Abstract
Scanning Electron Microscopy (SEM) has long been the standard in imaging the sub-micrometer surface ultrastructure of both hard and soft materials. In the case of biological samples, it has provided great insights into their physical architecture. However, three of the fundamental challenges in the SEM imaging of soft materials are that of limited imaging resolution at high magnification, charging caused by the insulating properties of most biological samples and the loss of subtle surface features by heavy metal coating. These challenges have recently been overcome with the development of the Helium Ion Microscope (HIM), which boasts advances in charge reduction, minimized sample damage, high surface contrast without the need for metal coating, increased depth of field, and 5 angstrom imaging resolution. We demonstrate the advantages of HIM for imaging biological surfaces as well as compare and contrast the effects of sample preparation techniques and their consequences on sub-nanometer ultrastructure.
The Scanning Electron Microscope (SEM) has become one of the fundamental tools used in the study of exterior morphology and structure for biological samples1. More recently it has become an essential tool in the study of connectomics and cellular interactions with advances in serial block-face imaging techniques, for example, using either an in situ microtome2 or focused-ion beam (FIB) milling3. The caveat, however, is that conventional SEM imaging requires samples to be conductive and vacuum friendly, both of which can be accomplished via drying of the sample and coating it with a heavy metal1. Although this method of sample preparation has been standard for many years, there are some instances where minute surface details can be obscured by the metal coating or where the sample is too delicate or small to survive the requisite fixation and drying procedures4,5,6. To combat this, numerous advances have been made to reduce charge as well as image samples in their hydrated form.
One of the most effective methods used to minimize the charging effects observed in insulating samples is to reduce the accelerating voltage of the electron beam, also known as low-voltage scanning electron microscopy (LVSEM)7. Lowering the voltage in conjunction with brighter field-emission sources and state of the art detectors has the advantage of higher resolution, increased topographical contrast and surface sensitivity as well as increased secondary electron generation. However, as the voltage decreases, chromatic aberration increases, and depth of field decreases with the necessary decrease in working distance, thus limiting the low voltage range of most conventional non aberration-corrected SEMs8,9,10. An alternative approach is to delay the dehydration of samples using variable and extended pressure imaging techniques11,12. By injecting nitrogen gas (variable pressure) or water vapor (extended pressure) into the imaging chamber, charge can be negated by creating secondary ionization products of the working gas molecules1. Beam stability is maintained in these systems by means of an In-Lens differential pumping system that allows the maintenance of a high vacuum in the electron gun with a low vacuum in the imaging chamber. Although useful, this technique is limited in resolution and signal to noise as a result of beam skirting13. In addition, there is an increased chance of beam-induced damage to the sample due to the higher accelerating voltages required for imaging within a gaseous environment14.
One of the most formidable developments in imaging biological surfaces in their most native form without any chemical fixation is Cryo-SEM15, which freezes or cryo-immobilizes the sample and transfers it to a chilled stage prior to imaging. Although it is possible to maintain gross morphology with this technique, samples can potentially sublimate under high vacuum conditions thus increasing the risk for charging. This often requires the frozen samples to be sputter coated or imaged in a variable pressure environment14,16.
Notwithstanding these advances in controlling charge and maintaining biological surface structure, they all still suffer from limitations in both resolution and contrast. Recently, an ultra-high brightness gas field ionization source (GFIS) has been developed for use as an ultra-high resolution and high contrast imaging system (Orion, Carl Zeiss, Peabody, MA)17. Despite this relatively contemporary innovation, the original concept of a Field Ion Microscope (FIM) based on helium ions was originally proposed by Erwin Müller in 195618. However, recent refinements to the design overcame previous limitations and now boast a reproducible and stable ion source over many days. The key to the systems performance is its source, which is shaped to a three-sided pyramidal tip consisting of only three atoms at the apex. When the tip is cryogenically cooled and exposed to a high voltage (30 kV) in the presence of helium, an ionized gas field is generated at the apex of the tip. This is due to field only being intense enough at those three atoms to efficiently ionize the helium gas. As such, the ion emission originates preferentially from those atoms at the apex, of which only one is selected for imaging. Thus imaging occurs using a single atom as the ion source. The extracted ions are accelerated down the column of the microscope in the form of a beam that is then shaped and raster scanned much in the same fashion as in an SEM. Due to the ultra high brightness of the GFIS a very small beam defining aperture may be used and therefore spherical and chromatic aberrations of the ion optical column are insignificant. The helium ion microscope therefore exhibits substantially higher resolution with a larger depth of field resulting in an estimated resolution that is five times greater than that of a modern FE-SEM17. Also, since the de Broglie wavelength of a helium ion is very small (below one picometer for a 30 kV accelerating voltage) the spot size of the scanned beam is not limited by diffraction aberration17. In addition to the increase in resolution, the increased mass of a helium ion results in a strong forward scattering of the ions causing the majority of the beam energy to be deposited in deeper areas of sample. This results in high contrast secondary electron emission deriving mostly from the primary beam (SE1 electrons), with typically 2–5 electrons produced per helium ion19. Because of the deeper penetration of the forward scattered ions in biological samples, there is a much lower propensity for beam-induced surface-damage due to lower sample densities as compared with crystalline materials such as silicon. The lack of deposition of energy at the sample surface results in minimal damage at 30 kV where currents used for imaging range from 0.1–0.4 pA19,20,21,22. Furthermore, the sputter rate of helium is two orders of magnitude lower than gallium at comparable beam energies. This results in biological samples being able to be imaged repetitively for a relatively long time before any noticeable beam damage occurs. Such effects may be visible as slight deformations or almost negligible material removal (a polishing effect) from the sample surface. For electrically insulating samples, positive charge resulting from the ion beam is easily compensated by directing a very low voltage electron beam (flood gun) at the sample. The use of such a device does not have any affect on the helium ion spot size and thus has no negative impact on resolution19. Also, the recent development of patterning scan engines (Fibics, Ottawa, Canada) allow the routine nanoscale milling and modification of samples17.
To date, helium ion microscopy has been primarily been used in high-contrast imaging of inorganic materials; however23,24, there have been some limited instances where it has been used in imaging of biological samples. These have included imaging nanofibers in hyaluronan-based pericellular matrix25, edge detection of colon cancer cells26, visualization of articular cartilage networks27 and the ultrastructure of Lepidoptera scales28. More recently, HIM has been used successfully to visualize the microvilli and glomerular structures in the rat kidney29 and nanoscale cuticle structures on Drosophila melanogaster30. In addition to using slow helium ions to image, research has been undertaken on the development of a Mega Electron Volt (MeV) microscope for whole cell density mapping31.
Despite these isolated demonstrations of HIM imaging, there has yet to be any significant interest demonstrated in developing HIM technology for biological applications. As such, the aim of this study was to undertake a set of experiments to test if the advantages described above would lead to improved insight on biological specimens. Therefore the HIM was tested on a traditionally difficult range of challenging biological samples. In addition, besides demonstrating the resolution and contrast abilities of the method, we also compared results to a modern FE-SEM as well as identify any potential consequences of sample preparation (critical point drying and sputter coating), as seen in the ultrastructure of the various specimens.
Results
To compare SEM to HIM, a set of biological samples were chosen where the unique aspects of the HIM system could provide advantages in their imaging. (i) Arabidopsis thaliana, the most favored model system for plant biology to study exterior nanoscale morphology (ii) HeLa cells, as an adherent cell type to study membrane surface details and filopodia, (iii) BoFeN1 iron-oxidizing bacteria to showcase the effects of drying and heavy metal coating on surface morphology and (iv) Pristionchus pacificus predator nematodes to illustrate the depth of field of the HIM system and its precision milling abilities.
Imaging parameters were chosen to provide a rational comparison between the two technologies. Since the overarching goal of the study was to investigate uncoated samples as well as to achieve high-resolution images at high magnifications, the FE-SEM was operated at a low voltage (0.7–1.0 kV for uncoated samples, and 3 kV for coated samples) with an aperture size of 30 μm. Both In-Lens (I-L) and Everhart-Thornley (E-T) detectors were used to image the samples as only secondary electrons were studied in the HIM. Alternatively, the HIM was operated at an optimal imaging voltage of 30 kV, an aperture size of 5 μm and a beam blanker current of 0.5 pA. An Everhart-Thornley (E-T) detector was used to image the samples. In both instances, working distance and tilt were slightly varied between images and samples to produce optimal imaging conditions of electron detection, depth of field, and charge compensation.
Care was taken during sample preparation to ensure that any artifacts were minimal. To reduce drying artifacts and to make the samples vacuum stable, proper fixation and critical point drying (CPD) was necessary. It was found that small plants and nematodes could be fixed with ice-cold ethanol and then directly critical point dried. Cells, however, were more sensitive to the CPD process and required the use of robust fixatives such as glutaraldehyde to maintain intact membrane structures. In addition all samples were kept in a desiccator or under vacuum at all times to minimize artifacts caused by rehydration of the tissues from native humidity.
Arabidopsis thaliana
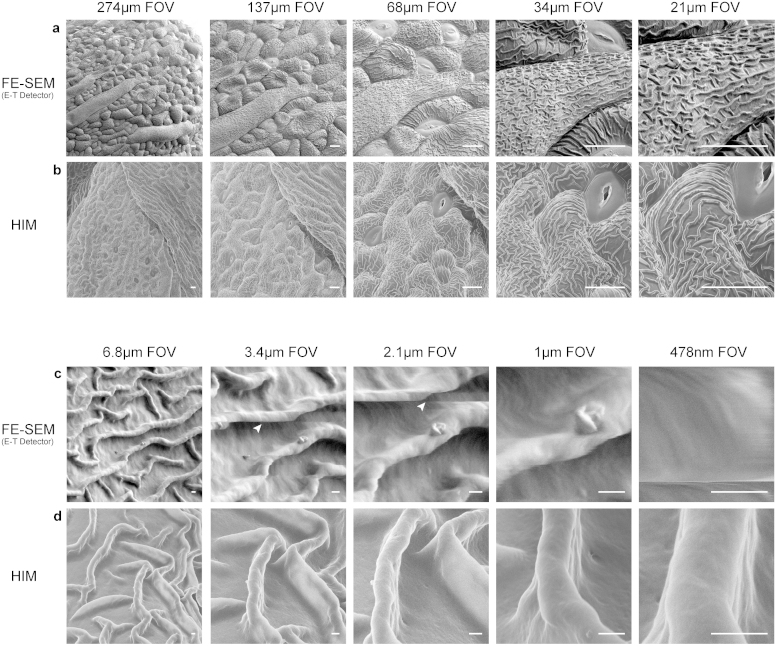
As a model plant, Arabidopsis was chosen as a stable and well-studied organism to compare the effects of imaging at ever increasing magnification ranges between the FE-SEM and HIM. Since the samples were critical point dried and uncoated, they were prone to charging and thus low voltage (<1 kV) imaging in the FE-SEM was required. When compared to the HIM, images in the SEM were similar at lower magnifications in both contrast, depth of field, and lack of charging (Figure 1a and b panels). However, despite low-voltage imaging in the FE-SEM, the sample exhibited charging effects starting at ~20 kX magnification (3.4 μm field of view), resulting in motion shown by streaks or bands within the image (Figure 1c, white arrowheads). In addition, at ~38 kX (2.1 μm field of view) fine textures on the sample surface and minute ridges on the cuticle could no longer be discerned. By 163 kX (478 nm field of view), the cuticle could no longer be clearly resolved in the FE-SEM as a result of either heavy charging or beam-induced thermal drift due to the very small probe size necessary to achieve such a magnification (Figure 1c far right panel). In contrast, charging in the HIM was completely eradicated via the use of the microscope's charge-compensation flood gun. As a result, higher magnifications up to 163 kX could be easily achieved without any form of charging, thermal drift and most importantly beam-induced damage, thus revealing fine surface structures, such as the defects in the wax on a single ridge of the plant's cuticle (Figure 1d panels).
Figure 1. Comparison of HIM and FE-SEM imaging in Arabidopsis thaliana.
(a) Low and (c) high magnification series of uncoated and critical point dried Arabidopsis thaliana sepal cuticle structures in the FE-SEM at low voltage (<1 kV) using an Everhart-Thornley (E-T) detector. At 22 kX magnification, charging artifacts become visible in the form of streaks or bands (white arrowheads) that go horizontally across the image. (b) Low and (d) high magnification series of the sepal cuticle structure in the HIM. Throughout the magnification range, no charging or imaging artifacts are apparent and cuticle structures are still visible at 163 kX. Scale Bars: (a,b) 10 μm (c,d) 200 nm.
HeLa cells
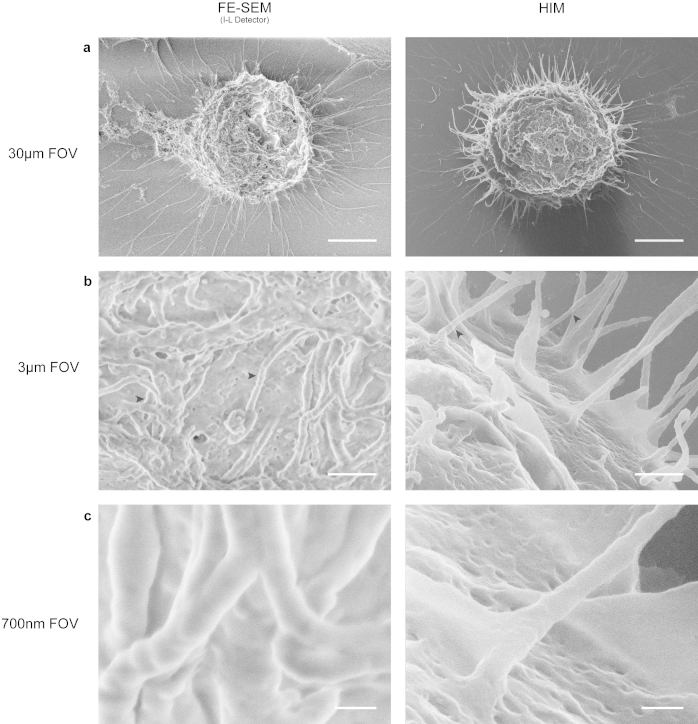
As a model cell line used in many cell biology applications, HeLa cells were chosen to demonstrate the effects of imaging fine structures such as filopodia and cell adhesion points in both HIM and SEM. The comparison of two cells in mitosis at low magnification (Figure 2a) clearly shows the efficient charge neutralization ability of the HIM over the FE-SEM. The intensity-saturated regions of the cell as well as the appearance of dark bands on the glass coverslip indicated the areas of charging in the FE-SEM. As the magnification was increased to 38 kX (3 μm field of view), two separate effects can be seen within each microscope. In the HIM, filopodia had a tendency to appear “ghosted” or somewhat translucent when there were additional features in close z-proximity (Figure 2b, right panel, black arrowheads). As described previously, this is likely due to the deeper penetration of the helium ions that pass through thinner regions of the sample, generating not only SE1 electrons on the surface closest to the source, but also secondary electrons on the surfaces immediately behind them31. This is due to the ion beam not changing geometry over that distance. In the SEM, the filopodia exhibited edge effects, a common artifact of low-voltage imaging of fine or filamentous structures (Figure 2b, left panel, black arrowheads). As magnification was increased a third time to 163 kX (700 nm field of view) (Figure 2c), the filopodia were easily recognizable in both microscopes, however, the surface features and textures were indistinguishable in the FE-SEM (Figure 2c, left panel).
Figure 2. Comparison of HIM and FE-SEM imaging of mitotic HeLa cells.
(a) Low magnification of uncoated critical point dried HeLa cells grown on glass coverslips in metaphase in both the FE-SEM at low voltage (<1 kV) with an In-Lens (I-L) detector (left panel) and HIM (right panel). Whilst the HIM exhibits high contrast and depth of field, the FE-SEM shows signs of charging in the form of saturated areas and dark and light banding on the glass coverslip. (b) At ~38 kX (3 μm field of view), the “ghosting” effect of some filopodia becomes apparent in the HIM (right panel, black arrowheads), while the FE-SEM exhibits a large amount of edge effects (left panel, black arrowheads). (c) High magnification of the same dividing cells shows no sign of loss of resolution in the HIM compared to the FE-SEM as depicted by the presence of cell membrane textures. Scale Bars: (a) 5 μm (b) 500 nm (c) 100 nm.
In addition to the comparison of the HIM to the SEM, the maximum overall resolution of the HIM was demonstrated in Figure 3. A random HeLa cell was chosen (Figure 3a) and from there, a single filopodia attachment point was recorded at increasing magnifications (Figure 3b–d) until a maximum magnification of 285 kX, or a field of view of 400 nm was achieved. At this magnification, the sample contrast between the cell and the glass coverslip was still high enough to discern the size of the attachment point (~5 nm). Remarkably, repeated imaging of these small and highly delicate structures caused no discernable beam damage.
Figure 3. Single filopodia attachment point of a HeLa cell as illustrated using the HIM.
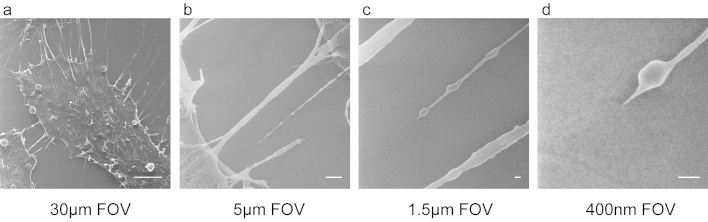
(a) A single cell was chosen that had minimal contact with neighboring cells and visible attachment points to the glass substrate. (b,c) The magnification was increased on a single adhesion point to a final magnification of (d) ~285 kX (400 nm field of view) while maintaining high enough contrast to depict an attachment width of ~5 nm. Scale Bars: (a) 5 μm (b) 1 μm (c,d) 50 nm.
Iron-oxidizing bacteria (BoFeN1)
The nitrate-reducing Acidovorax sp. strain BoFeN1, originally isolated from anoxic freshwater sediments of Lake Constance causes oxidation of Fe(II) either by an enzymatic process or by an abiotic reaction of Fe(II) with nitrite, which is an intermediate denitrification product32. It is known from numerous previous studies that, when grown in the presence of Fe(II), some cells tend to grow a crust of platelet-shaped Fe(III) minerals on the cell surface while others do not33.
The HIM images that were acquired at low magnification show three-dimensional cell mineral aggregates (Figure 4c and d, left panels). The images show a large depth of field in comparison to FE-SEM images that were acquired at low voltage at similar magnifications (Figure 4a and b, left panels). Low voltages were necessary in particular for imaging the uncoated samples by FE-SEM to avoid charging artifacts. The comparison of coated and uncoated samples by HIM at high resolution reveals two major artifacts caused by the sputter coating with platinum. Comparing the coated and non-coated critical point dried samples (Figure 4c and d) it appears that the crystallites attached to the cells end in rather sharp tips without coating, whereas mineral platelets on the cell surfaces of the platinum-coated samples appear to be relatively thick at the end and sometimes even show globule-like structures in the size-range of tens of nanometers (Figure 4c, right panel). A potential explanation for this change in mineral shape and structure might be a localized, preferential deposition of platinum due to charge-effects on the electrically isolating mineral structures. Additionally, the critical point dried samples show especially clean surfaces, whereas the surfaces of the minerals in plunge-frozen and freeze-dried samples appear to have a much smoother envelope in the case of the uncoated samples. We attribute this to a thin layer of extracellular polymeric substances (EPS) coating the whole aggregate. In contrast, we observed a relatively rough surface in the case of the platinum-coated samples (see Supplementary Fig. 1). This indicates that sputter coating with platinum introduces an artificial surface roughness.
Figure 4. Comparison of platinum coated and uncoated sample preparations critical point dried BoFeN1 bacteria in both the HIM and FE-SEM at low and high magnifications.
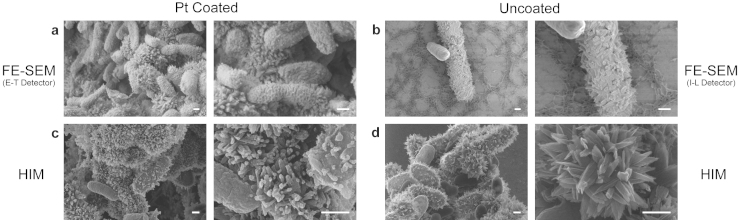
(a) Low and high magnifications of platinum coated (using an Everhart-Thornley (E-T) detector) and (b) uncoated bacterium (using an In-Lens (I-L) detector) in the FE-SEM reveal Fe(III) platelet structures that were on the order of tens of nanometers in size. Although the structures appeared much more flat in the uncoated sample, resolution was limited due to the necessity to use low-voltage imaging. (c) Low and high magnifications of platinum coated and (d) uncoated bacterium in the HIM reveal the coating induced artifact of globular structures and artificial textures on the platelets. The uncoated platelets in the HIM were for the first time revealed to be relatively flat with relatively sharp-tipped cellular attachments. Scale bars: 250 nm.
HIM imaging has, for the first time, allowed the identification of this coating artifact because of the higher spatial resolution in combination with the capability of analyzing non-conductive, uncoated samples. In addition, the existence of an EPS-envelope coating the minerals in plunge-frozen and freeze-dried samples could only be unambiguously shown in non-coated samples. Furthermore, sputter coating with platinum seemed only to affect the surface of the chemically fixed and critical point dried bacteria by creating a textured surface with network-like structures that created high secondary electron-signals (Figure 4c). We did not observe this effect when the cells were not coated with platinum (Figure 4d). We also did not observe these structures on plunge-frozen and freeze-dried samples, both with and without the heavy metal coating (see Supplementary Fig. 1). Since dehydration in solvents such as ethanol (a necessary step preceding critical point drying), can cause shrinkage in biological samples34, it is also possible that a similar effect could lead to a slightly textured cell surface that is potentially electrically insulating and thus, could favor localized deposition of platinum due to charge induced effects.
Predator nematode - pristionchus pacificus
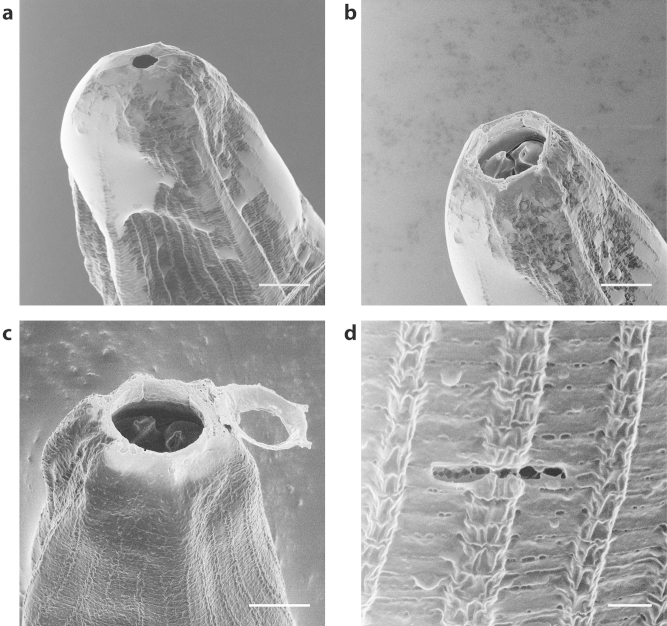
Finally the predator nematode, Pristionchus pacificus35 served as an ultrastructural imaging challenge in the sense that the interior of the mouth cavity, including teeth morphology, had never been successfully imaged. A sister nematode, Parasitodiplogaster laevigata, a parasite of fig wasps has previously been shown via SEM imaging to have a protruding tooth structure and a visible Dorsal Esophogeal Gland Orifice (DEGO) located at the base of the protruding tooth36. However, in the initial HIM imaging of Pristionchus pacificus, it was revealed that a membranous sheath obscured the primary tooth structure (Figure 5a). This afforded an opportunity to employ another unique aspect of the HIM system – precision nano-machining using focused noble gas ions. By switching out the working gas of the HIM from helium to neon, it was possible to delicately remove the tip of the membranous sheath, while leaving the interior of the mouth cavity intact (Figure 5c). This reveals the interior mouth structure as well as the primary tooth morphology and DEGO shape and size (Figure 5b). The very precise milling ability of the neon beam was effectively demonstrated by cutting the exterior of the nematode with a final dose of 0.3 nC/μm3 (Figure 5d). This proved intense enough to cut through the outer skin of the worm without damaging any of the surrounding tissue. For deeper cuts (such as the one shown in Figure 5c), the final cutting dose was increased to 30 nC/μm3 to penetrate the entire depth of the worm and efficiently mill through some of the tougher, more filamentous portions of the sample such as the mechanosensory fibers around the nematode head.
Figure 5. HIM imaging of Pristionchus pacificus.
(a) Native imaging of the nematode revealed the interior mouth cavity and tooth structures were obscured by a sheath-like structure. (b) neon milling of the outer sheath in the HIM revealed the primary tooth and DEGO structure of the nematode with minimal thermal damage to the cutting surface. (c) The removal of the outer sheath was performed in a very controlled manner with a final dose of 30 nC/μm3. (d) The delicate nature of the neon milling was demonstrated by the patterning of a small horizontal line using a dose of 0.3 nC/μm3. Despite being strong enough to penetrate the outer cuticle of the nematode, minimal ablation and thermal damage is visible on the surrounding area. Scale Bars: (a–c) 5 μm (d) 1 μm.
Discussion
Our results demonstrate both novel and exciting applications for helium ion microscopy in visualizing the surface ultrastructure of biological specimens at an unparalleled level of resolution and contrast. Specifically, we show that the common charging artifacts with the FE-SEM can be greatly reduced using the HIM approach. Moreover, the low sputtering rate of the helium ions beam for organic materials enables the repeated imaging of small and delicate surface features with no discernable beam damage. Also, we show that the heavy metal coating as generally required for SEM introduces clearly resolvable structural artifacts and HIM is a superior method in clearly revealing surface features at high magnification. Finally, we couple a neon ion beam with the HIM setup allowing us to mill away surface membranes and provide us with unprecedented access to sub-surface structures. The viability of this approach centers on the sputter rates of neon being four times lower than that of gallium19. As such, the neon ion beam is ideally suited for the precision milling of very small and delicate biological structures. In addition, it causes far less discernable sample damage, or metal ion deposition, as is sometimes the case when using liquid metal ion sources such as gallium37,38.
The resolution attained using HIM imaging is far greater than that obtained from a traditional FE-SEM approach. This higher resolution necessitates the development of new protocols for sample preparation, particularly to mitigate the possibility of fixation and drying artifacts. Cryo-immobilization offers a proven and reliable alternative for the current fixation and dehydration approaches. We suggest that combining this cryo-based sample preparation paradigm with HIM will allow routine imaging with little or no artifacts.
In conclusion we believe that this novel imaging method enabling the observation of completely unmodified structures on the nanometer scale will revolutionize our ability to interrogate both surface and sub-surface structures and provide deep scientific insights into the nanoscale architecture of biological specimens and soft materials.
Methods
Samples were grown/cultured and subsequently fixed and critical point/freeze dried using the following methods. After the drying stage, the samples were carefully removed and adhered to double-sided carbon tabs on aluminum stubs and stored in a desiccator. For SEM imaging, the Arabidopsis thaliana, HeLa cells and Pristionchus pacificus samples were imaged on a Zeiss Sigma VP FE-SEM (Carl Zeiss, Cambridge, UK) and the Acidovorax sp. samples were imaged on a Zeiss Crossbeam 1540XB (Carl Zeiss, Oberkochen, Germany) using the parameters mentioned above. All samples were subsequently imaged on a Zeiss Orion PLUS HIM (Carl Zeiss, Peabody, MA).
Arabidopsis thaliana
Growth
Wild type Arabidopsis were grown in soil to stage 6. Plants were trimmed by carefully pinching off regions of interest using two pairs of #5 forceps and the subsequent pieces were placed into ice-cold 100% EM grade ethanol for fixation.
Critical point drying
The ethanol fixed plant pieces were placed into a Teflon carrier for separation and into an automated critical point drier (Leica EM CPD300, Leica Vienna) which was set to perform 25 exchange cycles of CO2 at medium speed and 20% stirring. All additional fill, heating, and venting steps were performed at medium speed as well. After drying, the samples were carefully removed and adhered to double-sided carbon tabs on aluminum stubs and stored in a desiccator.
HeLa cells
Coverslip preparation
Cleaned 22 × 22 mm no. 1.5 coverslips were coated with a 1× solution of poly-L lysine for 15 minutes and air-dried at room temperature. Coated coverslips were then sterilized by UV light (NuAire cell culture cabinet) for 15 minutes on each side and stored until use.
Cell culture
HeLa cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum, 1.0% l-glutamine with 0.018% NaHCO3 and penicillin/streptomycin in 5% CO2 and 95% air in a Thermo Scientific Forma Series II Water Jacketed CO2 cell culture incubator at 37°C. Cells were allowed to grow for at least 36 hours from the previous passage until reaching 60–80% confluency before passage again. Cells were washed twice with 2–5 mls of room temperature 1× DPBS and the trypsinized with 1–2 mls of 0.05% Trypsin-EDTA (Life Technologies) for 5–10 minutes at 37°C. Trypsin was neutralized with 2–4 ml of 37°C complete media and collected in a 15 ml conical vial. 80,000–120,000 cells (per well) were plated in 6-well plates containing 3 mls of complete media and a single poly-L lysine coated coverslip. Cells were allowed to grow for 36–48 hours to ensure that the cells enter the exponential growth phase prior to fixation. Shorter periods of growth or over confluency yielded a much lower percentage of mitotic cells upon fixation.
Cell fixation
Plated cells were gently rinsed with 2 mls of 37°C DPBS and aspirated off prior to fixation. 3–5 mls of freshly prepared 37°C fixation solution containing 4% paraformaldehyde and 2% glutaraldehyde in DPBS was applied for 30 minutes at 37°C and swapped with fresh 37°C fixative to continue fixing for 1.5 hours at 37°C. Once the cells were fixed they washed briefly in DI water and were either critically point dried or were stored at 4°C in 1% paraformaldehyde and 2% glutaraldehyde in DPBS.
Critical point drying
Fixed coverslips were placed into an automated critical point drier (Leica EM CPD300, Leica Vienna) which was set to perform 25 exchange cycles of CO2 at medium speed and 20% stirring. All additional fill, heating, and venting steps were performed at medium speed as well. After drying, the samples were carefully removed and adhered to double-sided carbon tabs on aluminum stubs and stored in a desiccator.
Iron-oxidizing bacteria (BoFeN1)
Cell culture
Cultures of the nitrate-reducing Acidovorax sp. strain BoFeN1 were grown under anoxic conditions as described previously33. In brief, cells were grown in anoxic freshwater medium with initial concentrations of 10 mM Fe2+ and 10 mM nitrate and 5 mM acetate. Samples of the cell suspension were taken with syringes after 5 to 6 days of incubation when Fe(II) is completely oxidized39.
Critical point drying
1 mL of a centrifuged aliquot of the cell suspension were treated with 2.5% glutaraldehyde at 4°C for 1 hour. The samples were rinsed twice with deionized water and deposited onto 10 mm glass cover slips that were previously coated with a 8 nm Pt-layer and poly-l-lysine. After 20 minutes to ensure binding of cell-mineral aggregates to the poly-l-lysine layer, the samples were rinsed with de-ionized water und subsequently dehydrated in a series of ethanol dilutions (30%, 70%, 95% and 3× 100% dried on molecular sieve) with an equilibration step of 10 minutes each. The samples were then placed in a critical point dryer (Polaron E3000), wherein the ethanol was replaced by liquid CO2 at 1°C. After flushing of 3 × 200 g CO2 with 20 minute equilibration periods after each step the temperature was gradually increased to 38°C while the pressure was monitored and adjusted to reach and remain at 84 bars. This ensured bypassing the critical point when the pressure was released from 84 bars to atmospheric pressure. Half of the samples were then sputter-coated with a 4 nm platinum layer using a BALZERS-UNION MED 010 (BAL-TEC, Liechtenstein) sputter coater. After reaching a vacuum of 8*10−6 mbar, the sputtering process at was started in 2*10−2 mbar of Ar.
Rapid freezing followed by freeze-drying
To be able to compare samples treated after the standard sample preparation protocol of CPD with a more natural state of the cell-mineral aggregates, we decided to use a physical fixation (rapid-freezing) instead of chemical fixation, followed by freeze-drying under high vacuum. Since the cell-mineral aggregates of BoFeN1 are within the μm size ranges, high-pressure freezing was not required to avoid the formation of ice crystals, but plunge-freezing in liquid propane was sufficient. Therefore, droplets of the cell suspension were deposited onto formvar-coated 300 mesh TEM-grids. They were blotted with filter paper and immediately shot into liquid propane at liquid-nitrogen (LN2) temperature (<−196°C). The grids were stored in LN2 until freeze-drying. Freeze-drying was done on the cooling stage of a BAL-TEC BA 500 high vacuum coater. The grids were placed on the cooling stage at −120°C. Thermal conduction between the sample and the stage was ensured by a small droplet of liquid propane, which also prevented ice formation on the stage during sample transfer. The stage was gradually heated up to −82°C, while maintaining the pressure in the recipient below 5*10−5 mbar. The progress of freeze-drying was followed visually. Upon complete disappearance of the ice after approximately 35–45 minutes, the stage was gradually heated up to +25°C before the chamber was vented and the samples could be removed. Half of the samples were sputter-coated with a 4 nm Pt layer as described previously.
Predator nematode - pristionchus pacificus
Worm culture
Pristionchus pacificus specimens (strain identification: rlh24) were raised on agar plates coated with E. coli (OP50)40. At the young adult stage, worms were rinsed 3× in saline buffer (M9) and transferred to an uncoated agar plate. Worms then remained off food for 5 hours, providing sufficient time for P. pacificus to consume residual E. coli adherent to their exterior cuticle. This process is critical to produce a clean imaging sample. The predators were then fixed by placing them into a solution of ice-cold 100% EM grade ethanol.
Critical point drying
The ethanol fixed worms were placed into a 30 mm porous pot (Electron Microscopy Sciences (EMS) cat# 70187-20) and into an automated critical point drier (Leica EM CPD300, Leica Vienna) which was set to perform 25 exchange cycles of CO2 at medium speed and 20% stirring. All additional fill, heating, and venting steps were performed at medium speed as well. After drying, the samples were carefully removed and adhered to double-sided carbon tabs on aluminum stubs and stored in a desiccator.
Author Contributions
M.S.J., J.M.K. and Y.J.S. were responsible for the preparation of the plant and cell culture samples and M.S.J. and C.H. undertook the plant, nematode and cell HIM imaging experiments. C.H. and D.F. undertook the neon cutting of nematode experiments. F.Z., M.O. and C.B. were responsible for the preparation of the bacterial samples and the bacterial HIM imaging. Worm samples were prepared by K.C. and S.H.C. Study was conceived by B.G. and J.A.J.F., M.S.J. and J.A.J.F. prepared the figures and wrote the manuscript with contributions from M.O., C.B., L.A.S. and B.G.
Supplementary Material
Supplementary Information
Acknowledgments
We gratefully acknowledge financial support from the Waitt Advanced Biophotonics Center endowment (J.M.K., M.S.J. and J.A.J.F.), NCI P30 Cancer Center Support Grant CA014195-39 (Y.J.S. and J.A.J.F.), NINDS P30 Neuroscience Center Core Grant NS072031-01A1 (M.S.J. and J.A.J.F.) and DFG Emmy Noether grant OB 362/1-1 (F.Z. and M.O.). We would also like to thank the Ion Microscopy Innovation Center at Carl Zeiss Microscopy (Peabody, MA) for providing access to an Orion PLUS helium ion microscope, Carl Procko from the Chory lab at the Salk Institute for providing wild-type Arabidopsis Thaliana for HIM imaging experiments and Kat Kearny for critical reading of the manuscript and for helpful suggestions regarding organization and formatting.
Footnotes
The author(s) declare that Chuong Huynh, David Ferranti, Lewis Stern and Bernhard Goetze are employed by the manufacturer of the commercial Orion PLUS Helium Ion Microscope (Carl Zeiss Microscopy, LLC). No other competing financial interests apply.
References
- Bozzola J. J. & Russell L. D. Electron microscopy: principles and techniques for biologists. 2nd edn, (Jones and Bartlett, 1999). [Google Scholar]
- Denk W. & Horstmann H. Serial Block-Face Scanning Electron Microscopy to Reconstruct Three-Dimensional Tissue Nanostructure. PLoS Biol 2, e329, 10.1371/journal.pbio.0020329 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D. J., Morrissey F. & Lich B. H. A New Approach to Studying Biological and Soft Materials Using Focused Ion Beam Scanning Electron Microscopy (FIB SEM). J. Phys.: Conf. Ser. 26, 50 (2006). [Google Scholar]
- Crang F. E. & Klomparens K. L. Artifacts in biological electron microscopy. (Plenum Press, New York & London, 1988). [DOI] [PubMed] [Google Scholar]
- Pathan A. K., Bond J. & Gaskin R. E. Sample preparation for scanning electron microscopy of plant surfaces-Horses for courses. Micron 39, 1049–1061, 10.1016/j.micron.2008.05.006 (2008). [DOI] [PubMed] [Google Scholar]
- Timp W. & Matsudaira P. in Biophysical Tools for Biologists, Vol 2: In Vivo Techniques Vol. 89 Methods in Cell Biology (eds Correia J. J., & Detrich H. W.) 391–407 (Elsevier Academic Press Inc, 2008). [Google Scholar]
- Reimer L. Image formation in low-voltage scanning electron microscopy. (SPIE Optical Engineering Press, 1993). [Google Scholar]
- Bell D. C. & Erdman N. Low Voltage Electron Microscopy: Principles and Applications. First edn, Vol. 1 (John Wiley & Sons Ltd., 2013). [Google Scholar]
- Joy D. C. & Joy C. S. Low voltage scanning electron microscopy. Micron 27, 247–263 (1996). [Google Scholar]
- Pawley J. & Schatten H. Biological Low-Voltage Scanning Electron Microscopy. First edn, (Springer, 2008). [Google Scholar]
- Cazaux J. About the Mechanisms of Charging in EPMA, SEM, and ESEM with Their Time Evolution. Microsc Microanal 10, 670–684, 10.1017/S1431927604040619 (2004). [DOI] [PubMed] [Google Scholar]
- Stabentheiner E., Zankel A. & Pölt P. Environmental scanning electron microscopy (ESEM)–a versatile tool in studying plants. Protoplasma 246, 89–99, 10.1007/s00709-010-0155-3 (2010). [DOI] [PubMed] [Google Scholar]
- Bergmans L., Moisiadis P., Van Meerbeek B., Quirynen M. & Lambrechts P. Microscopic observation of bacteria: review highlighting the use of environmental SEM. Int Endod J 38, 775–788, 10.1111/j.1365-2591.2005.00999.x (2005). [DOI] [PubMed] [Google Scholar]
- Stokes D. J., Mugnier J. Y. & Clarke C. J. Static and dynamic experiments in cryo-electron microscopy: comparative observations using high-vacuum, low-voltage and low-vacuum SEM. J Microsc 213, 198–204, 10.1111/j.1365-2818.2004.01282.x (2004). [DOI] [PubMed] [Google Scholar]
- Echlin P. Low-temperature microscopy and analysis. (Plenum Press, 1992). [Google Scholar]
- Yu Z., Chen H., Tong Y. & Wu P. in Rice Protocols Vol. 956 Methods in Molecular Biology (ed Yinong Yang) Ch. 17, 243–248 (Humana Press, 2013). [Google Scholar]
- Ward B. W., Notte J. A. & Economou N. P. 6 edn 2871–2874 (AVS).
- Müller E. W. & Bahadur K. Field Ionization of Gases at a Metal Surface and the Resolution of the Field Ion Microscope. Phys Rev 102, 624–631 (1956). [Google Scholar]
- Hill R., Notte J. A. & Scipioni L. in Advances in Imaging and Electron Physics Vol. Volume 170 (ed Hawkes Peter W.) 65–148 (Elsevier, 2012). [Google Scholar]
- Bell D. Contrast Performance: Low Voltage Electrons vs. Helium Ions. Microsc Microanal 17, 660–661, 10.1017/S143192761100417X (2011). [Google Scholar]
- Bell D. C. Contrast Mechanisms and Image Formation in Helium Ion Microscopy. Microsc Microanal 15, 147–153, 10.1017/S1431927609090138 (2009). [DOI] [PubMed] [Google Scholar]
- Ramachandra R., Griffin B. & Joy D. A model of secondary electron imaging in the helium ion scanning microscope. Ultramicroscopy 109, 748–757, 10.1016/j.ultramic.2009.01.013 (2009). [DOI] [PubMed] [Google Scholar]
- Joy D. C. & Griffin B. J. Is Microanalysis Possible in the Helium Ion Microscope? Microsc Microanal 17, 643–649, 10.1017/s1431927611000596 (2011). [DOI] [PubMed] [Google Scholar]
- Postek M. T., Vladar A. E. & Bin M. Recent progress in understanding the imaging and metrology using the helium ion microscope. Proc SPIE-Int Soc Opt Eng 7378, 737808 (737810 pp.)–737808 (737810 pp.), 10.1117/12.824533 (2009). [Google Scholar]
- Gerlach D. et al. Nanofibers in a hyaluronan-based pericellular matrix. Matrix Biol 29, 664–667, 10.1016/j.matbio.2010.08.006 (2010). [DOI] [PubMed] [Google Scholar]
- Bazou D., Behan G., Reid C., Boland J. J. & Zhang H. Z. Imaging of human colon cancer cells using He-Ion scanning microscopy. J Microsc 242, 290–294, 10.1111/j.1365-2818.2010.03467.x (2011). [DOI] [PubMed] [Google Scholar]
- Vanden Berg-Foels W. S., Scipioni L., Huynh C. & Wen X. Helium ion microscopy for high-resolution visualization of the articular cartilage collagen network. J Microsc 246, 168–176, 10.1111/j.1365-2818.2012.03606.x (2012). [DOI] [PubMed] [Google Scholar]
- Boden S. A., Asadollahbaik A., Rutt H. N. & Bagnall D. M. Helium ion microscopy of Lepidoptera scales. Scanning 34, 107–120, 10.1002/sca.20267 (2012). [DOI] [PubMed] [Google Scholar]
- Rice W. L. et al. High resolution helium ion scanning microscopy of the rat kidney. PloS one 8, 10.1371/journal.pone.0057051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boseman A., Nowlin K., Ashraf S., Yang J. & Lajeunesse D. Ultrastructural analysis of wild type and mutant Drosophila melanogaster using helium ion microscopy. Micron 51, 26–35, 10.1016/j.micron.2013.06.005 (2013). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Whole-Cell Imaging at Nanometer Resolutions Using Fast and Slow Focused Helium Ions. Biophys J 101, 1788–1793, 10.1016/j.bpj.2011.08.028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klueglein N. & Kappler A. Abiotic oxidation of Fe(II) by reactive nitrogen species in cultures of the nitrate-reducing Fe(II) oxidizer Acidovorax sp BoFeN1 - questioning the existence of enzymatic Fe(II) oxidation. Geobiology 11, 180–190, 10.1111/gbi.12019 (2013). [DOI] [PubMed] [Google Scholar]
- Kappler A., Schink B. & Newman D. K. Fe(III) mineral formation and cell encrustation by the nitrate-dependent Fe(II)-oxidizer strain BoFeN1. Geobiology 3, 235–245, 10.1111/j.1472-4669.2006.00056.x (2005). [Google Scholar]
- Montesinos E., Esteve I. & Guerrero R. Comparison between direct methods for determination of microbial cell-volume - Electron-Microscopy and Electronic Particle Sizing. Appl Environ Microbiol 45, 1651–1658 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumbarger D. J., Riebesell M., Rodelsperger C. & Sommer R. J. System-wide Rewiring Underlies Behavioral Differences in Predatory and Bacterial-Feeding Nematodes. Cell 152, 109–119, 10.1016/j.cell.2012.12.013 (2013). [DOI] [PubMed] [Google Scholar]
- Giblin-Davis R. M. et al. Stomatal ultrastructure, molecular phylogeny, and description of Parasitodiplogaster laevigata n. sp (Nematoda: Diplogastridae), a parasite of fig wasps. J Nematol 38, 137–149 (2006). [PMC free article] [PubMed] [Google Scholar]
- Tondare V. N. Quest for high brightness, monochromatic noble gas ion sources. J Vac Sci Technol A 23, 1498–1508, 10.1116/1.2101792 (2005). [Google Scholar]
- Drobne D., Milani M., Zrimec A., Leser V. & Berden Zrimec M. Electron and ion imaging of gland cells using the FIB/SEM system. J Microsc 219, 29–35, 10.1111/j.1365-2818.2005.01490.x (2005). [DOI] [PubMed] [Google Scholar]
- Miot J. et al. Iron biomineralization by neutrophilic nitrate-reducing iron-oxidizing bacteria. Geochim Cosmochim Ac 73, A884–A884 (2009). [Google Scholar]
- Sommer R. J. Pristionchus pacificus., WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.7.1 (2006), http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information