−/− mice were injected with blocking antibodies against IL-12, the IFN-α receptor (IFNαR1), or both before infection. By percentage, Ly49H+ NK cell expansion was significantly reduced in the spleen and liver of mice treated with blocking antibodies compared with control Nfil3−/− mice treated with PBS (Fig. 4, A and B). The absolute numbers of Ly49H+ NK cells in antibody-treated Nfil3−/− mice were reduced by >10-fold compared with control animals (Fig. 4, C and D). Reduced numbers were similarly detected in all lymphoid and nonlymphoid tissues of antibody-treated mice (unpublished data). Interestingly, disruption of either IL-12 or type-I IFN signaling results in a defect comparable to ablation of both cytokine pathways, suggesting an absolute requirement for each of these cytokines in mediating the NK cell expansion during MCMV infection.
Figure 4.
Proinflammatory cytokines IL-12 and type I IFNs drive proliferation of Ly49H+ NK cells in Nfil3−/− mice during MCMV infection. Nfil3−/− mice were pretreated with the indicated blocking antibodies (or PBS as a control) 24 h before MCMV infection, and the percentage of Ly49H+ NK cells in spleen (A) and liver (B) was determined by flow cytometry at day 7 PI. (C and D) Absolute numbers of Ly49H+ NK cells at day 7 PI were determined in spleen (C) and liver (D). Error bars for all graphs show SEM and data are representative of n = 3 mice per group, and the experiments were performed twice.
Nfil3−/− NK cells become long-lived memory cells after MCMV infection
MCMV infection generates a population of memory Ly49H+ NK cells that persists for several months after viral clearance (Sun et al., 2009a). To determine whether Nfil3 influences the formation and persistence of memory NK cells, we measured the number of Ly49H+ NK cells in Nfil3−/− mice at various time points after MCMV infection. Ly49H+ NK cells were detected in the blood, liver, and spleen of Nfil3−/− mice for >60 d after infection (Fig. 5 A and not depicted). Thus, the stages of NK cell expansion, contraction, and memory persistence after MCMV infection could be clearly observed in Nfil3−/− mice (Fig. 5 B), similar to previous observations in WT mice (Sun et al., 2009a). Interestingly, although optimal numbers of effector and memory Nfil3−/− NK cells were only observed after infection with MCMV, but not MCMVΔm157, a small population of NK cells appeared in Nfil3−/− mice infected with MCMVΔm157 at day 14 PI (Fig. 5 B and not depicted), suggesting that ligand-independent signals, such as inflammation, are sufficient to drive a limited degree of NK cell expansion in Nfil3−/− mice.
Figure 5.
MCMV infection of Nfil3−/− mice produces long-lived memory NK cells. (A) Nfil3−/− mice were infected with MCMV (6 × 102 pfu), and percentage of Ly49H+ NK cells in spleen was determined by flow cytometry at the indicated time points. (B) Nfil3−/− mice were infected with MCMV or MCMV-Δm157, and Ly49H+ NK cell percentages were enumerated in spleen at the indicated time points. (C) Ly49H+ NK cells from day 7 MCMV-infected WT (CD45.1) and Nfil3−/− (CD45.2) were transferred into Ly49h−/− mice and frequency of transferred cells was determined at 12 h (top plot) or 14 d (bottom plot) after transfer. Error bars for all graphs show SEM and data are representative of n = 3–4 mice per group, and experiments were repeated three times.
To confirm that the persisting NK cells in MCMV-infected Nfil3−/− mice truly represent long-lived memory cells, and not newly generated cells egressing from the bone marrow at later time points, equal numbers of WT (CD45.1) and Nfil3−/− (CD45.2) Ly49H+ NK cells from mice infected with MCMV (day 7 PI) were adoptively transferred into naive Ly49h−/− hosts (Ly49h−/− mice do not express the Ly49H receptor but have normal numbers of NK cells). Donor WT NK cells were derived from NK cells originally transferred to Ly49h−/− host mice before MCMV infection, to compare NK cell populations that had undergone a similar fold expansion. The absence of CD69 up-regulation on the lymphocytes of recipient mice after adoptive transfer confirmed that virus had not been co-injected along with the NK cells into the naive Ly49h−/− mice (unpublished data). WT and Nfil3−/− Ly49H+ NK cells were detectable in peripheral blood at 12 h after transfer and were recovered at similar ratios 14 d later (corresponding to 21 d from initial virus exposure; Fig. 5 C), suggesting survival and maintenance of memory NK cell populations occur independent of Nfil3.
Survival of effector and memory NK cells is IL-15 dependent
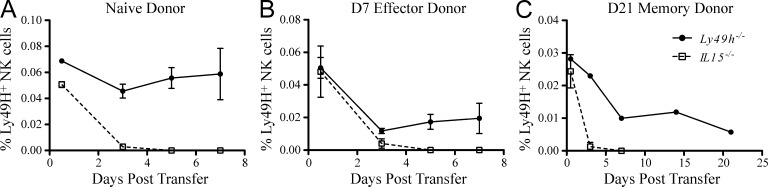
Our data demonstrates that NK cell effector and memory cell generation occurs independent of Nfil3. Because Nfil3 has been suggested to act downstream of IL-15 receptor signaling (Gascoyne et al., 2009), we hypothesized that memory NK cells may not have the same requirement for IL-15 as resting NK cells. To test this, resting NK cells (Fig. 6 A) or effector Ly49H+ NK cells isolated from MCMV-infected WT donor mice at day 7 PI (Fig. 6 B) were transferred into Il15−/− or Ly49h−/− mice. Whereas survival of both the transferred WT NK cell populations was observed in Ly49h−/− mice, resting and effector NK cells transferred into the Il15−/− hosts rapidly disappeared (Fig. 6, A and B). Similarly, memory NK cells (purified at day 21 PI) were also found to be dependent on IL-15, as cells transferred into Il15−/− hosts disappeared within 3 d (Fig. 6 C). Thus, the maintenance of memory NK cells after MCMV infection appears to be an IL-15–dependent but Nfil3-independent process. These results indicate the existence of an Nfil3-independent signaling pathway that facilitates the long-term survival of IL-15–dependent NK cells.
Figure 6.
IL-15 requirement of resting, effector, and memory NK cells. NK cells purified from WT mice were adoptively transferred into Il15−/− or Ly49h−/− mice. Percentages of naive mature NK cells (from uninfected mice; A), day 7 PI effector NK cells (B), or day 21 PI memory NK cells (C) were determined at indicated time points after transfer. (Transferred effector and memory NK cells were originally isolated from Ly49h−/− mice receiving Ly49H+ NK cells and infected 7 or 21 d earlier, respectively.) Error bars for all graphs show SEM and data are representative of n = 3–4 mice per group, and experiments were independently repeated three times.
Survival and homeostasis of mature NK cells is Nfil3 independent
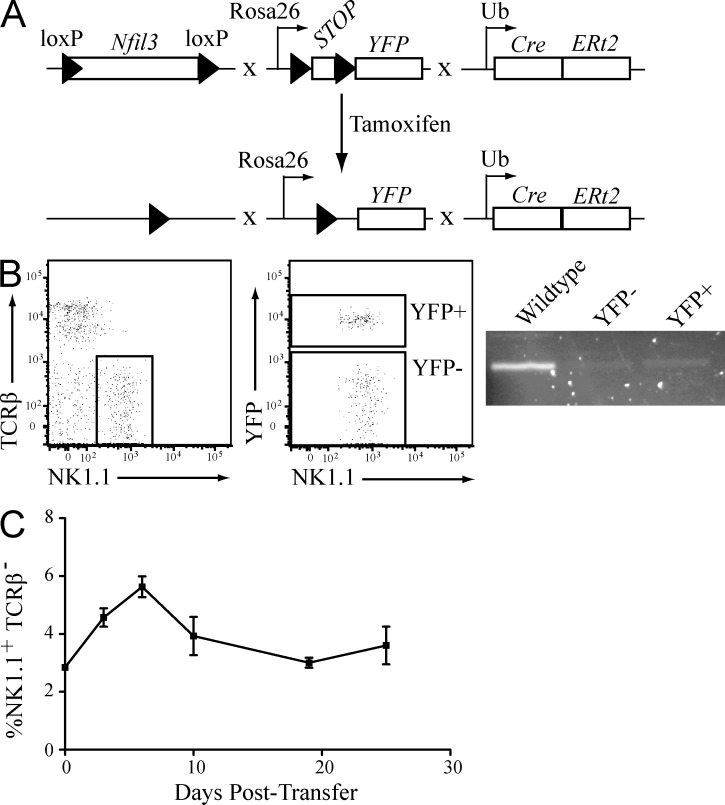
To determine whether mature NK cells continued to require Nfil3 for lineage survival in the absence of infection or inflammation, we generated mice in which Nfil3 could be conditionally deleted in mature resting NK cells. We generated mice in which the Nfil3 gene is flanked by loxP sites (Nfil3fl/fl; Motomura et al., 2011), a tamoxifen-inducible Cre cassette is driven by the Ubiquitin promoter (UbCre-ERt2; Ruzankina et al., 2007), and the Rosa26 promoter drives a floxed “STOP” with YFP reporter cassette (R26Y; Srinivas et al., 2001). These mice (Nfil3fl/fl × R26Y × UbCreERt2) allow for the inducible deletion of Nfil3 upon tamoxifen treatment, and Cre-recombinase activity monitored by tracking the YFP+ population (Fig. 7 A). To address the impact of Nfil3 ablation on the homeostasis of mature NK cells, NK cells from Nfil3fl/fl × R26Y × UbCreERt2 mice (CD45.2) were adoptively transferred into WT recipient mice (CD45.1). Upon tamoxifen treatment, ablation of Nfil3 in both YFP+ and YFP− NK cells was confirmed by PCR (Fig. 7 B), suggesting that YFP expression underreports Cre activity and excision of the floxed Nfil3 gene. Interestingly, transferred NK cells were readily detectable 25 d after tamoxifen treatment (Fig. 7 C). Thus, genetic deletion of Nfil3 in mature NK cells had no effect on homeostatic survival and maintenance.
Figure 7.
Nfil3 expression is dispensable in mature NK cells. (A) Schematic of the Nfil3fl/fl × R26Y × UbCre-ERt2 mouse system for tamoxifen-inducible, Cre/loxP-mediated deletion of the Nfil3 gene. (B) Nfil3 gene deletion after tamoxifen treatment was confirmed by PCR in sorted YFP+ or YFP− NK cells. WT NK cells were sorted as a positive control for Nfil3 gene detection. (C) Nfil3fl/fl × R26Y × UbCre-ERt2 mice were treated with tamoxifen and mature NK cells were sorted and transferred into WT recipient mice. The presence of transferred cells determined at indicated time points after transfer. Error bars for all graphs show SEM and data are representative of n = 3–4 mice per group, and experiments were independently repeated three times.
Requirement for Nfil3 restricted to NK cell progenitors
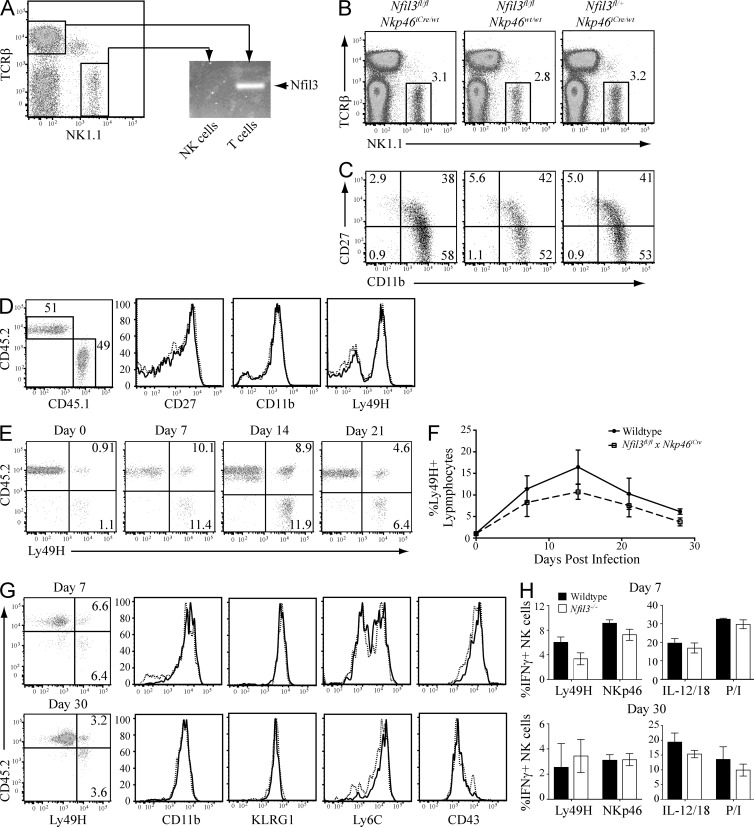
We next investigated whether Nfil3 is required during the developmental stage when NK cell progenitors first acquire their activating receptors (e.g., NKp46). We crossed the recently developed Nkp46iCre mouse, which expresses Cre-recombinase under control of the Ncr1 (NKp46) gene promoter (Narni-Mancinelli et al., 2011), to the Nfil3fl/fl mice. Expression of NKp46 is thought to occur immediately after acquisition of the NK lineage-defining markers CD122 and NK1.1 (Narni-Mancinelli et al., 2011). Deletion of Nfil3 was confirmed in NK cells but not other immune cell lineages from Nfil3fl/fl × Nkp46iCre mice by PCR analysis of genomic DNA from sorted NK cells and T cells obtained from the same mouse (Fig. 8 A). Surprisingly, Nfil3fl/fl × Nkp46iCre mice contained a comparable number of mature NK cells to littermate control mice (Fig. 8 B). Moreover, surface expression of common NK cell maturation markers CD11b and CD27 did not differ significantly between NK cells from Nfil3fl/fl × Nkp46iCre mice and littermate controls (Fig. 8 C). We next generated mixed 1:1 WT:Nfil3fl/fl × Nkp46iCre bone marrow chimeric mice to evaluate the ability of the Nfil3fl/fl × Nkp46iCre NK cells to develop in a competitive environment with WT NK cells. We found that NK cells that have ablated Nfil3 early in development (CD45.2) reconstituted similarly to WT NK cells (CD45.1), suggesting that Nfil3 is exclusively required during the progenitor stages, but not once developing NK cells acquire their first activating receptors (Fig. 8 D).
Figure 8.
Nfil3 is not required for NK cell maturation and function beyond the early NK cell progenitor stage. (A) NK cells and T cells were sorted from Nfil3fl/fl × Nkp46iCre mice and Nfil3 expression was analyzed by PCR. (B) Percentage of mature NK cells was determined in Nfil3fl/fl × Nkp46iCre mice and littermate controls. (C) Expression of maturation markers CD27 and CD11b was determined on NK cells from Nfil3fl/fl × Nkp46iCre mice and littermate controls. (D) Mixed bone marrow chimeric mice were generated using a 1:1 ratio of bone marrow from WT (CD45.1, solid lines) and Nfil3fl/fl × Nkp46iCre (CD45.2, dotted lines) donor mice. Percentages of donor cells and expression of CD27, CD11b, and Ly49H was determined by flow cytometry at 8 wk after irradiation and reconstitution. (E and F) Splenic NK cells were purified from chimeric mice generated in D and adoptively transferred to Ly49h−/− hosts. Percentages of Ly49H+ NK cells were determined at indicated time points after MCMV infection. (G) Ly49H+ NK cells from MCMV-infected WT (solid lines) and Nfil3fl/fl × NKp46iCre (dotted lines) mice were analyzed for expression of the indicated molecules at days 7 (top) and 30 (bottom) PI. (H) The NK cells from G were stimulated for 5 h with anti-Ly49H, anti-NKp46, IL-12 + IL-18, or PMA + Ionomycin (P/I) and evaluated for IFN-γ production by flow cytometry. Error bars for all graphs show SEM and all data are representative of n = 3–4 mice per group per time point, repeated in three independent experiments.
Lastly, we adoptively transferred the WT (CD45.1) and Nfil3fl/fl × Nkp46iCre (CD45.2) NK cells from the bone marrow chimeras into Ly49h−/− mice to investigate their ability to respond to MCMV infection. Both NK cell populations expanded robustly in response to infection and contracted to form comparable memory cell populations (Fig. 8, E and F). The effector and memory Nfil3fl/fl × Nkp46iCre NK cells were phenotypically and functionally similar to their WT counterparts (Fig. 8, G and H). Altogether, these results confirm that Nfil3 is not required for the survival or maintenance of NK cells beyond the earliest stages of development and lineage commitment.
DISCUSSION
IL-15 and Nfil3 are both factors critical for the development of a mature and functional NK cell pool in the periphery, and mice deficient in Nfil3 or components of the IL-15 signaling machinery lack NK cells at steady-state. However, despite stringent requirement for IL-15 in NK cell development, recent data indicate that viral infection of NK cell–deficient mice (e.g., Il15−/−, Il15ra−/−, Rag2−/− × Il2rb−/−, and Rag2−/− × Il2rg−/−) drives out a substantial population of mature NK cells that are functionally and phenotypically similar to those generated in infected WT mice (Sun et al., 2009b). These findings suggested that ligation of activating receptors (e.g., Ly49H) and signals from proinflammatory cytokines such as IL-12 can bypass the requirement for IL-15 during viral infection (Sun et al., 2009b). Our data indicates that although Nfil3 is indispensable for early stage NK cell development, it is not essential when NK cell maturation and functionality are driven by viral infection. We have established that Ly49H ligation plays a large role in the generation of Nfil3-deficient effector and memory NK cells, as do the proinflammatory cytokines IL-12 and type I IFN. Interestingly, a recent study demonstrated that cross-presenting CD8α+ DCs, which are also dependent on Nfil3 for their development (Kashiwada et al., 2011), could be induced to expand in short-term bone marrow reconstitution settings in the absence of Nfil3, and were phenotypically similar to WT DCs (Seillet et al., 2013). Together with our findings in NK cells, these results highlight that under specific conditions, certain lineage-determining transcriptional regulators are dispensable for subsequent lineage maintenance and function.
The precise source of these virally expanded NK cells in Nfil3−/− mice remains elusive. NK cells differentiate from CLP cells in the bone marrow, maturing through several stages under the influence of specific cytokines (especially IL-15) and transcription factors to result in mature, functional cells (Yokoyama et al., 2004; Martín-Fontecha et al., 2011). Similar to NK cell–deficient mice where IL-15 signals are ablated, the minute number of peripheral NK cells present in Nfil3−/− mice may be contributing to the robust NK cell numbers after MCMV infection. However, it is also possible that immature NK cells newly expressing Ly49H in the bone marrow are rapidly recruited to peripheral organs during viral infection, even though NK cell development in Nfil3−/− mice is thought to be arrested at the early NK cell progenitor (NKP) stage (Gascoyne et al., 2009; Kamizono et al., 2009). We have previously demonstrated that adoptive transfer of as few as 104 Ly49H+ NK cells into Ly49H-deficient recipient mice, followed by MCMV infection, results in a detectable effector population at day 7 PI (Sun et al., 2009a). Therefore, to determine whether the MCMV-expanded Nfil3−/− Ly49H+ NK cells originated from the small percentage of peripheral NK cells, whole splenocytes from Nfil3−/− mice were adoptively transferred into Ly49h−/− recipient mice and infected with MCMV. No effector Ly49H+ NK cells could be detected at day 7 PI (unpublished data), suggesting that our whole spleen preparation contained <104 Ly49H+ NK cells. Although the origin of the MCMV-driven NK cells remains to be determined, perhaps a combination of the immature bone marrow–derived NK cells and peripheral NK cells contribute to the significant population of Ly49H+ NK cells measured in Nfil3−/− mice at day 7 PI.
Given that NK cells could be induced to expand in mice lacking IL-15 signaling machinery, it is perhaps not surprising that NK cells could similarly expand in Nfil3−/− mice in response to viral infection. Unexpectedly however, the MCMV-expanded Nfil3−/− NK cells persisted in mice for a prolonged period after initial infection, even though Nfil3 has been suggested to function downstream of the IL-15 receptor (Gascoyne et al., 2009). Although mature, resting NK cells depend on IL-15 signals for persistence (Kennedy et al., 2000), it was previously unknown whether memory NK cells have the same requirement for IL-15. Our data demonstrates that like resting NK cells, memory NK cells remain fully dependent on IL-15, as memory NK cells transferred into IL-15–deficient hosts disappeared with similar kinetics to both resting and effector NK cells. Together with data demonstrating that tamoxifen-induced deletion of Nfil3 in mature NK cells did not affect NK cell survival, our results predict the existence of an Nfil3-independent prosurvival pathway downstream of IL-15 that permits the maintenance of peripheral NK cells. Because memory CD8+ T cells also depend on IL-15 signals for survival (Ma et al., 2006), we tested whether generation of this lymphocyte population depended on Nfil3 in like manner to developing NK cells. After MCMV infection of mixed WT:Nfil3−/− bone marrow chimeric mice, we observed a comparable percentage of antigen-specific WT and Nfil3−/− CD8+ T cells at memory time points (unpublished data), indicating that this IL-15–dependent, but Nfil3-independent, pathway is not restricted to the NK cell lineage.
Strikingly, the coincident deletion of Nfil3 with NKp46 expression during early development resulted in normal peripheral NK cell numbers, suggesting that the activity of Nfil3 is likely critical only at the early transitional stages from CLP to NKP. Along with the tamoxifen-induced deletion of Nfil3 in mature NK cells, these data together are in contrast to the loss of certain lymphocyte subsets and function during deletion of lineage-determining transcription factors. One of the best characterized examples is regulatory T cell dependence on continuous Foxp3 expression, not only during development but also for lineage maintenance and function (Williams and Rudensky, 2007). Among the transcription factors that govern NK cell development, induced deletion of Eomes in peripheral NK cells has recently been shown to result in de-differentiation of mature NK cells (Gordon et al., 2012), further suggesting that such lineage-defining transcription factors play a continuous role in the maintenance of lymphocyte identity. Thus, future studies will determine why this is not the case for Nfil3, and how Nfil3 initiates programming of the NK cell lineage but then is dispensable for lineage maintenance and function. Furthermore, a greater understanding of the interplay between IL-15 signaling and Nfil3 activity during NK development for the generation of a mature peripheral NK cell pool will inform our efforts to harness this potent effector immune cell for therapeutics and vaccines against pathogen infection and cancer.
MATERIALS AND METHODS
Mice.
WT C57BL/6 (B6) and congenic (CD45.1) mice were obtained from Taconic. Nfil3−/− (Kashiwada et al., 2010), Nfil3fl/fl (Motomura et al., 2011), Nkp46iCre (Narni-Mancinelli et al., 2011), UbCre-ERt2 × R26Y, B2m−/−, Ly49h−/−, and Rag2−/− × Ly49h−/− mice were bred and maintained at MSKCC. Il15−/− mice were obtained from The Jackson Laboratory. All mice are on the C57BL/6 background. Bone marrow chimeric mice were generated, as previously described (Sun et al., 2009a). Mice were housed and maintained according to Memorial Sloan-Kettering Cancer Center (MSKCC) guidelines, and all animal experiments were performed in accordance with MSKCC Institutional Animal Care and Use Committee approval and institutional guidelines.
Viruses.
A salivary gland stock of MCMV (Smith strain) was administered to WT mice via i.p. injection at 6 × 103 plaque forming units (pfu) per mouse. Nfil3−/− mice were administered 6 × 102 pfu MCMV to induce NK cell expansion without associated spleen and liver pathology observed at higher doses. MCMV-Δm157 (provided by U. Koszinowski, Ludwig-Maximilians-University, Munich, Germany; Bubić et al., 2004) was injected at 4.5 × 104 pfu. Recombinant vesicular stomatitis virus expressing the MCMV m157 protein (VSV-m157) was generated as previously described (Kim et al., 1998) and administered by i.v. injection at a dose of 107 pfu. The recombinant vaccinia virus–OVA (VACV-OVA) was provided by J. Yewdell and J. Bennink (National Institute of Allergy and Infectious Disease, National Institutes of Health, Bethesda, MD). To generate the recombinant vaccinia virus expressing m157 (VACV-m157), a cDNA encoding a fusion protein of the CD8-derived signal sequence, followed by a FLAG-tag and MCMV-m157, was cloned into VACV transfer plasmid pRB21 containing a functional F13L gene, a VACV synthetic early/late promoter, and VACV genomic flanking sequences for homologous recombination and integration into the F13L-deficient VACV vRB12. Identification of recombinant viruses and plaque purification was performed on BSC-1 cells. Recombinant viruses were propagated, purified, and titrated using standard methodology, and 107 pfu were injected i.p.
Tamoxifen treatment of mice.
Mice were administered 8 mg tamoxifen dissolved in 200 µl olive oil by oral gavage on days 0, 1, and 3. In treated Nfil3fl/fl × R26Y × UbCreERt2 mice, effectiveness of tamoxifen treatment was evaluated by monitoring peripheral blood lymphocytes for the appearance of a YFP+ population corresponding to the loss of Nfil3, measured by PCR for Nfil3 DNA and qRT-PCR for Nfil3 mRNA.
In vivo blockade of cytokine signaling.
In vivo blockade of IL-12 and type I IFN signaling during MCMV infection was accomplished by i.p. injection of purified antibodies for IL-12 (clone C17.8) and IFN-α receptor (clone MAR1-5A3) 1 d before MCMV infection. Each mouse was treated with 1 mg of antibody.
Cell sorting and flow cytometry.
NK cells from spleens were enriched using negative selection with magnetic beads (QIAGEN), or by sorting on an Aria II flow cytometer (BD). Purified NK cells were then transferred into Ly49h−/− or Il15−/− mice by i.v. injection and monitored by congenic markers and expression of the Ly49H receptor. Single cell suspensions were generated from indicated organs and incubated with the anti-Fc receptor antibody 2.4G2 before staining with indicated mAbs (BioLegend, eBioscience, and BD) for 20 min on ice. Samples were acquired using an LSRII flow cytometer with FACSDiva software (BD). Analysis was performed with the FlowJo 9.6 software package (Tree Star).
Ex vivo stimulation of NK cells.
Whole splenocytes were obtained from MCMV-infected mice on specified days, and stimulation of NK cells by plate-bound antibodies against activating receptors along with Brefeldin A was performed as described previously (Sun et al., 2012). In brief, 96-well plates were coated with 10 µg/ml of purified antibodies against Ly49H or NKp46. PBS-treated control wells, and wells containing 50 ng/ml PMA and 1 µg/ml ionomycin, or 20 ng/ml IL-12 and 10 ng/ml IL-18 were included. Cells were stained for intracellular IFN-γ and CD107a (LAMP-1).
In vivo and in vitro cytotoxicity assays.
In vivo cytotoxicity was assessed by i.v. injection of target cells (WT or B2m−/−) labeled with different concentrations of CFSE at a 1:1 ratio into indicated mice. Spleens were collected at indicated time points and examined for percentage of each target cell population. For in vitro cytotoxicity assays, enriched NK cells were incubated with 51Cr-labeled YAC-1 target cells in triplicate. Supernatants were harvested and assayed for 51Cr release after 4 h of incubation at 37°C, and percentage of specific lysis was determined.
Quantitative RT-PCR.
CD8+ T cells, CD4+ T cells, and NK cells were purified (>99%) from total splenocytes using an Aria II sorter (BD), and lysed in Tri-Reagent (Ambion). RNA purification and cDNA synthesis were performed using the RNeasy kit (with DNase I treatment; QIAGEN), and MuLV reverse transcription and Oligo dT(16) primers (Applied Biosystems). iQ Sybr Green SuperMix (Bio-Rad Laboratories) was used for qRT-PCR, and data were normalized to β-actin and expressed as relative target abundance via the ΔΔCT method. Relevant primer sequences are as follows: Nfil3 forward, 5′-ATGCCCAAGAAATCCAGAAA-3′; Nfil3 reverse, 5′-GGGAGAGTGCTTGATGACTG-3′; β-actin forward, 5′-TGCGTGACATCAAAGAGAAG-3′; and β-actin reverse, 5′-CGGATGTCAACGTCACACTT-3′.
Statistical analysis.
Results are expressed as mean ± SE. Data were analyzed using one-way ANOVA, and unpaired Students t test for multiple comparisons. All analyses were performed using Prism 5.0b (GraphPad Software), and differences were considered significant when P ≤ 0.05.
Acknowledgments
We thank members of the Sun laboratory for technical support and experimental assistance, and members of the MSKCC NK club for insightful comments and helpful discussions. Lewis Lanier, Sasha Rudensky, Ulrich Koszinowski, Bernie Moss, Jonathan Yewdell, and Jack Bennink provided reagents critical to this study. We thank the ImmGen consortium for providing the microarray data used in this study.
M.A. Firth is supported by a fellowship from the Lucille Castori Center for Microbes, Inflammation, and Cancer. S. Madera is supported by a pre-doctoral Cancer Research Institute training grant. A.M. Beaulieu is supported by a NIH T32 award (CA009149). G. Gasteiger is supported by an Irvington Fellowship of the Cancer Research Institute. E. Vivier is supported by the THINK ERC Advanced Grant. J.C. Sun is supported by the Searle Scholars Program, the Cancer Research Institute, and grants from the NIH (AI085034 and AI100874).
The authors declare no financial conflicts of interest.
Footnotes
Abbreviations used:
- CLP
- common lymphoid progenitor
- MCMV
- mouse cytomegalovirus
- PI
- post infection
References
- Arase H., Mocarski E.S., Campbell A.E., Hill A.B., Lanier L.L. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326 10.1126/science.1070884 [DOI] [PubMed] [Google Scholar]
- Björkström N.K., Lindgren T., Stoltz M., Fauriat C., Braun M., Evander M., Michaëlsson J., Malmberg K.-J., Klingström J., Ahlm C., Ljunggren H.-G. 2011. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J. Exp. Med. 208:13–21 10.1084/jem.20100762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubić I., Wagner M., Krmpotić A., Saulig T., Kim S., Yokoyama W.M., Jonjić S., Koszinowski U.H. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536–7544 10.1128/JVI.78.14.7536-7544.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.A., Bush J.E., Fehniger T.A., VanDeusen J.B., Waite R.E., Liu Y., Aguila H.L., Caligiuri M.A. 2002. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 100:3633–3638 10.1182/blood-2001-12-0293 [DOI] [PubMed] [Google Scholar]
- Cooper M.A., Elliott J.M., Keyel P.A., Yang L., Carrero J.A., Yokoyama W.M. 2009. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA. 106:1915–1919 10.1073/pnas.0813192106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels K.A., Devora G., Lai W.C., O’Donnell C.L., Bennett M., Welsh R.M. 2001. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 194:29–44 10.1084/jem.194.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Chiesa M., Falco M., Podestà M., Locatelli F., Moretta L., Frassoni F., Moretta A. 2012. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood. 119:399–410 10.1182/blood-2011-08-372003 [DOI] [PubMed] [Google Scholar]
- Dokun A.O., Kim S., Smith H.R.C., Kang H.-S.P., Chu D.T., Yokoyama W.M. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2:951–956 10.1038/ni714 [DOI] [PubMed] [Google Scholar]
- Foley B., Cooley S., Verneris M.R., Pitt M., Curtsinger J., Luo X., Lopez-Vergès S., Lanier L.L., Weisdorf D., Miller J.S. 2012. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 119:2665–2674 10.1182/blood-2011-10-386995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne D.M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., Brady H.J.M. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10:1118–1124 10.1038/ni.1787 [DOI] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., Reiner S.L. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 36:55–67 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima S., Inukai T., Inaba T., Nimer S.D., Cleveland J.L., Look A.T. 1997. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl. Acad. Sci. USA. 94:2609–2614 10.1073/pnas.94.6.2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono S., Duncan G.S., Seidel M.G., Morimoto A., Hamada K., Grosveld G., Akashi K., Lind E.F., Haight J.P., Ohashi P.S., et al. 2009. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206:2977–2986 10.1084/jem.20092176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M., Levy D.M., McKeag L., Murray K., Schröder A.J., Canfield S.M., Traver G., Rothman P.B. 2010. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc. Natl. Acad. Sci. USA. 107:821–826 10.1073/pnas.0909235107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M., Pham N.-L., Pewe L.L., Harty J.T., Rothman P.B. 2011. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. 117:6193–6197 10.1182/blood-2010-07-295873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.K., Glaccum M., Brown S.N., Butz E.A., Viney J.L., Embers M., Matsuki N., Charrier K., Sedger L., Willis C.R., et al. 2000. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med. 191:771–780 10.1084/jem.191.5.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-K., Reed D.S., Olson S., Schnell M.J., Rose J.K., Morton P.A., Lefrançois L. 1998. Generation of mucosal cytotoxic T cells against soluble protein by tissue-specific environmental and costimulatory signals. Proc. Natl. Acad. Sci. USA. 95:10814–10819 10.1073/pnas.95.18.10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Matsuoka K., Sheikh S.Z., Elloumi H.Z., Kamada N., Hisamatsu T., Hansen J.J., Doty K.R., Pope S.D., Smale S.T., et al. 2011. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J. Immunol. 186:4649–4655 10.4049/jimmunol.1003888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodolce J.P., Boone D.L., Chai S., Swain R.E., Dassopoulos T., Trettin S., Ma A. 1998. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 9:669–676 10.1016/S1074-7613(00)80664-0 [DOI] [PubMed] [Google Scholar]
- Lopez-Vergès S., Milush J.M., Schwartz B.S., Pando M.J., Jarjoura J., York V.A., Houchins J.P., Miller S., Kang S.-M., Norris P.J., et al. 2011. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 108:14725–14732 10.1073/pnas.1110900108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Koka R., Burkett P. 2006. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu. Rev. Immunol. 24:657–679 10.1146/annurev.immunol.24.021605.090727 [DOI] [PubMed] [Google Scholar]
- Martín-Fontecha A., Lord G.M., Brady H.J.M. 2011. Transcriptional control of natural killer cell differentiation and function. Cell. Mol. Life Sci. 68:3495–3503 10.1007/s00018-011-0800-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura Y., Kitamura H., Hijikata A., Matsunaga Y., Matsumoto K., Inoue H., Atarashi K., Hori S., Watarai H., Zhu J., et al. 2011. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat. Immunol. 12:450–459 10.1038/ni.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Chaix J., Fenis A., Kerdiles Y.M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E., et al. 2011. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA. 108:18324–18329 10.1073/pnas.1112064108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary J.G., Goodarzi M., Drayton D.L., von Andrian U.H. 2006. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 7:507–516 10.1038/ni1332 [DOI] [PubMed] [Google Scholar]
- Paust S., Gill H.S., Wang B.-Z., Flynn M.P., Moseman E.A., Senman B., Szczepanik M., Telenti A., Askenase P.W., Compans R.W., von Andrian U.H. 2010. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat. Immunol. 11:1127–1135 10.1038/ni.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y., Pinzon-Guzman C., Asare A., Ong T., Pontano L., Cotsarelis G., Zediak V.P., Velez M., Bhandoola A., Brown E.J. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 1:113–126 10.1016/j.stem.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Jackson J.T., Markey K.A., Brady H.J., Hill G.R., Macdonald K.P., Nutt S.L., Belz G.T. 2013. CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood. 121:1574–1583 10.1182/blood-2012-07-445650 [DOI] [PubMed] [Google Scholar]
- Smith H.R., Heusel J.W., Mehta I.K., Kim S., Dorner B.G., Naidenko O.V., Iizuka K., Furukawa H., Beckman D.L., Pingel J.T., et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 99:8826–8831 10.1073/pnas.092258599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.-S., William C.M., Tanabe Y., Jessell T.M., Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Lanier L.L. 2011. NK cell development, homeostasis and function: parallels with CD8+ T cells. Nat. Rev. Immunol. 11:645–657 10.1038/nri3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., Lanier L.L. 2009a. Adaptive immune features of natural killer cells. Nature. 457:557–561 10.1038/nature07665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Ma A., Lanier L.L. 2009b. Cutting edge: IL-15-independent NK cell response to mouse cytomegalovirus infection. J. Immunol. 183:2911–2914 10.4049/jimmunol.0901872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Lopez-Verges S., Kim C.C., DeRisi J.L., Lanier L.L. 2011. NK cells and immune “memory”. J. Immunol. 186:1891–1897 10.4049/jimmunol.1003035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Madera S., Bezman N.A., Beilke J.N., Kaplan M.H., Lanier L.L. 2012. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med. 209:947–954 10.1084/jem.20111760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L.M., Rudensky A.Y. 2007. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8:277–284 10.1038/ni1437 [DOI] [PubMed] [Google Scholar]
- Yokoyama W.M., Kim S., French A.R. 2004. The dynamic life of natural killer cells. Annu. Rev. Immunol. 22:405–429 10.1146/annurev.immunol.22.012703.104711 [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang J., Kornuc M., Kwan K., Frank R., Nimer S.D. 1995. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol. Cell. Biol. 15:6055–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]