Abstract
Using N-(2-Aminoethyl)maleimide-cysteine(StBu) (Mal-Cys) as a medium, protein thiols were converted into N-terminal cysteines. After a biocompatible condensation reaction between the N-terminal cysteine and fluorescent probe 2-cyanobenzothiazole-Gly-Gly-Gly-fluorescein isothiocyanate (CBT-GGG-FITC), a new fluorogenic structure Luciferin-GGG-FITC was obtained. The latter exhibits near one order of magnitude (7 folds) enhanced fluorescence emission compared to the precursor moiety due to fluorescence resonance energy transfer (FRET) effect between the newly formed luciferin structure and the FITC motif. Theoretical investigations revealed the underlying mechanism that satisfactorily explained the experimental results. With this method, enhanced fluorescence imaging of thiols on proteins, outer membranes of living cells, translocation of membrane proteins, and endothelial cell layers of small arteries was successfully achieved.
Site-specific labeling of proteins is an important approach for direct visualization of protein expression, association, and translocation, understanding the spatial and temporal underpinnings of life inside cells1,2,3. For the past decade, genetically encoded fluorescent proteins have been ideal reagents to label their fusion proteins with absolute specificity and spatial resolution. Although they have revolutionized cell biology, fluorescent proteins (FPs) have shortcomings. The 235-amino-acid proteins are large enough to interfere with the localization, structure and/or activity of the proteins to which they are fused4. Furthermore, the barrel-like structure of FPs isolates the chromophore from the cellular environment, making them insensitive to the environmental cues like hydrophobicity, ion concentrations, etc1. To circumvent these problems, chemical labeling is used where a receptor protein is often used to bind or react with a ligand tagged with a fluorophore5,6,7,8. Alternatively, small tags on the targeted proteins, such as short peptides, are labeled by selective binding with fluorogenic dyes or by enzymatic ligation to fluorescent probes9,10,11,12,13,14,15,16,17. Biorthogonal, water-compatible reactions between proteins and chemical probes are also applied to improve the labeling efficiency. These reactions include Staudinger ligation between azides and triphenylphosphane18,19,20, the Huisgen cycloaddition or “click reaction” between azides and alkynes21,22,23,24, or reactions between aldehydes (or ketones) and aminooxy-containing reagents (or hydrazides)25,26,27.
Recently, Rao and co-workers developed a biocompatible condensation reaction between the 1,2-aminothiol group of cysteine (Cys) and the cyano group of 2-cyanobenzothiazole (CBT) which could be controlled by pH, reduction, and protease28,29,30. Kinetic study of this condensation reaction revealed that it has a second-order reaction rate of 9.19 M−1s−1, significantly larger than that of a biocompatible click reaction (7.6 × 10−2 M−1s−1)28,31. Besides its promising applications such as imaging protease activities in living cells, designing smart optical and MRI probes, and controlling the self-assembly of nanoparticles29,32,33, this condensation reaction was also successfully applied to label N-terminal Cys residues on proteins and cell membranes28. However, due to the rare occurrences of N-terminal Cys residues in natural proteins, it is necessary to hydrolyze natural proteins to artificially generate N-terminal Cys residues. It is also possible to genetically express proteins with N-terminal Cys residues for subsequent labeling of the proteins using the abovementioned condensation reaction. This indirect labeling of N-terminal Cys limits the applications of this condensation reaction. Unlike N-terminal Cys residues, thiols exist in almost all proteins, either in the free form or oxidized disulfide bond form for maintaining the secondary structure of a protein. An excess or lack of specific biological thiols can serve as evidence of many diseased states, such as leucocyte loss, psoriasis, liver damage, cancer, and AIDS34,35. Therefore, exact and effective labeling of thiols on biomolecules is necessary and important. As maleimide readily reacts with the thiol group at physiological conditions, many methods based on maleimide derivatives for labeling thiols have been developed36,37,38.
Inspired by these pioneering studies, as shown in Fig. 1, we developed a new method for labeling protein thiols using the abovementioned condensation reaction with sevenfold enhanced fluorescence emission. Briefly, thiols on proteins react with the maleimide motif of Mal-Cys at pH 7.4, followed by disulfide bond reduction by tris(2-carboxyethyl)-phosphine (TCEP) to generate a N-terminal Cys motif. The N-terminal Cys on the protein then condenses with the fluorescent probe CBT-GGG-FITC and thereafter labeling of the thiols on the protein is achieved. Compared with the thiazole structure in CBT motif, “double thiazoles” (DT) structure in the newly formed Luciferin motif (i.e., obtained after condensation) tends to attract two protons from the solvent environment and evolves into the Luciferin(2H+) structure which can be effectively excited by photons from 350 to 450 nm, rendering the possibility of FRET between Luciferin(2H+) and FITC. Thus, the fluorescence emission of the probe is greatly enhanced after thiol labeling (7.1 folds, 4096 vs. 579, Fig. 2a). Therefore, with the combination of these two biocompatible reactions (nucleophilic addition between thiol and maleimide, condensation between CBT and N-terminal Cys), a new method was developed for more effectively labeling thiols than conventional maleimide methods.
Figure 1. Schematic illustration of a new method for effectively labeling thiols with enahnced emission induced by FRET.
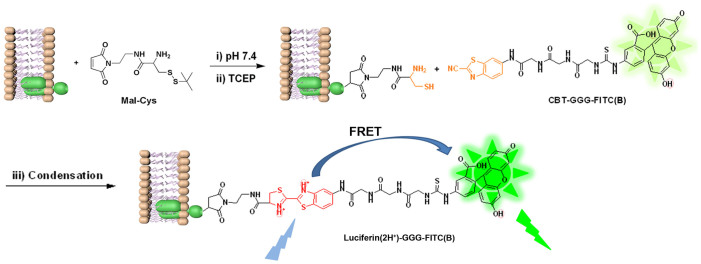
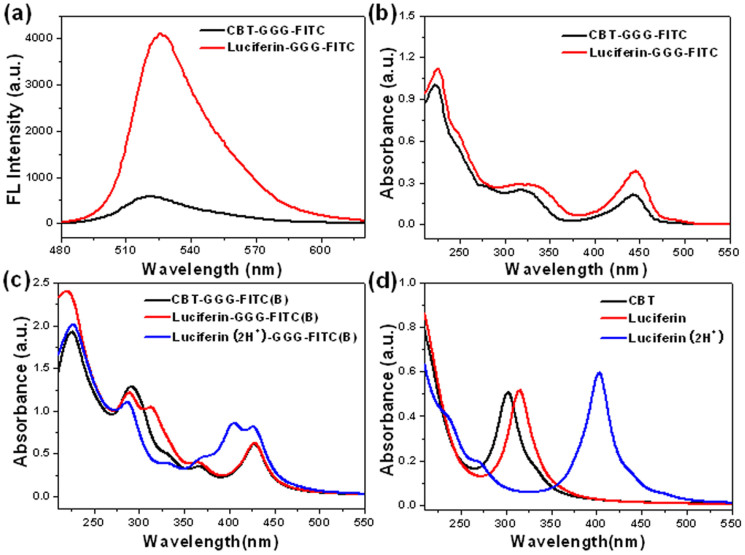
Figure 2. Intramolecular FRET of Luciferin-GGG-FITC and theoretical study.
(a) Fluorescent spectra of CBT-GGG-FITC (black) and Luciferin-GGG-FITC (red) at 15 μM in PBS (pH 7.4) in the presence of 2% dimethylsulfoxide (v/v). Excitation: 465 nm. (b) UV-vis spectra of CBT-GGG-FITC (black) and Luciferin-GGG-FITC (red) at 10 μM in PBS (pH 7.4) in the presence of 2% dimethylsulfoxide (v/v). (c) Simulated photon-absorption spectra of CBT-GGG-FITC(B), Luciferin-GGG-FITC(B), and Luciferin(2H+)-GGG-FITC(B). (d) Simulated photo-absorption spectra of CBT, Luciferin, and Luciferin(2H+).
Results
Syntheses and rationale of the design
As shown in Supplementary Scheme S1–S3, we began the study with the syntheses of the key intermediate Mal-Cys, fluorophores CBT-GGG-FITC and Luciferin-GGG-FITC. The syntheses for these three compounds are simple and straightforward. We designed Mal-Cys to convert the thiols on proteins to N-terminal cysteines. CBT-GGG-FITC was designed to offer CBT motif to covalently label the N-terminal Cys with the condensation reaction. After labeling, the CBT motif on CBT-GGG-FITC is converted to the luciferin structure which has FRET effect with the FITC group and thereby greatly enhances its fluorescence. We also synthesized Luciferin-GGG-FITC to validate the mechanism of condensation-induced fluorescence enhancement as mentioned hereinafter.
Model reaction of cysteine with Maleimide-Cys(Fmoc)(StBu) (A)
Besides the nucleophilic addition reaction, there is a possible side reaction between thiol and Mal-Cys (i.e., reduction of the disulfide bond of Mal-Cys by the thiol). If the side reaction is faster than nucleophilic addition, intramolecular nucleophilic addition among those reduced Mal-Cys themselves will be prior to the intermolecular nucleophilic addition between Mal-Cys and thiols on proteins, causing the conversion of the thiols into N-terminal Cys infeasible. Herein a model reaction using Cys to replace thiol for in vitro study was designed (Supporting Information). As shown in Supplementary Fig. S5, one new peak appeared on the HPLC trace after 2 h incubation of Cys with A. From its 1HNMR and HR-MALDI-MS spectra (Supplementary Figs. S6 and S7), we identified that this new peak was exactly the desired nucleophilic addition product (i.e., D). This result indicates that the nucleophilic addition between Cys and Mal-Cys is effective and clean, suggesting that Mal-Cys could be used as a medium to convert the protein thiols into N-terminal cysteines.
Intramolecular FRET of Luciferin-GGG-FITC and theoretical study
After that, we studied the fluorescence difference of CBT-GGG-FITC before and after labeling thiols (i.e., condensation with Cys to form Luciferin-GGG-FITC). As shown in Fig. 2a, fluorescence emission of Luciferin-GGG-FITC at 525 nm is sevenfold of that of CBT-GGG-FITC (4096 vs. 579, 7.1 folds) at same concentration in phosphate buffered saline (PBS, 0.01 M, pH = 7.4) excited at same wavelength of 465 nm. To explain this, we conducted theoretical investigations. Molecular models were built for CBT-GGG-FITC and Luciferin-GGG-FITC, for which geometries, photo absorption and emission properties were simulated with the density function theory (DFT) and time-dependent DFT (TDDFT) using the Gaussian09 program, at the B3LYP/6-31++G** level. The water solvent effect was considered with the polarizable continuum model (PCM).
It is indubitable that the photo fluorescence at around 520 nm in our experiment could be ascribed to the FITC group. However, there are two possible isomeric FITC structures at neutral pH, as labeled with FITC (A) and (B) in Supplementary Fig. S8a. The energy difference between these two structures is only 3.47 kcal/mol implying easy transformation even under environmental fluctuation. The fluorescence centered at about 520 nm observed in our experimental is mostly likely contributed to FITC(B) with calculated photo-emissions at ~525 nm, instead of FITC(A) with fluorescence emission at ~426 nm in simulation. Moreover, the computed absorption spectra in Supplementary Fig. S8b suggest that FITC(A) could hardly be excited by laser light at 465 nm used in our experiment, while FITC(B) has effective photo-absorptions from 400 to 500 nm. Thus we knew that it is FITC(B) active in this fabricated system.
It was found in our experiments (Fig. 2b) that the Luciferin-GGG-FITC has stronger photo-absorption ability for photons from 400 to 500 nm than CBT-GGG-FITC, which could be one of the causes of its fluorescence enhancement at 525 nm induced by 465 nm excitation. However, the simulated photo-absorptions at 400 ~ 500 nm are almost identical for CBT-GGG-FITC(B) and Luciferin-GGG-FITC(B) structures (Fig. 2c). And the computed spectra of isolated CBT and Luciferin in Fig. 2d do not show any absorption in the 400 ~ 500 nm regime. These suggest the existence of other Luciferin structure accounting for the extra photo-absorption.
As shown in Supplementary Fig. S9, after condensation, the thiazole structure on the CBT motif of CBT-GGG-FITC changes to form a “double thiazoles” (DT) structure on the Luciferin motif of Luciferin-GGG-FITC. Our DFT calculations found that the DT has two electronegative nitrogen atoms (~0.5e charges), which tends to attract two protons from the solvent environment and evolves into the Luciferin(2H+) system (Fig. S9). Our DFT calculations also indicated that Luciferin(2H+) is more stable than one isolated Luciferin with two protons in water solutions, with a lower energy of 9.67 kcal/mol. From simulations, we then found that Luciferin(2H+) can be effective excited by photons from 350 to 450 nm, as indicated in Fig. 2c&d. This satisfactorily explains the absorption enhancements (from 400 to 500 nm) of Luciferin-GGG-FITC compared to CBT-GGG-FITC in our experiments (Fig. 2b). From the experimental absorption spectra in Fig. 2b, we can calculate that the content fractions of Luciferin-GGG-FITC(B) and Luciferin(2H+)-GGG-FITC(B) is about 35.6% and 64.4% for the absorption measurement, respectively.
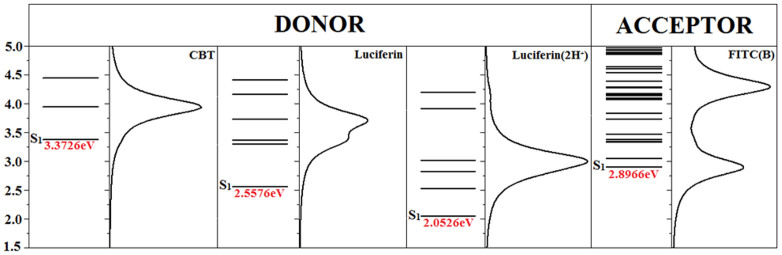
From calculations, we found the almost same electronic transition densities (wavefunction) for the photon absorptions and emissions of CBT, Luciferin, and Luciferin(2H+) (Supplementary Figs. S10–S11). Therefore, the energy differences of their excited states should be responsible for the shift of absorption and fluorescence wavelength. The energetic structures of the donor groups (CBT, Luciferin, and Luciferin(2H+)) and acceptor (FITC(B)) for the FRET process, together with corresponding transition probabilities at room temperature (RT), are schematically shown in Fig. 3.
Figure 3. Simulated electronic states and transition probabilities for three donor groups of CBT, Luciferin, and Luciferin(2H+), and the acceptor group of FITC(B) for the FRET process at room temperature.
Applying Fermi's golden rule39, we may write the FRET rate (probability per unit time) KDA from the donor to acceptor as:
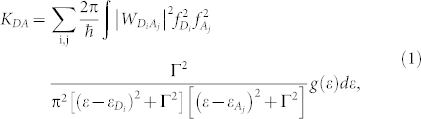 |
in which 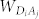 represents the electronic dipole-dipole interaction matrix element between the i-th and j-th transitions in the donor and acceptor, respectively, and can be calculated using the dipole approximation.
represents the electronic dipole-dipole interaction matrix element between the i-th and j-th transitions in the donor and acceptor, respectively, and can be calculated using the dipole approximation.
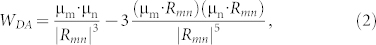 |
where μm(μn) stands for the transition dipole of the molecular fragment m (n), and Rmn is the distance vector from m to n. 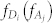 and
and 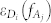 stand for the transition probability and energy of the donor (acceptor), which were obtained from TDDFT calculations. Applying the Lorentzian function to broaden the transition probability with the full width at half maximum (FWHM) Γ as 0.2 eV (corresponding to 30 nm width at around 450 nm), we calculated the transition probability spectra of three donors and one acceptor, as plotted aside to energy state levels in Fig. 3. The energy distribution of the incident laser light centered at the frequency of ν with the FWHM of σ can be described with the Gaussian function of
stand for the transition probability and energy of the donor (acceptor), which were obtained from TDDFT calculations. Applying the Lorentzian function to broaden the transition probability with the full width at half maximum (FWHM) Γ as 0.2 eV (corresponding to 30 nm width at around 450 nm), we calculated the transition probability spectra of three donors and one acceptor, as plotted aside to energy state levels in Fig. 3. The energy distribution of the incident laser light centered at the frequency of ν with the FWHM of σ can be described with the Gaussian function of
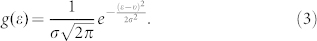 |
Labeling thiols on protein
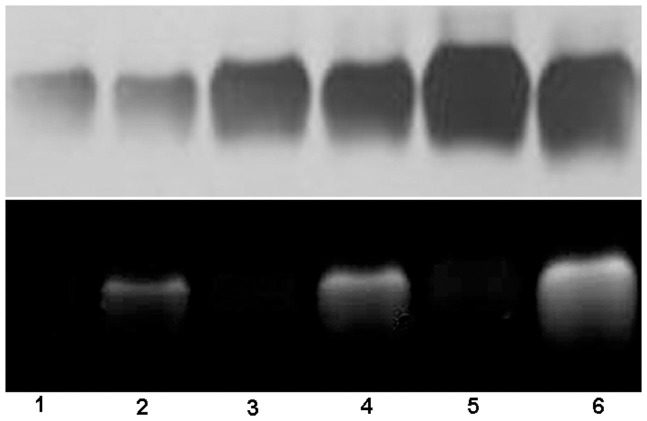
Bovine serum albumin (BSA, 200 μM) in PBS was sequentially incubated with 14 equiv. of TCEP for 10 min, with (or without) 16 equiv. of Mal-Cys for 30 min, and 14 equiv. of TCEP with 20 μM CBT-GGG-FITC for 45 min at RT. The incubation mixture was then diluted and applied to SDS-PAGE for separation and imaging (Fig. 4). As shown in Fig. 4, thiols on BSA were successfully labeled with CBT-GGG-FITC via this condensation reaction and the fluorescence intensity of the labeled BSA bands increases along with the increase of BSA concentration (lanes 2, 4, and 6 in Fig. 4). In contrast, BSA whose thiols were not converted into N-terminal Cys (i.e., without treatment of Mal-Cys) did not show any fluorescence at any concentration (lanes 1, 3, and 5 in Fig. 4).
Figure 4. Labeling thiols on BSA.
BSA (200 μM) in PBS was incubated with 14 equiv. of TCEP for 10 min, with (or w/o) 16 equiv. of Mal-Cys for 30 min, and 14 equiv. of TCEP with 20 μM CBT-GGG-FITC for 45 min at RT. The incubation mixture was then diluted and applied to SDS-PAGE. Lane 1, 3, and 5: BSA at 12.5, 25, and 50 μM without Mal-Cys respectively. Lane 2, 4, and 6: BSA at 12.5, 25, and 50 μM with Mal-Cys respectively. The size of BSA did not show obvious change after labeling. Upper panel: white-light image of the gel after silver staining. Lower panel: fluorescence image of the gel. Excitation: 365 nm.
Labeling protein thiols on the outer cellular membranes of living HepG-2 cells and imaging the translocation of the membrane proteins
After thorough studies of different conditions for incubation prior to imaging (concentration-dependent study of TCEP, Mal-Cys, and CBT-GGG-FITC respectively, incubation time study of CBT-GGG-FITC, and temperature-dependent study) (Supplementary Figs. S14–S18), we successfully labeled the thiols on the outer membrane proteins of living HepG-2 cells. We denote step 1 as the incubation of TCEP (100 μM) with cells for 15 min, step 2 as the incubation of Mal-Cys (200 μM) with cells in culture media without serum for 3 h at 37°C, step 3 as the incubation of TCEP (100 μM) and CBT-GGG-FITC (15 μM) with cells in culture media without serum for 1 h at 37°C. The cells were washed with PBS for three times prior to imaging. The concentration of TCEP used here should not have any cytotoxicity according to the literatures reported40. As shown in Fig. 5a, when the cells were sequentially treated from step 1 to 3 in turn, a clear outer membrane staining by the probe was observed. At step 3, if CBT-GGG-FITC is added in the absence of TCEP for incubation, the disulfide bond on the Mal-Cys at step 2 will not be reduced, disabling the condensation reaction. Consequently, the cells are uniformly stained by CBT-GGG-FITC probably due to the nonspecific binding of the probe (Fig. 5b). When the cells were treated with step 3 only (i.e., disulfide bonds on the membrane protein were reduced and only the thiols of N-terminal Cys could be labeled by the probe directly), circular fluorescence images which represent the shapes of outer membranes of the cells are hardly observed (Fig. 5c). This suggests that N-terminal Cys account for a very tiny proportion of total Cys of the membrane proteins. At step 3, when the CBT-GGG-FITC was replaced by FITC at the same concentration, cells have a similar staining pattern to that in Fig. 4b due to the nonspecific binding of FITC (Fig. 5d). In another control experiment, cells were treated with steps 1 and 2, and followed by step 3 without probe. No fluorescence signal was observed (Fig. S19). Propidium iodide staining indicated that these 3-step labeling did not result in membrane disruption to the cells (Supplementary Fig. S20). We also labeled thiols on the outer membrane of living HepG-2 cells and monitored the endocytosis process of our probe accompanying the translocation of the membrane protein. As shown in the Supplementary Fig. S21, the fluorophore penetrated the cell outer membrane and was internalized inside the cells within 25 min.
Figure 5. Labeling thiols on the outer membrane of living HepG-2 cells.
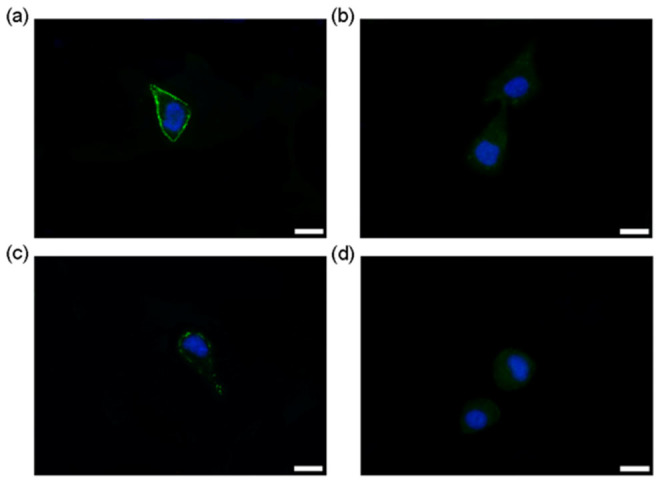
(a) Fluorescence image of HepG-2 cells (EGFP channel and DAPI channel) after incubation with TCEP (100 μM) for 15 min (step 1), Mal-Cys (200 μM) in culture media without serum for 3 h at 37°C (step 2), TCEP (100 μM) and CBT-GGG-FITC (15 μM) in culture media without serum for 1 h at 37°C, then treated with 1 μg/mL Hoechst 3342 and 20 μg/mL propidium iodide for 6 min at RT prior to imaging (step 3). (b) Fluorescence image of cells (EGFP channel and DAPI channel) after treatment of steps 1 and 2, and step 3 without TCEP. (c) Fluorescence image of cells (EGFP channel and DAPI channel) treated only with step 3. (d) Fluorescence image of cells (EGFP channel and DAPI channel) after treatment of steps 1, 2, and step 3 with CBT-GGG-FITC being replaced by FITC. Scale bar: 20 μm.
Labeling thiols on brain tissue
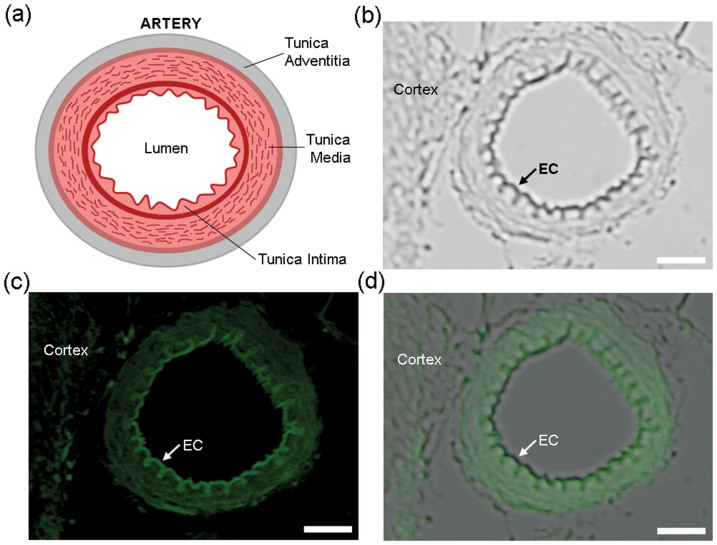
As illustrated in Fig. 6a, the arterial wall can be divided into three layers41: the innermost layer tunica intima, the middle layer tunica media, and the outermost layer tunica adventitia. The innermost of tunica intima is the layer of endothelial cells (ECs). Besides free Cys, ECs contain a high concentration of tripeptide glutathione, which has a Cys content of 39.4%. Even endothelin, an important 21-amino acid peptide produced in ECs, has a Cys content of 10%. Compared with other tissues in a small artery such as smooth muscle cells (SMCs), collagen, and elastic fiber, ECs contain the highest content of Cys42. Following the protocol to get a cerebral cortex section of 2 μm thickness, we incubated the section with TCEP (14 mM) for 10 min, Mal-Cys (16 mM) for 30 min, and TCEP (7 mM) with CBT-GGG-FITC (50 μM) for 2 h at RT in turn and imaged it under a fluorescence microscope. As shown in Fig. 6b, near the cerebral cortex, we observed a transverse section of a small cerebral artery with an outer diameter of 47 μm and inner diameter of 31 μm. The innermost, rugate layer of EC could be easily discerned. When the tissue section was exposed to an excitation light of 470–490 nm, clearly we observed that the EC layer has the strongest fluorescence emission among all the tissues of the artery (Fig. 6c). An overlay of previous two images undoubtedly indicates that the strongest fluorescence emission comes from the EC layer (Fig. 6d).
Figure 6. Labeling thiols on brain tissue.
(a) Cartoon illustration of the structure of a typical artery. (b) DIC image of a tissue slice from a rat brain after incubation with TCEP (14 mM) for 10 min, Mal-Cys (16 mM) for 30 min, and TCEP(7 mM) with CBT-GGG-FITC (50 μM) for 2 h at RT. Left part: cerebral cortex. Right: small cerebral artery. Arrow indicates the innermost layer (single layer of endothelial cell) of the small artery. (c) Fluorescence image of the tissue slice in b. Arrow indicates that the single layer of endothelial cell has the brightest fluorescence emission among the tissues of the small artery. (d) Overlay of images in b and c. Scale bar: 10 μm.
Discussion
Theoretical investigations revealed that Luciferin(2H+) and FITC(B) group can be effectively excited by photons at around 400 nm (3.1 eV) or 425 nm (2.9 eV) respectively, suggesting strong possibility of FRET effect between them. As Fig. 3 shown, the dominant transition peak centered at about 3.0 eV of Luciferin(2H+) has the strongest overlap with the transition peaks of FITC(B), so that it serves as the best one for the FRET process among three donor candidates.
Using the abovementioned formula, the FRET rate of three Donor-Acceptor systems excited by 465 nm laser light are computed and listed in Supplementary Table S1, which shows that the FRET probability of Luciferin(2H+)-GGG-FITC(B) is about 10 and 7 times higher than the CBT-GGG-FITC(B) and Luciferin-GGG-FITC(B) system, respectively. Using the content fractions of Luciferin-GGG-FITC(B) and Luciferin(2H+)-GGG-FITC(B) deduced from the measured photo-absorption spectra, we can calculate the ratio of the FRET rates of our system before and after condensation treatment as 1:7.56 ~ 10.08, which agrees very well with the fluorescence enhancement observed in our experiment (i.e., 7.1 folds).
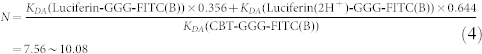 |
The details for the computational process of FRET rate and KDA's distribution are shown in Supplementary Information.
We firstly tested this thiol-labeling method on pure protein BSA. As shown in Fig. 4, after TCEP reduction and nucleophilic addition of Mal-Cys, thiols on BSA were successfully labeled with CBT-GGG-FITC via this condensation reaction. In contrast, BSA whose thiols were not converted into N-terminal Cys (i.e., without treatment of Mal-Cys) could neither condense with CBT-GGG-FITC nor be labeled by it. This result proves the feasibility of our method for labeling thiols on protein.
As shown in Fig. 5a, when the living cells were sequentially treated from step 1 to 3 in turn, a clear outer membrane staining by the probe was observed. These results convincingly proved that these three steps are necessary for labeling the total thiols on the cellular outer membranes. Moreover, labeling thiols on the outer membrane proteins was employed to monitor the endocytosis process of our probe accompanying the translocation of the membrane protein (Supplementary Fig. S21). And the result suggests that our method could be applied to imaging the translocation process of the membrane proteins.
The reason that we chose small artery for thiol labeling is that its diameter (normally 37 μm) is suited for microscopic observation for one reason. For another reason, the structure of the arterial wall is anisotropic, multi-storey and nonlinearly elastic, having different Cys content in different layers. An overlay of Fig. 6b and Fig. 6c undoubtedly indicates that the strongest fluorescence emission comes from the EC layer (Fig. 6d), echoing that ECs contains the highest content of Cys. Since the morphological abnormality of EC layer is closely associated with the occurrence of vascular diseases such as aneurysm, hypertension, and stroke43, our method of thiol-labeling on EC layer of small artery has potential applications in clinical diagnoses.
In conclusion, using Mal-Cys as a medium and combining two biocompatible reactions (nucleophilic addition between thiol and maleimide, and condensation between CBT and N-terminal Cys), we successfully developed a new method for effectively labeling thiols on proteins, on the outer membrane of living cells, and on EC layer of small arteries with sevenfold enhanced fluorescence emission induced by intramolecular FRET. Theoretical investigations satisfactorily explained the FRET mechanism between the protonated luciferin structure (i.e., Luciferin(2H+)) and FITC(B) structure. Moreover, using Fermi's golden rule, we also calculated the ratio of the FRET rates of our system to be 7.56 ~ 10.08, which agrees very well with the fluorescence enhancement observed in our experiment. Since the protonated Luciferin(2H+)-GGG-FITC(B) structure only accounts for about 64.4% of the total probe at pH 7.4 in this work, we can envision that at lower pH value this probe could have enhanced fluorescence more than sevenfold after thiol-labeling. Compared with those strategies for thiol or N-terminal Cys labeling, our method has several obvious advantages. First, our method could be applied to labeling not only thiols (Fig. 5a) but also N-terminal Cys (Fig. 5c). In contrast, those methods for N-terminal Cys labeling (e.g., thioester-forming ligation44,45 and thiazolidine-forming ligation46) have not been extended for thiol-labeling. Second, once the Mal-Cys is replaced with 3-azidopropyl-1-maleimide or N-propargylmaleimide, the widely used “click reaction” between azides and alkynes could be easily employed to our method. This shows the versatility of our method. Third, compared to those methods which need to genetically express proteins with N-terminal Cys residues or hydrolyze natural proteins to artificially generate N-terminal Cys residues28, our method offers a facile and simple method for artificially generating N-terminal Cys residues. Four, after the optimizations of the labeling conditions, our method was developed as a new strategy for specifically labeling outer cellular membrane. Nevertheless, our method needs three steps for thiol labeling. Thus, it consumes more time than one-step methods do. Due to the tight association between the morphological abnormality of EC layer and the occurrence of vascular diseases such as aneurysm, hypertension, and stroke43, our method might find its broad applications for clinical diagnoses in the future.
Methods
General methods
All the starting materials were obtained from Adamas or Sangon Biotech. Commercially available reagents were used without further purification, unless noted otherwise. All other chemicals were reagent grade or better. PBS buffer (0.01 M, pH 7.4) was prepared with pills purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Ultrapure water (18.2 MΩ. cm) was used throughout the experiment. HepG-2 human liver cancer cells were supplied by the Molecular Biology Laboratory of Anhui Medical University. The spectra of electrospray ionization-mass spectrometry (ESI-MS) were recorded on a LCQ Advantage MAX ion trap mass spectrometer (Thermo Fisher). High resolution ESI/MS spectra were obtained on a GCT premier mass spectrometer (Waters). MALDI-TOF/TOF mass spectra were obtained on a time-of-flight Ultrflex II mass spectrometer (Bruker Daltonics). HPLC analyses were performed on an Agilent 1200 HPLC system equipped with a G1322A pump and in-line diode array UV detector using a YMC-Pack ODS-AM column with CH3OH (0.1% of TFA) and water (0.1% of TFA) as the eluent. 1H-NMR spectra were obtained on a 300 MHz Bruker AV300. White-light and fluorescence gel images were taken under EC3 imaging system (UVP, Upland, CA) equipped with a BioChemi HR camera (UVP, EC3 Chemi HR 410 imaging system). Fluorescence microscopic images were taken under a fluorescence microscope OLMPUS IX71 (Japan). Cells were routinely cultured in Dul-becco's modified Eagle's medium (DMEM, Hycolon) supplemented with 10% fetal bovine serum at 37°C, 5% CO2, and humid atmosphere.
Labeling thiols on protein
50 μL of bovine serum albumin (BSA, 1 mM) dissolved in PBS (pH 7.4) was incubated with 50 μL of TCEP (14 mM) for 10 min at RT (step 1). Then 50 μL of Mal-Cys (16 mM) was added into the mixture and incubated for 30 min (step 2). At the last step, 50 μL of TCEP (14 mM) together with 50 μL CBT-GGG-FITC (100 μM) were added into the mixture and incubated for 45 min (step 3). The final concentration of BSA was 200 μM. The control experiment was carried out at the same conditions except the Mal-Cys was replaced with PBS at step 2. Before running sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), the reaction mixture was diluted with sample buffer until a final concentration of BSA at 50 μM, 25 μM, or 12.5 μM was obtained respectively.
Labeling thiols on brain tissue
All animal treatments were performed strictly in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Animal studies were conducted with the approval of the Institutional Animal Care and Use Committee. All rats were supplied by Lab Animal Center of Anhui Medical University. Adult Sprague-Dawley Rats were deeply anesthetized with urethane and perfused with physiological saline until the liver became pale, immediately followed by 0.1 M chilled PBS containing 4% (w/v) paraformaldehyde for 1 h. Brains were removed and stored in the same paraformaldehyde solution overnight. Then the samples were washed with water, dehydrated in ethanol, transparentized with xylene, and embedded in paraffin. Coronal sections of 2 μm thickness were cut and mounted for staining on microscope slides. The sections were deparaffinized in xylene, hydrated through a graded series of ethanol (from 100%, 95%, 90%, to 70%), and incubated twice in distilled water and each time for 10 min. After being washed with PBS, the sections were soaked in TCEP (14 mM) for 10 min, washed with PBS again for 3 times (5 min for each time). The sections were then incubated with Mal-Cys at 16 mM for 30 min at RT. After the incubation solution being discarded, the sections were washed with PBS for three times and then incubated with TCEP at 7 mM and CBT-GGG-FITC at 50 μM for 2 hours. Then the sections were washed with PBS for three times and placed under a fluorescence microscope (OLMPUS IX71, Japan) for imaging.
Author Contributions
G.L. designed the project and wrote the manuscript; Y.Y., D.Z., A.T. and L.A. did the syntheses, characterizations, and cell imaging; X.W. and J.J. conducted the theoretical investigations; B.M. did the protein labeling and tissue imaging; X.H. helped with the project design and manuscript preparation.
Supplementary Material
Labeling Thiols on Proteins, Living Cells, and Tissues with Enhanced Emission Induced by FRET
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 21175122, 91127036, 91221104), the Fundamental Research Funds for Central Universities (Grant WK2060190018), and the Construction Project for Guizhou Provincial Key Laboratories (ZJ[2011]4007).
References
- Tsien R. Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998). [DOI] [PubMed] [Google Scholar]
- Marks K. M. & Nolan G. P. Chemical labeling strategies for cell biology. Nat. Methods 3, 591–596 (2006). [DOI] [PubMed] [Google Scholar]
- Fernandez-Suarez M. & Ting A. Y. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 9, 929–943 (2008). [DOI] [PubMed] [Google Scholar]
- Lisenbee C. S., Karnik S. K. & Trelease R. N. Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic 4, 491–501 (2003). [DOI] [PubMed] [Google Scholar]
- Keppler A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotech. 21, 86–89 (2003). [DOI] [PubMed] [Google Scholar]
- Miller L. W., Sable J., Goelet P., Sheetz M. R. & Cornish V. W. Methotrexate conjugates: A molecular in vivo protein tag. Angew. Chem. Int. Ed. 43, 1672–1675 (2004). [DOI] [PubMed] [Google Scholar]
- Los G. V. et al. HatoTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- Mizukami S., Watanabe S., Hori Y. & Kikuchi K. Covalent Protein Labeling Based on Noncatalytic beta-Lactamase and a Designed FRET Substrate. J. Am. Chem. Soc. 131, 5016–5017 (2009). [DOI] [PubMed] [Google Scholar]
- Griffin B. A., Adams S. R. & Tsien R. Y. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269–272 (1998). [DOI] [PubMed] [Google Scholar]
- Hauser C. T. & Tsien R. Y. A hexahistidine-Zn2+-dye label reveals STIM1 surface exposure. Proc. Natl. Acad. Sci. USA 104, 3693–3697 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignet E. G., Hovius R. & Vogel H. Reversible site-selective labeling of membrane proteins in live cells. Nat. Biotechnol. 22, 440–444 (2004). [DOI] [PubMed] [Google Scholar]
- Ojida A. et al. Oligo-Asp Tag/Zn(II) complex probe as a new pair for labeling and fluorescence imaging of proteins. J. Am. Chem. Soc. 128, 10452–10459 (2006). [DOI] [PubMed] [Google Scholar]
- Halo T. L., Appelbaum J., Hobert E. M., Balkin D. M. & Schepartz A. Selective Recognition of Protein Tetraserine Motifs with a Cell-Permeable, Pro-fluorescent Bis-boronic Acid. J. Am. Chem. Soc. 131, 438–439 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I., Howarth M., Lin W. Y. & Ting A. Y. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods 2, 99–104 (2005). [DOI] [PubMed] [Google Scholar]
- Yin J., Liu F., Li X. H. & Walsh C. T. Labeling proteins with small molecules by site-specific posttranslational modification. J. Am. Chem. Soc. 126, 7754–7755 (2004). [DOI] [PubMed] [Google Scholar]
- Clarke K. M., Mercer A. C., La Clair J. J. & Burkart M. D. In vivo reporter labeling of proteins via metabolic delivery of coenzyme A analogues. J. Am. Chem. Soc. 127, 11234–11235 (2005). [DOI] [PubMed] [Google Scholar]
- Gauchet C., Labadie G. R. & Poulter C. D. Regio- and chemoselective covalent immobilization of proteins through unnatural amino acids. J. Am. Chem. Soc. 128, 9274–9275 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxon E. & Bertozzi C. R. Cell surface engineering by a modified Staudinger reaction. Science 287, 2007–2010 (2000). [DOI] [PubMed] [Google Scholar]
- Cohen A. S., Dubikovskaya E. A., Rush J. S. & Bertozzi C. R. Real-Time Bioluminescence Imaging of Glycans on Live Cells. J. Am. Chem. Soc. 132, 8563–8565 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel S. S., van Eldijk M. B. & van Hest J. C. M. Staudinger Ligation as a Method for Bioconjugation. Angew. Chem. Int. Ed. 50, 8806–8827 (2011). [DOI] [PubMed] [Google Scholar]
- Kolb H. C., Finn M. G. & Sharpless K. B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40, 2004–2021 (2001). [DOI] [PubMed] [Google Scholar]
- Link A. J. & Tirrell D. A. Cell surface labeling of Escherichia coli via copper(I)-catalyzed 3 + 2 cycloaddition. J. Am. Chem. Soc. 125, 11164–11165 (2003). [DOI] [PubMed] [Google Scholar]
- Agard N. J., Prescher J. A. & Bertozzi C. R. A strain-promoted 3 + 2 azide-alkyne cycloaddition for covalent modification of blomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047 (2004). [DOI] [PubMed] [Google Scholar]
- Guo J., Wang S., Dai N., Teo Y. N. & Kool E. T. Multispectral labeling of antibodies with polyfluorophores on a DNA backbone and application in cellular imaging. Proc. Natl. Acad. Sci. USA 108, 3493–3498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahal L. K., Yarema K. J. & Bertozzi C. R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 276, 1125–1128 (1997). [DOI] [PubMed] [Google Scholar]
- Rush J. S. & Bertozzi C. R. New Aldehyde Tag Sequences Identified by Screening Formylglycine Generating Enzymes in Vitro and in Vivo. J. Am. Chem. Soc. 130, 12240–12241 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Ramya T. N. C., Dirksen A., Dawson P. E. & Paulson J. C. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 6, 207–209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H. J. et al. A Biocompatible Condensation Reaction for the Labeling of Terminal Cysteine Residues on Proteins. Angew. Chem. Int. Ed. 48, 9658–9662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Ren H. & Rao J. A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat. Chem. 2, 54–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D., Liang G., Ma M. L. & Rao J. Controlling Intracellular Macrocyclization for the Imaging of Protease Activity. Angew. Chem. Int. Ed. 50, 2275–2279 (2011). [DOI] [PubMed] [Google Scholar]
- Baskin J. M. et al. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA 104, 16793–16797 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Chen Y., Wu F., Deng Y. & Liang G. Caspase-3 controlled assembly of nanoparticles for fluorescence turn on. Chem. Commun. 47, 10320–10322 (2011). [DOI] [PubMed] [Google Scholar]
- Yuan Y. et al. Detection of Glutathione in Vitro and in Cells by the Controlled Self-Assembly of Nanorings. Anal. Chem. 85, 1280–1284 (2013). [DOI] [PubMed] [Google Scholar]
- Lim S., Escobedo J. O., Lowry M., Xu X. & Strongin R. Selective fluorescence detection of cysteine and N-terminal cysteine peptide residues. Chem. Commun. 46, 5707–5709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhou Y., Peng X. & Yoon J. Fluorescent and colorimetric probes for detection of thiols. Chem. Soc. Rev. 39, 2120–2135 (2010). [DOI] [PubMed] [Google Scholar]
- Springer J. W. et al. Biohybrid Photosynthetic Antenna Complexes for Enhanced Light-Harvesting. J. Am. Chem. Soc. 134, 4589–4599 (2012). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. New method for effectively and quantitatively labeling cysteine residues on chicken eggshell membrane. Org. Biomol. Chem. 10, 8082–8086 (2012). [DOI] [PubMed] [Google Scholar]
- Shen B. Q. et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat. Biotechnol. 30, 184–189 (2012). [DOI] [PubMed] [Google Scholar]
- Beljonne D., Curutchet C., Scholes G. D. & Silbey R. J. Beyond Förster Resonance Energy Transfer in Biological and Nanoscale Systems. J. Phys. Chem. B 113, 6583–6599 (2009). [DOI] [PubMed] [Google Scholar]
- Follmann W. & Wober J. Investigation of cytotoxic, genotoxic, mutagenic, and estrogenic effects of the flame retardants tris-(2-chloroethyl)-phosphate (TCEP) and tris-(2-chloropropyl)-phosphate (TCPP) in vitro. Toxicol. Lett. 161, 124–134 (2006). [DOI] [PubMed] [Google Scholar]
- Lusis A. J. Atherosclerosis. Nature 407, 233–241 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Shireman P. K., Hampton B., Burgess W. H. & Greisler H. P. The Cysteine-Free Fibroblast Growth Factor 1 Mutant Induces Heparin-Independent Proliferation of Endothelial Cells and Smooth Muscle Cells. J. Surg. Res. 92, 255–260 (2000). [DOI] [PubMed] [Google Scholar]
- Lee R. M. K. W. Morphology of cerebral arteries. Pharmacol. Therapeut. 66, 149–173 (1995). [DOI] [PubMed] [Google Scholar]
- Schuler B. & Pannell L. K. Specific Labeling of Polypeptides at Amino-Terminal Cysteine Residues Using Cy5-benzyl Thioester. Bioconjugate Chem. 13, 1039–1043 (2002). [DOI] [PubMed] [Google Scholar]
- Gentle I. E., De Souza D. P. & Baca M. Direct Production of Proteins with N-Terminal Cysteine for Site-Specific Conjugation. Bioconjugate Chem. 15, 658–663 (2004). [DOI] [PubMed] [Google Scholar]
- Tolbert T. J. & Wong C. H. New Methods for Proteomic Research: Preparation of Proteins with N-Terminal Cysteines for Labeling and Conjugation. Angew. Chem. Int. Ed. 41, 2171–2174 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Labeling Thiols on Proteins, Living Cells, and Tissues with Enhanced Emission Induced by FRET