Abstract
In both mammalian and insect models of ethanol intoxication, high doses of ethanol induce motor impairment and eventually sedation. Sensitivity to the sedative effects of ethanol is inversely correlated with risk for alcoholism. However, the genes regulating ethanol sensitivity are largely unknown. Based on a previous genetic screen in Drosophila for ethanol sedation mutants, we identified a novel gene, tank (CG15626), the homolog of the mammalian tumor suppressor EI24/PIG8, which has a strong role in regulating ethanol sedation sensitivity. Genetic and behavioral analyses revealed that tank acts in the adult nervous system to promote ethanol sensitivity. We localized the function of tank in regulating ethanol sensitivity to neurons within the pars intercerebralis that have not been implicated previously in ethanol responses. We show that acutely manipulating the activity of all tank-expressing neurons, or of pars intercerebralis neurons in particular, alters ethanol sensitivity in a sexually dimorphic manner, since neuronal activation enhanced ethanol sedation in males, but not females. Finally, we provide anatomical evidence that tank-expressing neurons form likely synaptic connections with neurons expressing the neural sex determination factor fruitless (fru), which have been implicated recently in the regulation of ethanol sensitivity. We suggest that a functional interaction with fru neurons, many of which are sexually dimorphic, may account for the sex-specific effect induced by activating tank neurons. Overall, we have characterized a novel gene and corresponding set of neurons that regulate ethanol sensitivity in Drosophila.
Introduction
Alcohol is one of the most widely used and abused drugs in the world. Although lower doses of ethanol act as a stimulant, higher doses act as a depressant (Pohorecky, 1977). The depressant effects of ethanol manifest in humans as depressed mood, fatigue, and cognitive and motor impairment (Babor et al., 1983; Miller et al., 2009); animal models also exhibit motor incoordination and ultimately sedation (Pohorecky, 1977). Genetic factors influence susceptibility to alcohol use disorders (AUDs), but few specific genes have been identified (Mayfield et al., 2008). Several studies have suggested that susceptibility to AUDs is correlated with decreased sensitivity to the depressant effects of ethanol (Schuckit, 1994; Morean and Corbin, 2010). In addition, several genes regulate both acute ethanol sensitivity and ethanol consumption in rodent models (e.g., Thiele et al., 1998; Hodge et al., 1999). Characterizing the genes that mediate acute ethanol sensitivity may therefore provide insight into alcohol addiction.
The fruit fly Drosophila melanogaster is an established model for studying the genes underlying acute ethanol responses (Kaun et al., 2012). As in humans and rodents, lower doses of ethanol stimulate locomotor activity in flies (Wolf et al., 2002), whereas higher doses induce motor incoordination and sedation (Moore et al., 1998; Rothenfluh et al., 2006). Many evolutionarily conserved genes regulate acute ethanol responses in both flies and mammals, such as protein kinase A (Moore et al., 1998; Thiele et al., 2000), epidermal growth factor receptor (EGFR) (Corl et al., 2009), and neuropeptide F/Y (NPF/NPY) (Thiele et al., 1998; Wen et al., 2005). Drosophila also offers powerful tools for studying neural circuits (Kaun et al., 2012); however, the neurons mediating ethanol sedation in flies are largely unknown. NPF-expressing cells promote sedation sensitivity (Wen et al., 2005), as do cells expressing the neural sex determination factor fruitless (fru; Devineni and Heberlein, 2012), whereas insulin signaling in cells within the pars intercerebralis (PI) promotes sedation resistance (Corl et al., 2005). Additional studies have identified the neurons in which specific genes function to regulate ethanol sedation, but most of these studies did not determine whether the activity of these neurons acutely regulates ethanol sedation (Urizar et al., 2007; Corl et al., 2009; Eddison et al., 2011).
In this study, we characterize the novel Drosophila gene tank, a tumor suppressor homolog that functions in the adult nervous system to promote sensitivity to ethanol sedation. We show that tank is expressed in a limited set of neurons and that manipulating the activity of these neurons affects ethanol sensitivity in a partially sex-specific manner. We localize the function of tank to neurons within the PI and show that the activity of these neurons in particular regulates ethanol sensitivity. Finally, we examine whether the sexually dimorphic effect of activating tank neurons is due to expression of fru, which mediates many sexual dimorphisms. We find that although tank-expressing cells do not express fru, they form likely synaptic connections with fru-expressing neurons, suggesting that sexual dimorphisms in downstream fru neurons may mediate the sex-specific effect of tank neuron activation.
Materials and Methods
Fly stocks and maintenance.
Flies were generally reared at 25°C and 70% relative humidity on standard cornmeal/molasses food. Flies for TrpA1 and shits experiments were reared at 22°C and flies for Gal80ts experiments were reared at 18°C and shifted to 29°C for 4–5 d during adulthood. w Berlin was used as the control strain. All lines used for behavior were outcrossed into this background for at least five generations. All assays were performed on 3- to 5-d-old flies that were generally nonvirgin.
The tank4-12 P element mutant was generated in our laboratory, as were the seven Gal4 lines used to target the PI. The tank4-12 precise excision line was generated by remobilization of the P element using delta2–3 transposase (Robertson et al., 1988) and was verified by DNA sequencing. tank RNA interference (RNAi) lines were obtained from the Vienna Drosophila RNAi Center: GD7844 (tankRNAi) and KK106205 (tankRNAi-2). The UAS-GFP line used contains two insertions, a membrane-targeted UAS-CD8-GFP and a cytoplasmic-targeted UAS-T2-GFP. The C155 insertion of elavGal4 was used. Additional lines used were as follows: dilp2Gal4 (Rulifson et al., 2002), SIFaGal4 (Terhzaz et al., 2007), DMSGal4 and DSKGal4 (Park et al., 2008), OK107 and c747 (Connolly et al., 1996), fruGal4 (Manoli et al., 2005), frulex (Mellert et al., 2010), and fruFLP and UAS>STOP>CD8-GFP (Yu et al., 2010).
Behavioral assays.
Ethanol sedation assays were performed in the “booz-o-mat,” an eight-chambered apparatus that delivers a specified concentration of ethanol vapor (Wolf et al., 2002). Sedation was assayed manually as described previously (Corl et al., 2009) using 67% ethanol vapor unless otherwise specified. Briefly, flies were tested at 5 min intervals by applying a mechanical stimulus (twirling each vial within the booz-o-mat chamber), causing flies to lose balance, and flies that were unable to right themselves were scored as sedated. The experimenter was blinded to the identity of each genotype. The time required for 50% of flies to sedate (ST50) was determined for each vial by linear interpolation. Twenty flies per vial were assayed, and n refers to the number of vials tested. Assays were conducted at ∼25° with the exception of the TrpA1 and shits experiments, which were conducted at the specified temperatures. The temperature used to activate TrpA1 varied between 27°C and 30°C depending on the Gal4 line, because we observed that TrpA1 activation using some Gal4 lines induced general behavioral defects when the temperature was raised too high.
Measurement of internal ethanol concentration.
Internal ethanol concentration was assayed in fly extracts as described previously (Devineni et al., 2011) using the ethanol assay kit from Diagnostic Chemicals. Ethanol concentration within flies was calculated based on the water content of flies, which was measured previously to be 0.60 μl for males and 0.93 μl for females (Devineni and Heberlein, 2012). Because sedated flies show an increased rate of ethanol absorption compared with nonsedated flies (U.H., unpublished data), ethanol concentration was measured in flies that were exposed to a nonsedating dose of ethanol, generally 10–15 min at 47% ethanol (except for the experiments shown in Fig. 1, in which sedation times were equalized between groups).
Figure 1.
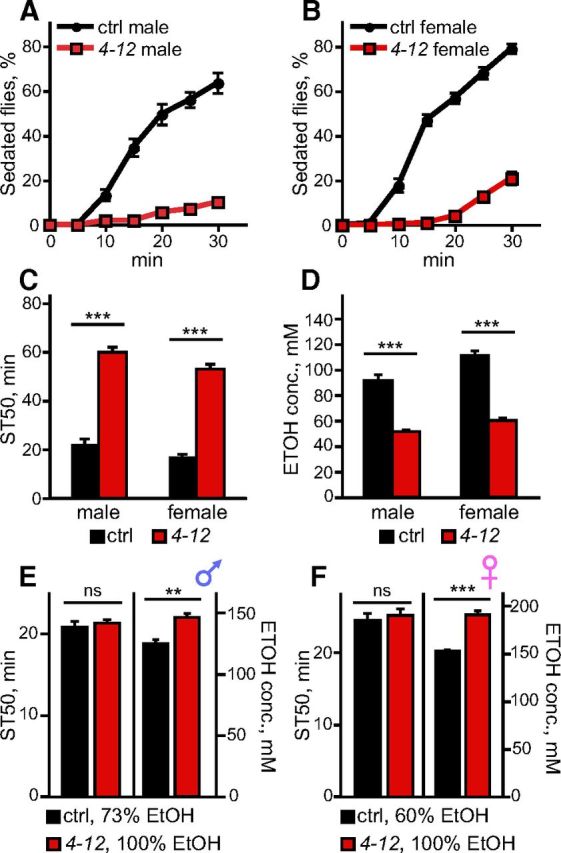
4-12 mutants show altered ethanol sensitivity. A, B, Sedation curves of control and 4-12 mutant males (A) or females (B) exposed to 67% ethanol vapor (n = 10). C, ST50 values calculated from the sedation curves in A and B, with each sample being assayed for 30 min or until reaching 50% sedation if <50% of flies were sedated at 30 min (n = 10). D, Internal ethanol concentration of flies exposed to 47% ethanol for 12 min (n = 4). E, Control males tested at 73% ethanol sedated at the same rate as 4-12 males tested at 100% ethanol, with both groups showing an ST50 of ∼21 min (n = 8). When assayed after 21 min of ethanol exposure under these conditions, 4-12 males contained more ethanol than control males (n = 4). F, Control females tested at 60% ethanol sedated at the same rate as 4-12 females tested at 100% ethanol, with both groups showing an ST50 of ∼25 min (n = 8). When assayed after 25 min of ethanol exposure under these conditions, 4-12 females contained more ethanol than control females (n = 4). **p < 0.01, ***p < 0.001. For C, D, two-way ANOVA followed by Bonferroni's post tests were used; for E, F, unpaired t test was used.
Immunohistochemistry.
Immunohistochemistry was performed essentially as described previously (Joseph et al., 2009). Samples were dissected in PBS and fixed for 20 min in 4% paraformaldehyde in PBS. frulex/4-12Gal4 colocalization and GRASP experiments were performed in fixed, unstained brains. For characterization of Gal4 expression patterns, brains were stained with rabbit anti-green fluorescent protein (anti-GFP; 1:200) and mouse nc82 (1:50), followed by the secondary antibodies Cy3-coupled goat anti-mouse antibody (1:500) and FITC-coupled goat anti-rabbit antibody (1:500); ventral nerve cords (VNCs) were stained with chicken anti-GFP (1:1000) and mouse nc82 (1:30), followed by the secondary antibodies Alexa Fluor 488 goat anti-chicken antibody (1:200) and Alexa Fluor 633 goat anti-mouse antibody (1:200). Confocal images were analyzed using ImageJ.
Molecular biology.
The genomic DNA flanking the 4-12 insertion was isolated using inverse PCR and sequenced. UAS-tankRA was generated by cloning EST RE16861, which encodes tank-RA, into the pUAST vector. The transgene was verified by DNA sequencing and injected into w Berlin flies.
qRT-PCR was performed as described previously (Tsai et al., 2004) except that we used the SYBR Green protocol (Applied Biosystems). RNA was extracted from fly heads and cDNA was prepared using reverse transcription reagents. tank primers were designed using Primer Express, targeted to exon junctions to avoid amplifying genomic DNA, and verified by BLAST to be specific to tank. tank transcript levels were normalized to levels of the control transcript rp49, and relative RNA abundance was calculated using the standard curve method. The tank primers used were as follows: 5′-ATCAGCAAGCACTGGCACAG-3′ and 5′-CGGGTGGACAGACCTTAGTTCA-3′ (tank-RA); 5′-CCCCCGATCAGGAACTAAGG-3′ and 5′-TTCTTTATCGCCGCCATCTC-3′ (tank-RB).
Statistical analyses.
Statistical analyses were performed using Prism Version 4 software (GraphPad). Experimental and control flies were always tested simultaneously; experiments in which each sedation “run” contained one sample of each genotype were considered to be paired/matched experiments, whereas unpaired tests were used for all other experiments. All graphs represent mean ± SEM.
For Gal4/UAS experiments, experimental flies were compared with both the Gal4/+ and UAS/+ controls. Only experimental lines that differed from both controls in the same direction were considered to have a phenotype. In the figures, significant differences that are biologically relevant are depicted in black, whereas all other comparisons (i.e., comparisons in which the experimental value lies between the two controls or is not significantly different from both controls) are depicted in gray.
Results
4-12 mutants show increased resistance to ethanol sedation
In Drosophila, high doses of ethanol induce sedation, which can be measured using a loss-of-righting assay (Rothenfluh et al., 2006). The mutation 4-12 was isolated previously from a genetic screen for sensitivity to ethanol sedation (Corl et al., 2009; Devineni et al., 2011). When exposed to a high concentration of 67% ethanol vapor, 4-12 mutant males and females showed very strong resistance to ethanol sedation compared with control males and females, respectively (Fig. 1A,B). We also noted that control females were more sensitive to sedation than control males (Fig. 1A–C), as described previously (Devineni and Heberlein, 2012). Sensitivity to ethanol sedation can be quantified as the ST50, the time required for 50% of flies to sedate. We compared the ST50 values of control and 4-12 males and females by two-way ANOVA, which revealed a significant effect of both genotype (F(1,36) = 409.1, p < 0.001) and sex (F(1,36) = 10.9, p = 0.002), but no significant interaction (F(1,36) = 0.2, p = 0.642), indicating that the 4-12 mutation has a comparable effect in males and females (Fig. 1C).
To determine whether the altered ethanol sensitivity in 4-12 mutants could be due to a difference in ethanol absorption or metabolism, we measured the internal ethanol concentration of 4-12 mutant and control flies. In both sexes, 4-12 mutants contained a lower ethanol concentration than controls (Fig. 1D), which likely contributes to their sedation resistance. We also observed that control females contained a higher ethanol concentration than control males, as described previously (Devineni and Heberlein, 2012). Again, two-way ANOVA revealed a significant effect of both genotype (F(1,12) = 203.0, p < 0.001) and sex (F(1,12) = 20.0, p < 0.001) on ethanol concentration, but no significant interaction (F(1,12) = 3.0, p = 0.110), indicating that the 4-12 mutation has a similar effect on internal ethanol levels in both sexes.
We next investigated whether factors other than decreased internal ethanol levels also contribute to the sedation resistance of 4-12 mutants. To do so, we conducted dose–response experiments to find a pair of ethanol doses at which 4-12 mutant and control flies sedated at the same rate, and we then measured their internal ethanol concentration. For both males and females, when the ST50 values of 4-12 mutant and control flies were equalized by adjusting the relative ethanol doses (t test; p = 0.564 for males, p = 0.617 for females), 4-12 mutants always contained more ethanol than controls (Fig. 1E,F; t test, p = 0.004 for males, p < 0.001 for females). Because 4-12 mutants require a higher internal ethanol concentration to show the same sedation sensitivity as control flies, we conclude that the 4-12 mutation must induce adaptations promoting sedation resistance that are independent of its effect on ethanol levels. Therefore, the increased sedation resistance induced by the 4-12 mutation has both a pharmacokinetic component (resistance due to reduced ethanol absorption and/or enhanced ethanol metabolism) and a pharmacodynamic component (resistance independent of ethanol concentration resulting from alterations in the physiological effects of ethanol).
4-12 disrupts expression of the novel gene tank
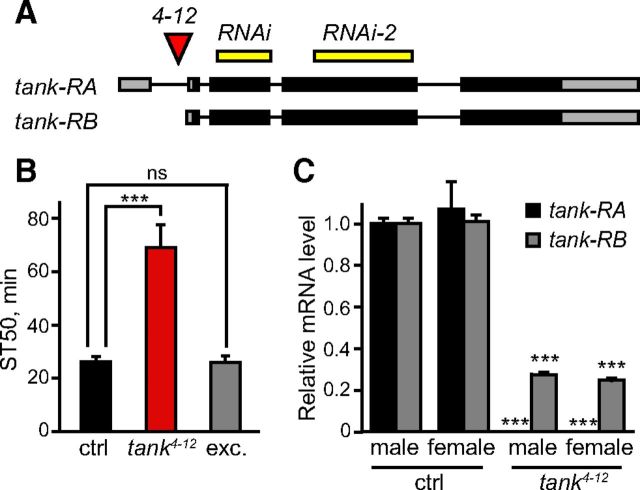
The 4-12 mutant carries a P element insertion that we mapped to a novel gene, CG15626. We decided to name this gene tank because its mutation allows flies to imbibe high levels of ethanol before becoming sedated. The 4-12 mutant will henceforth be referred to as tank4-12. tank encodes two transcripts, tank-RA and tank-RB, which are predicted to encode a single protein (Fig. 2A). The mammalian homolog of tank, EI24 (also known as PIG8), encodes a tumor suppressor that regulates apoptosis and has been linked to human cancer (Gu et al., 2000; Gentile et al., 2001; Zhao et al., 2005). tank bears significant homology to human EI24 in the central region of the protein (43% identity and 65% similarity; Blast e value = 2e-77). Like EI24, tank is a predicted six-transmembrane domain protein but contains no additional identifiable protein domains.
Figure 2.
tank regulates ethanol sensitivity. A, Diagram of the tank transcription unit. Black boxes represent coding regions and gray boxes represent untranslated regions. Shown are the insertion site of 4-12 and the regions targeted by the primary and secondary RNAi lines used. Results using the secondary line (RNAi-2) are reported in the text but not shown in the figures. B, Precise excision of tank4-12 restored normal ethanol sensitivity in males (one-way ANOVA followed by Newman–Keuls post tests, n = 8). C, Expression of both tank transcripts was reduced in the heads of tank4-12 male and females compared with their respective controls (unpaired t test, n = 4 replicates). ***p < 0.001.
Precise excision of the P element present in tank4-12 flies restored normal ethanol sensitivity, confirming that the phenotype is due to the P element insertion (Fig. 2B; F(2,21) = 22.6, p < 0.001 for effect of genotype, p > 0.05 comparing excision line to control). To verify that tank4-12 disrupts tank expression, we measured tank transcript levels in fly heads using qRT-PCR. tank4-12 males and females showed a strong reduction in levels of both tank transcripts compared with controls (t test, p < 0.001): tank-RA levels were nearly undetectable in the mutants, whereas tank-RB levels were reduced by ∼75% (Fig. 2C). These results indicate that tank4-12 is a loss-of-function mutation and suggest that tank normally acts in both sexes to promote sedation sensitivity.
tank functions in adult 4-12Gal4-expressing cells to regulate ethanol sensitivity
We next investigated whether tank function is necessary and sufficient to regulate ethanol sensitivity, focusing primarily on males for these experiments. First, we tested whether downregulation of tank promotes ethanol resistance, similar to the tank4-12 mutation. To downregulate tank, we used the Gal4/UAS system to express an RNAi transgene that targets both tank transcripts for degradation (tankRNAi). Ubiquitous expression of tankRNAi (tubGal4/UAS-tankRNAi) caused developmental lethality, so we restricted tankRNAi expression to adulthood by using Gal80ts, a temperature-sensitive Gal4 repressor that inhibits Gal4 function at 18° but not at 29° (McGuire et al., 2003). Adult-specific downregulation of tank in all cells (tubGal4,Gal80ts/UAS-tankRNAi) decreased the expression of tank-RA and tank-RB by 56% and 31%, respectively, and increased ethanol resistance (Fig. 3A; F(2,17) = 7.4, p = 0.005). This result confirms that tank is necessary to promote ethanol sensitivity, in accordance with the mutant data, and also indicates that tank is required during adulthood. Downregulation of tank exclusively in neurons (elavGal4/UAS-tankRNAi) also increased sedation resistance, indicating that tank functions in neurons to promote ethanol sensitivity (Fig. 3B; F(2,33) = 9.2, p < 0.001). Neuronal downregulation of tank did not significantly alter the internal ethanol concentration of flies, confirming that tank can regulate ethanol sensitivity independently of its effect on ethanol pharmacokinetics (Fig. 3C; F(2,9) = 11.8, p = 0.003 for effect of genotype, p > 0.05 comparing experimental and UAS-tankRNAi/+ flies).
Figure 3.
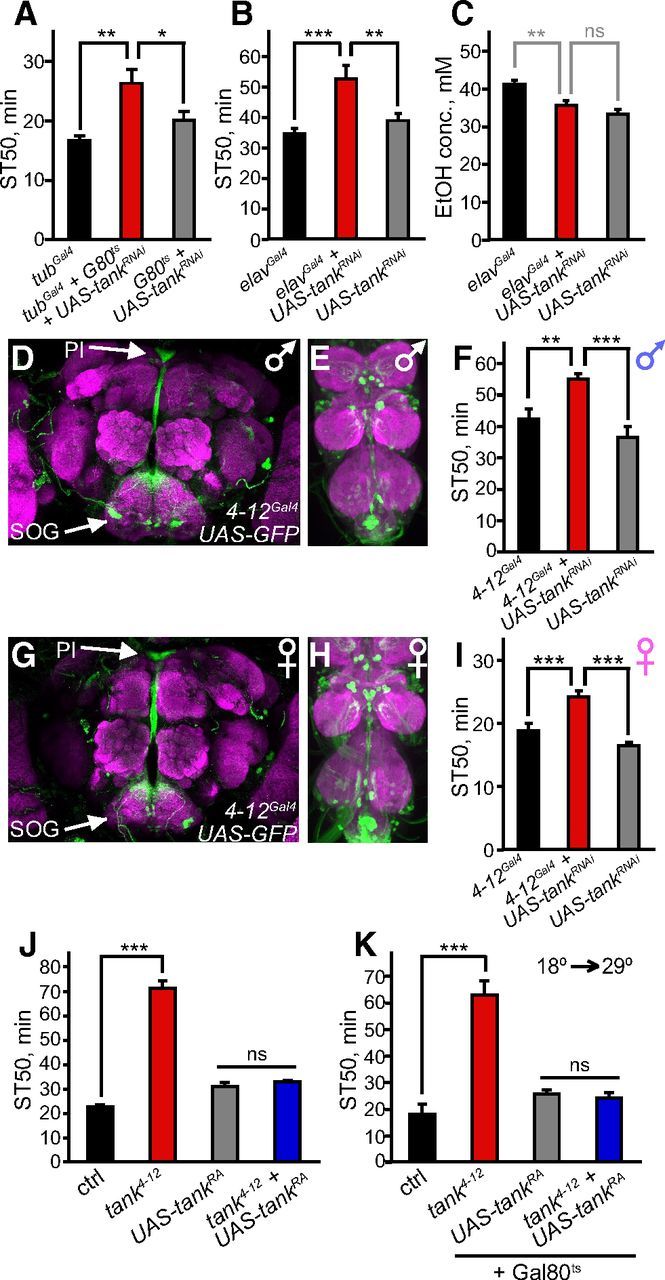
tank functions in adult 4-12Gal4-expressing neurons to regulate ethanol sensitivity. A, Adult-specific downregulation of tank in all cells (tubGal4,Gal80ts/UAS-tankRNAi) increased ethanol resistance in males (n = 6–8). Expression of tankRNAi was restricted to adulthood by growing flies at 18° (Gal80ts active) and shifting them to 29° during adulthood (Gal80ts inactive). B, Pan-neuronal downregulation of tank (elavGal4/UAS-tankRNAi) increased sedation resistance in males (n = 12). C, Pan-neuronal downregulation of tank in males (elavGal4/UAS-tankRNAi) had no significant effect on internal ethanol concentration after 15 min of exposure to 47% ethanol; experimental flies did not differ significantly from UAS-tankRNAi/+ control flies (n = 4). D, E, G, H, Visualization of the 4-12Gal4 expression pattern in the adult male brain (D), male VNC (E), female brain (G), and female VNC (H) using UAS-GFP (green), along with nc82 counterstaining of neuropil (magenta). Flies were heterozygous for 4-12Gal4. F, I, Downregulation of tank in 4-12Gal4-expressing cells (4-12Gal4/UAS-tankRNAi) increased ethanol resistance in males (F) and females (I) (n = 8–9). Flies were heterozygous for 4-12Gal4. J, The sedation resistance of tank4-12 mutant males was rescued by expression of tank-RA in 4-12Gal4-expressing cells (n = 8). tank4-12 homozygotes differed significantly from the background control, whereas no significant difference was observed between tank4-12;UAS-tankRA/+ rescue flies and UAS-tankRA/+ controls. K, The sedation resistance of tank4-12 mutant males was rescued by adult-specific expression of tank-RA in 4-12Gal4-expressing cells (n = 8). Flies were reared at 18° and shifted to 29° during adulthood to induce adult-specific tank-RA expression. A significant difference was observed between tank4-12;Gal80ts/+ and the background control, but not between tank4-12;UAS-tankRA/Gal80ts rescue flies and UAS-tankRA/Gal80ts controls. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA followed by Newman–Keuls post tests. Repeated-measures ANOVA was used in F and K.
Because tank acts in neurons to regulate ethanol sensitivity, we analyzed the likely expression pattern of tank within the CNS. The P element inserted in tank4-12 flies is a P{GawB} enhancer trap, in which Gal4 is expressed in cells likely to express endogenous tank. We visualized this expression pattern using GFP under Gal4 control (UAS-GFP). We observed GFP expression in a limited number of cells in the nervous system of adult males. Strong expression was observed in neurons within the PI that project ventrally to the subesophageal ganglion (SOG) (Fig. 3D). GFP expression was also observed in cell bodies in the SOG and lateral brain, very weakly in the mushroom body, and in scattered cells in the VNC (Fig. 3D,E). Downregulation of tank in 4-12Gal4-expressing cells (4-12Gal4/UAS-tankRNAi) increased ethanol resistance in males, confirming that tank is required in these cells to promote ethanol sensitivity (Fig. 3F; F(2,16) = 14.1, p < 0.001). We verified this result using an additional tankRNAi line (tankRNAi-2) targeted to a nonoverlapping region of the gene (Fig. 2A; data not shown; F(2,24) = 6.5, p = 0.006). The 4-12Gal4 expression pattern in females was similar to that in males (Fig. 3G,H), and downregulation of tank in 4-12Gal4-expressing cells of females increased ethanol resistance, as observed in males (Fig. 3I; F(2,21) = 18.5, p < 0.001).
To demonstrate conclusively that the ethanol resistance observed in tank4-12 mutants is due to decreased tank expression in 4-12Gal4-expressing cells, we tested whether this phenotype could be rescued by restoring tank expression within these cells. Indeed, expression of tank-RA in 4-12Gal4-expressing cells restored normal ethanol sensitivity to tank4-12 mutant males (Fig. 3J; F(3,28) = 149.1, p < 0.001 for effect of genotype, p > 0.05 comparing tank4–12;UAS-tankRA/+ rescue flies and UAS-tankRA/+ controls). Furthermore, using Gal80ts to express tank-RA in 4-12Gal4 cells exclusively during adulthood similarly restored normal ethanol sensitivity to tank4-12 mutant males (Fig. 3K; F(3,21) = 65.3, p < 0.001 for effect of genotype, p > 0.05 comparing tank4–12;UAS-tankRA/Gal80ts rescue flies and UAS-tankRA/Gal80ts controls). Overall, our results indicate that tank function in 4-12Gal4-expressing cells is both necessary and sufficient to regulate ethanol sensitivity, strongly suggesting that these cells express endogenous tank. Furthermore, tank function is required specifically during adulthood, implying that tank regulates some aspect of neuronal function rather than development.
tank-expressing neurons regulate ethanol sensitivity
Having shown that tank functions in the adult nervous system to promote ethanol sensitivity, and, based on its Gal4 marker, that it is expressed in a limited subset of neurons, we next investigated whether the activity of tank-expressing neurons acutely regulates ethanol sensitivity. To activate these neurons inducibly, we used the heat-activated cation channel TrpA1, which causes neuronal depolarization (Hamada et al., 2008). To silence tank neurons acutely, we used shits, a temperature-sensitive dynamin allele that depletes synaptic vesicles (Kitamoto, 2001). In preliminary experiments, we found that most genotypes tended to sedate more quickly at the high temperatures required for TrpA1 and shits experiments (27°–30°), showing steep sedation curves that made it difficult to obtain reliable ST50 values (data not shown). We therefore decreased the ethanol concentration from 67% to 60% for experiments conducted at >25° and statistically compared experimental and control flies tested within each condition, but not flies tested at different temperatures.
We first conducted activation and silencing experiments in males. We found that TrpA1 activation of 4-12Gal4 neurons at 30° increased ethanol sensitivity in males (Fig. 4A; F(2,24) = 20.8, p < 0.001 at 30°; F(2,14) = 3.0, p = 0.084 at 22°). Surprisingly, silencing 4-12Gal4 neurons in males using shits at 30° had the same effect, causing increased sedation sensitivity (Fig. 4B; F(2,21) = 41.9, p < 0.001 at 30°; F(2,14) = 1.1, p = 0.363 at 22°). These results indicate that the activity of 4-12Gal4 neurons acutely modulates ethanol sedation sensitivity, but does not do so in a straightforward manner. There are several possible explanations for these results (see Discussion), such as an inverted-U model in which a precise level of 4-12Gal4 neuron activity is required for normal ethanol resistance.
Figure 4.
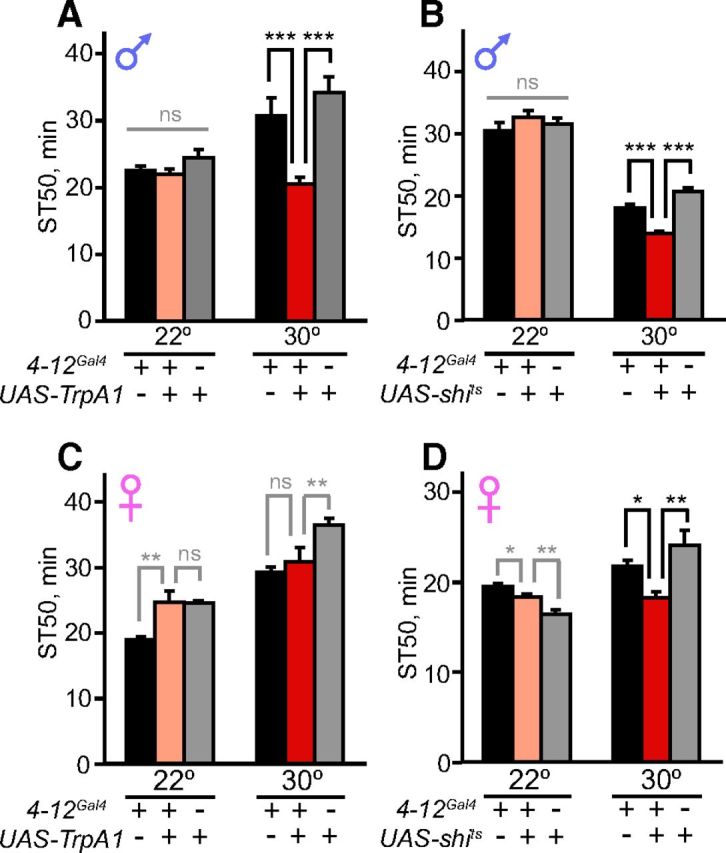
Acute manipulations of tank neuron activity alter ethanol sensitivity. A, Activation of 4-12Gal4 neurons (4-12Gal4/UAS-TrpA1) at 30° increased ethanol sensitivity in males (n = 13); no effect was observed at 22° when TrpA1 was inactive (n = 8). B, Silencing 4-12Gal4 neurons (4-12Gal4/UAS-shits) at 30° increased ethanol sensitivity in males, but no effect was observed at 22° when shits was inactive (n = 8). C, Activation of 4-12Gal4 neurons (4-12Gal4/UAS-Trp) at 30° did not affect ethanol sensitivity in females, nor was an effect observed at 22°; in each experiment, the ST50 of experimental flies did not differ from that of both controls (n = 8). D, Silencing 4-12Gal4 neurons (4-12Gal4/UAS-shits) at 30° increased ethanol sensitivity in females (n = 12). No effect was observed at 22° when shits was inactive; the ST50 of experimental flies was between the values of the two controls (n = 8). *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA followed by Newman–Keuls post tests. Repeated-measures ANOVA was used for all analyses except for the 30° experiments in B and D. In all experiments, flies were heterozygous for 4-12Gal4. For experiments conducted at 30°, the ethanol concentration was decreased to 60% instead of 67% because flies sedated more quickly at higher temperatures. Note that the magnitude of the temperature effect on sedation varied from experiment to experiment due to unknown factors.
We next investigated whether activation and silencing of tank-expressing neurons in females produced the same effects as in males. Interestingly, silencing 4-12Gal4 neurons using shits increased sedation sensitivity in females, as in males, but TrpA1 activation of these neurons in females had no significant effect (Fig. 4C,D; F(2,33) = 7.3, p = 0.002 for shits at 30°; F(2,14) = 19.7, p < 0.001 for shits at 22°; F(2,14) = 9.9, p = 0.002 for TrpA1 at 30°, p > 0.05 comparing experimental and 4-12Gal4/+ flies; F(2,14) = 11.2, p = 0.001 for TrpA1 at 22°, p > 0.05 comparing experimental and UAS-TrpA1/+ flies). Overall, our silencing experiments suggest that endogenous activity of tank neurons is required for normal sedation sensitivity in both males and females. However, the effect of activating tank neurons appears to be male-specific, suggesting the presence of sexual dimorphism in either these neurons themselves or in the downstream circuit. Because we observed no obvious sex differences in the number or morphology of 4-12Gal4 neurons (Fig. 3), potential dimorphisms in these neurons likely arise from their functional properties or fine connectivity.
tank functions in PI neurons projecting to the SOG
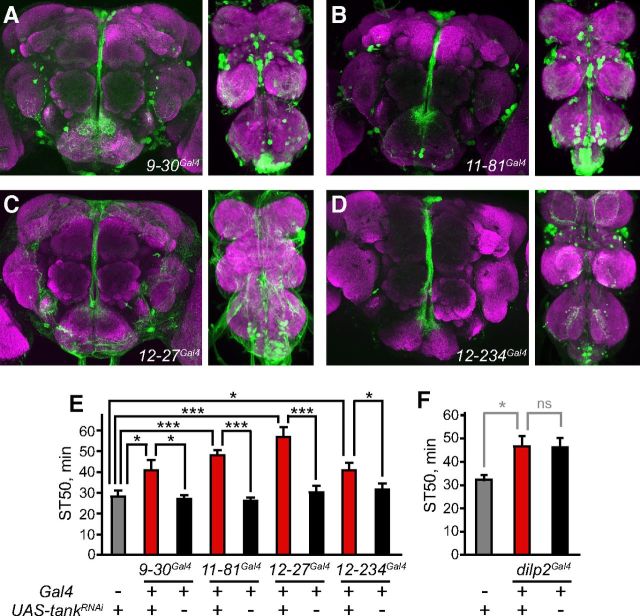
Our results indicate that tank function in 4-12Gal4-expressing neurons is necessary and sufficient to regulate ethanol sensitivity, and that the activity of these neurons acutely affects ethanol sedation. Although the 4-12Gal4 expression pattern is fairly limited within the nervous system, we wished to define more precisely the neurons in which tank functions. Because 4-12Gal4 expression appears most prominent in PI neurons projecting to the SOG, we asked whether tank is specifically required in these cells to promote ethanol sensitivity. We selectively downregulated tank in PI neurons of males by combining UAS-tankRNAi with several Gal4 lines that drive expression primarily in the PI (Fig. 5A–D). Downregulation of tank using four of seven PI-expressing Gal4 lines increased ethanol resistance, strongly suggesting that tank functions within the PI to promote ethanol sensitivity (Fig. 5E; F(2,21) = 4.9, p = 0.018; F(2,21) = 24.2, p < 0.001; F(2,21) = 17.6, p < 0.001; and F(2,33) = 4.2, p = 0.023 using 9-30Gal4, 11-81Gal4, 12-27Gal4 and 12-234Gal4, respectively). 4-12Gal4 also showed weak expression in the mushroom body (visible in individual sections, though not obvious in the full brain stacks shown in Fig. 3D, G). However, downregulation of tank using two different Gal4 lines expressed broadly in the mushroom body (OK107 and c747) did not affect ethanol sensitivity (data not shown). Because the Gal4 lines used to target the PI also showed some expression in the VNC (Fig. 5A–D), we cannot entirely rule out the possibility that tank is required in VNC neurons. However, given that the four Gal4 lines that produced a phenotype with tankRNAi showed varying expression patterns in the VNC (Fig. 5A–D), some of which were fairly sparse (e.g., 12-234Gal4), it seems unlikely that all four of these lines are coexpressed in a set of VNC cells that also express tank, and more likely that the sedation resistance results from loss of tank in the PI.
Figure 5.
tank functions in PI neurons projecting to the SOG. A–D, Expression patterns of the Gal4 lines used to drive tankRNAi expression in the PI were visualized in the adult male brain (left) and VNC (right) with UAS-GFP (green), along with nc82 counterstaining (magenta). E, Using the Gal4 lines shown in A–D, downregulation of tank in the PI increased ethanol resistance in males (n = 8–12). F, Downregulation of tank in insulin-producing cells using dilp2Gal4 had no significant effect on ethanol sedation in males; ST50 of experimental flies did not differ significantly from that of the dilpGal4/+ control (n = 8). *p < 0.05, ***p < 0.001, one-way ANOVA followed by Newman–Keuls post tests.
The fact that some PI-expressing Gal4 lines did not affect ethanol sensitivity when driving UAS-tankRNAi suggests that tank may function in a specific subset of neurons in the PI, a heterogenous region containing several classes of neuropeptidergic cells (Park et al., 2008). In particular, insulin-producing cells in the PI have been previously implicated in regulating ethanol sensitivity (Corl et al., 2005). However, RNAi downregulation of tank in insulin-producing cells using dilp2Gal4 (Rulifson et al., 2002) did not affect ethanol sensitivity (Fig. 5F; F(2,21) = 5.0, p = 0.017 for the effect of genotype, p > 0.05 comparing experimental and dilp2Gal4/+ flies). Similarly negative results were observed upon downregulation of tank in PI cells expressing any of three other classes of neuropeptides (SIFamide, dromyosuppressin, or drosulfakinin; data not shown). tank is therefore likely to function in a different subset of PI neurons that we did not test.
Having shown that the activity of tank-expressing neurons affects ethanol sensitivity (Fig. 4) and that tank functions in PI neurons to regulate this behavior (Fig. 5), we next investigated whether specifically manipulating the activity of PI neurons alters ethanol sedation. We activated PI neurons by driving TrpA1 expression with two of the PI-expressing Gal4 lines (9-30Gal4 and 11-81Gal4) that affected sedation when driving UAS-tankRNAi (Fig. 5E) and therefore label PI neurons in which tank functions. Interestingly, activation of these PI neurons increased sedation sensitivity in males, but not females, mimicking 4-12Gal4 neuron activation (Fig. 6A–D; for 9-30Gal4 and 11-81Gal4, respectively: F(2,13) = 30.1, p < 0.001 and F(2,13) = 61.8, p < 0.001 for males at 27°; F(2,14) = 46.0, p < 0.001 and F(2,14) = 37.0, p < 0.001 for males at 22°; F(2,21) = 99.8, p < 0.001 and F(2,21) = 34.4, p < 0.001 for females at 30°; F(2,14) = 44.8, p < 0.001 and F(2,14) = 18.9, p < 0.001 for females at 22°). These results suggest that tank functions in a subset of PI neurons, and the activity of these neurons regulates ethanol sensitivity in a sexually dimorphic manner.
Figure 6.
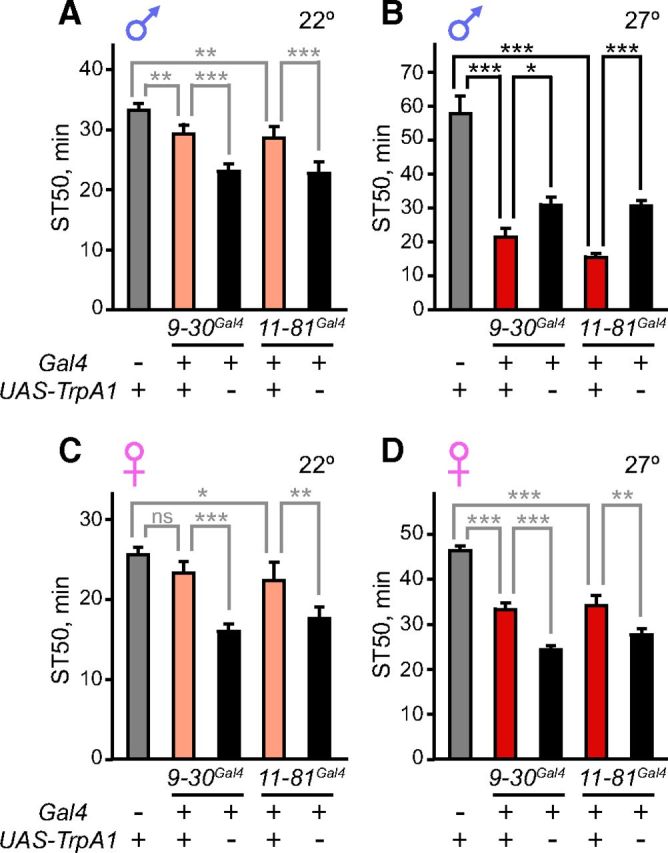
Activation of PI neurons enhances ethanol sensitivity in males. A, TrpA1 expression in PI neurons of males using either 9-30Gal4 or 11-81Gal4 did not affect ethanol sensitivity at 22° when TrpA1 was inactive; ST50 values of experimental flies were between those of the two controls (n = 8). B, TrpA1 activation of PI neurons at 27° using either 9-30Gal4 or 11-81Gal4 increased ethanol sensitivity in males (n = 4–6). C, D, TrpA1 expression in PI neurons of females using either 9-30Gal4 or 11-81Gal4 did not affect ethanol sensitivity at 22° (C) or 27° D); ST50 values of experimental flies were between those of the two controls (n = 8). *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA followed by Newman–Keuls post tests. Repeated-measures ANOVA was used in A and C. For experiments conducted at 27°, the ethanol concentration was decreased to 60% instead of 67%, as explained in the text.
tank- and fru-expressing neurons form likely synaptic connections
Our results suggest that although the endogenous activity of tank-expressing neurons is required for normal ethanol sensitivity in both sexes, tank neurons also show a sex-specific function; their activation enhances ethanol sedation in males, but not females. This sex-specific effect is likely attributable to tank-expressing neurons in the PI, because activating PI neurons in which tank functions produces a similar male-specific effect on sedation. Most sexually dimorphic behavior in flies is mediated by the gene fru, which encodes male-specific protein due to sex-specific splicing (Ryner et al., 1996; Demir and Dickson, 2005; Vrontou et al., 2006). fru has been recently implicated in the regulation of ethanol sedation sensitivity, and activation of fru-expressing neurons in males enhances ethanol sensitivity, similar to the effect of activating tank neurons (Devineni and Heberlein, 2012). We therefore hypothesized that the male-specific effect of tank neuron activation could be due to sexual dimorphisms in these neurons generated by fru, especially given that fru is strongly expressed in the PI (Manoli et al., 2005). To address this possibility, we tested whether fru is expressed in tank-expressing neurons. Although markers for both tank (4-12Gal4) and fru (frulex) were strongly expressed in the PI, we did not observe any cells coexpressing both markers (Fig. 7A–C). Within the median bundle, which contains projections from the PI to the SOG, 4-12Gal4-positive and fru-positive projections segregated to medial versus lateral fibers, respectively.
Figure 7.
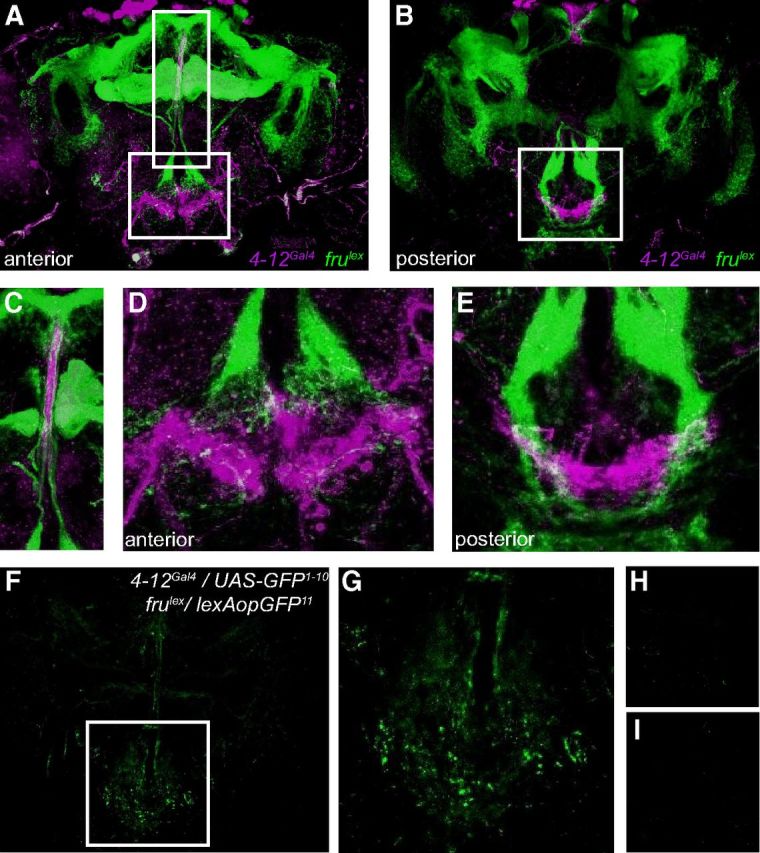
tank- and fru-expressing neurons form likely synaptic connections. A–C, Expression of 4-12Gal4 (magenta) and frulex (green) was visualized simultaneously in the adult male brain using UAS-RFP and lexAop-GFP, respectively. No coexpression was observed in either the anterior (A) or posterior (B) brain. A magnified view of the median bundle in A is depicted in C, showing 4-12Gal4 and frulex markers segregating to different fibers. D, E, Magnified views of the SOG from A and B showing that projections of 4-12Gal4- and frulex-expressing neurons were highly intermingled in the anterior (D) and posterior (E) SOG. F, G, GRASP experiment using male 4-12Gal4/+;frulex/UAS-GFP1-10,lexAop-GFP11 flies. Many GFP puncta were observed, almost all of which were localized to the SOG (magnified in G). H, I, GFP puncta were not observed in negative control flies expressing either of the individual GFP halves: 4-12Gal4/+;UAS-GFP1-10,lexAop-GFP11 (H) or frulex/UAS-GFP1-10,lexAop-GFP11 (I). Brains were imaged at equal or greater gain as used in F.
We corroborated this lack of coexpression using an intersectional FLP-out method. We generated males carrying 4-12Gal4, fruFLP, and UAS>STOP>GFP transgenes. In these flies, GFP is expressed only in cells expressing both fru and 4-12Gal4: an FRT-flanked transcriptional stop cassette is excised by FLP recombinase (Golic and Lindquist, 1989) only in fru-expressing cells, producing UAS-GFP, which can only be expressed in 4-12Gal4-expressing cells. We observed no GFP expression in these flies, although GFP was observed in positive control flies expressing the pan-neuronal marker elavGal4 in place of 4-12Gal4 (data not shown). These results confirm that the male-specific phenotype caused by activating 4-12Gal4 neurons is not due to expression of fru in these cells.
Although fru was not expressed in tank-expressing cells, we noticed that the projections of tank and fru neurons appeared highly intermingled in the SOG, suggesting that they might form synaptic connections (Fig. 7D,E). To test this possibility more directly, we used GRASP (GFP Reconstitution Across Synaptic Partners; Feinberg et al., 2008; Gordon and Scott, 2009). This method relies on expression of two halves of GFP on the membrane of distinct sets of neurons such that full-length GFP is reconstituted at sites of cell contact, including synapses. GRASP between 4-12Gal4- and frulex-expressing neurons revealed a large number of GFP puncta almost exclusively localized to the SOG (Fig. 7F,G). These puncta were not present in control flies expressing either of the individual halves of GFP (Fig. 7H,I). Although we cannot rule out the possibility that the GFP signal represents nonsynaptic cell contact, our results suggest that subsets of tank- and fru-expressing neurons likely form synaptic connections in the SOG. Because most of the projections of 4-12Gal4 neurons within the SOG appear to arise from the PI (Fig. 4), it seems likely that tank-expressing PI neurons, which regulate ethanol sensitivity (Fig. 5, Fig. 6), represent the cells that interact anatomically with fru neurons. Overall, these results raise the possibility that the male-specific effect of tank neuron activation may result from sexual dimorphisms in fru neurons acting postsynaptically, since many fru neurons show sex differences in their number and morphology (Cachero et al., 2010; Yu et al., 2010).
Discussion
In this study, we characterize the novel gene tank, a tumor suppressor homolog that promotes ethanol sensitivity in Drosophila. We identify a loss-of-function mutation of tank, 4-12, that causes strong resistance to ethanol sedation, a phenotype attributable to both pharmacokinetic and pharmacodynamic processes. We show that tank acts within the adult nervous system to promote ethanol sensitivity. Neuronal downregulation of tank increases sedation resistance without altering internal ethanol levels, suggesting that the pharmacokinetic effect of tank arises outside of the nervous system. Within the CNS, tank is expressed in a limited set of neurons, and we localize its function in regulating ethanol sedation to cells within the PI. Inducible manipulations of neuronal activity revealed that these neurons acutely regulate ethanol sensitivity, and do so in a sexually dimorphic manner: activation of all tank neurons, or of PI neurons in particular, increases sedation sensitivity in males, but not females. We suggest that the male-specific effect of tank neuron activation may be due to sexual dimorphisms in downstream fru neurons, since tank and fru neurons form likely synaptic connections, and fru neurons also regulate ethanol sensitivity (Devineni and Heberlein, 2012). In summary, we have characterized a novel gene that regulates ethanol sensitivity, identified a role for tank-expressing neurons in modulating ethanol sedation, and localized both of these effects to a subset of PI neurons not implicated previously in ethanol behaviors.
Our data suggest that tank neurons regulate ethanol sedation in a complex, nonlinear manner; either activation or silencing of 4-12Gal4 neurons enhanced ethanol sensitivity in males. Several possible explanations could account for these counterintuitive results. For example, a certain level of neuronal activity or a specific temporal pattern of activity could be required for normal sedation resistance. In both cases, either silencing neurotransmission or artificially driving activity could disrupt neuronal function and result in increased ethanol sensitivity. Alternatively, feedback within a neural circuit might oppose the effect of transgenic manipulations of neural activity, causing opposite manipulations to in fact have the same effect on some part of the circuit. Finally, it is possible that some 4-12Gal4 neurons promote sedation sensitivity whereas others promote sedation resistance, so manipulating all of these neurons simultaneously might cause a nonlinear effect depending on the relationships between these neurons.
Interestingly, the increased ethanol sensitivity induced by activating tank neurons was male-specific. This result is surprising, given that the effect of silencing tank neurons was not sex-specific, nor is the function of tank itself. These data suggest a model in which the normal function of tank and the endogenous activity of tank neurons (or a subset thereof) are necessary for normal ethanol sensitivity in both sexes; however, driving high levels of activity in these neurons reveals a male-specific sedation phenotype due to sex differences either within or downstream of tank neurons. Because we observed no obvious sex differences in the number or morphology of 4-12Gal4-expressing neurons, the male-specific phenotype could result from sex differences in the physiological properties of tank neurons or from anatomical or functional dimorphisms in the downstream neural circuit. fru, a key regulator of sexually dimorphic neuronal differentiation that has also been implicated in regulating ethanol sensitivity (Devineni and Heberlein, 2012), is a good candidate for mediating these dimorphisms. We showed that although tank neurons do not express fru, they form likely synaptic connections with fru-expressing neurons. Therefore, one plausible model is that the male-specific effect of tank neuron activation arises from sex differences in downstream fru neurons, because many fru neurons are sexually dimorphic (Cachero et al., 2010; Yu et al., 2010) and activating fru neurons in males enhances ethanol sensitivity (Devineni and Heberlein, 2012). Future studies will be needed to identify the specific tank and fru neurons showing an anatomical interaction and to confirm whether they show a functional interaction in regulating ethanol sensitivity.
We have localized tank function to a subset of neurons in the PI, the activity of which modulates ethanol sensitivity in males. The PI is a major locus of peptidergic neurons (Park et al., 2008). Neuropeptides modulate neuronal activity or other cellular properties and have been linked to many types of behavior, including ethanol responses (Corl et al., 2005; Wen et al., 2005; Nässel and Winther, 2010). We observed that downregulating tank in four different classes of peptidergic cells within the PI, including insulin-producing cells that were implicated previously in ethanol sensitivity (Corl et al., 2005), did not affect ethanol sedation. Although one possibility is that the strength of the downregulation was not sufficient to produce a phenotype, it is likely that tank functions in an unidentified class of PI neurons that has not been implicated previously in regulating ethanol sensitivity. It will be interesting to characterize the identity of these neurons and to determine whether they are defined by expression of a particular neuropeptide or by a different molecular or cellular feature.
The mammalian homolog of tank, EI24, encodes a transcriptional target of p53 that induces apoptosis in response to DNA damage (Gu et al., 2000; Mork et al., 2007). EI24 is a clinically important tumor suppressor; its inactivation has been associated with breast and cervical cancers (Gentile et al., 2001; Zhao et al., 2005; Mazumder Indra et al., 2011). It is interesting that several cancer-related genes have been identified in Drosophila as regulators of ethanol sensitivity. For example, the oncogenes EGFR and Anaplastic lymphoma kinase (Alk) regulate ethanol sensitivity in both flies and mammals (Morris et al., 1994; Pedersen et al., 2001; Corl et al., 2009; Lasek et al., 2011). In addition, the oncogenic PI3K/Akt pathway promotes ethanol sensitivity in flies, whereas PTEN, a tumor suppressor and negative regulator of the pathway, promotes ethanol resistance (Fresno Vara et al., 2004; Eddison et al., 2011). The implication that overlapping molecular pathways regulate oncogenesis and acute ethanol sensitivity, two processes that occur on very different timescales, is somewhat surprising. It will be interesting to determine whether these pathways regulate both processes through the same cellular mechanisms. For example, the EGFR and PI3K/Akt pathways regulate cytoskeletal dynamics, which play an important role in both ethanol sensitivity and oncogenesis (Rodriguez-Viciana et al., 1997; Wells et al., 1998; Hall, 2009; Rodan and Rothenfluh, 2010).
Future studies will be needed to determine whether, like EI24, tank regulates apoptosis and functions downstream of p53. Although we did not directly investigate the role of tank in apoptosis, we observed that tank4-12 mutants are resistant to antennal blackening induced by ethanol toxicity, a process that involves apoptosis of olfactory neurons (French and Heberlein, 2009), and this resistance was not dependent on altered ethanol pharmacokinetics (data not shown). These results suggest that tank is required for ethanol-induced apoptosis and may have a more general apoptotic function as well. In addition to its role in apoptosis, EI24 also regulates autophagy, a catabolic process involving lysosomal degradation of the cytosol (Tian et al., 2010; Zhao et al., 2012), representing another potential function for tank. In considering possible cellular functions of tank, it is important to note our data showing that adult tank function is necessary and sufficient to regulate ethanol sensitivity, indicating that although tank may participate in developmental processes, its regulation of ethanol sensitivity derives from an adult-specific effect on neuronal function. Given that the activity of tank neurons regulates ethanol sedation, another possible cellular function of tank is the regulation of neuronal activity (either direct or indirect). Physiological recordings from control and tank mutant flies could help to address this possibility.
In addition to determining whether tank shares the molecular and cellular functions related to EI24, it will be important to ascertain whether EI24 shares the behavioral function of tank in regulating ethanol sensitivity. This task may be facilitated by the recent development of a mouse line bearing a conditional deletion of EI24, which could be tested for ethanol-related behaviors (Zhao et al., 2012). Constitutive deletion of EI24 in the nervous system causes developmental abnormalities and motor deficits (Zhao et al., 2012), suggesting that an adult-specific nervous system deletion of EI24 may be required to test its role in regulating ethanol responses. Because sensitivity to the sedative effects of ethanol is inversely correlated with susceptibility to AUDs (Schuckit, 1994), a role for EI24 in regulating acute ethanol sensitivity could have clinical relevance for predicting or even treating these disorders.
Footnotes
This work was supported by the National Science Foundation (predoctoral fellowship to A.V.D) and the National Institute on Alcohol Abuse and Alcoholism–National Institutes of Health (grant to U.H.). We thank Bruce Baker, Barry Dickson, Paul Taghert, Devanand Manoli, the Bloomington Stock Center, and the Vienna Drosophila RNAi Center for providing fly strains, Melanie Raymundo for assistance with fly husbandry, Amy Lasek for designing the qPCR primers, and all members of the Heberlein laboratory for advice and helpful discussions regarding this project.
The authors declare no competing financial interests.
References
- Babor TF, Berglas S, Mendelson JH, Ellingboe J, Miller K. Alcohol, affect, and the disinhibition of verbal behavior. Psychopharmacology (Berl) 1983;80:53–60. doi: 10.1007/BF00427496. [DOI] [PubMed] [Google Scholar]
- Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GS. Sexual dimorphism in the fly brain. Curr Biol. 2010;20:1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Corl AB, Rodan AR, Heberlein U. Insulin signaling in the nervous system regulates ethanol intoxication in Drosophila melanogaster. Nat Neurosci. 2005;8:18–19. doi: 10.1038/nn1363. [DOI] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Devineni AV, Heberlein U. Acute ethanol responses in Drosophila are sexually dimorphic. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1218850110. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devineni AV, McClure KD, Guarnieri DJ, Corl AB, Wolf FW, Eddison M, Heberlein U. The genetic relationships between ethanol preference, acute ethanol sensitivity, and ethanol tolerance in Drosophila melanogaster. Fly (Austin) 2011;5:191–199. doi: 10.4161/fly.5.3.16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddison M, Guarnieri DJ, Cheng L, Liu CH, Moffat KG, Davis G, Heberlein U. arouser reveals a role for synapse number in the regulation of ethanol sensitivity. Neuron. 2011;70:979–990. doi: 10.1016/j.neuron.2011.03.030. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- French RL, Heberlein U. Glycogen synthase kinase-3/Shaggy mediates ethanol-induced excitotoxic cell death of Drosophila olfactory neurons. Proc Natl Acad Sci U S A. 2009;106:20924–20929. doi: 10.1073/pnas.0910813106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30:193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Gentile M, Ahnström M, Schön F, Wingren S. Candidate tumour suppressor genes at 11q23–q24 in breast cancer: evidence of alterations in PIG8, a gene involved in p53-induced apoptosis. Oncogene. 2001;20:7753–7760. doi: 10.1038/sj.onc.1204993. [DOI] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gilbert DJ, Valentine VA, Jenkins NA, Copeland NG, Zambetti GP. The p53-inducible gene EI24/PIG8 localizes to human chromosome 11q23 and the proximal region of mouse chromosome 9. Cytogenet Cell Genet. 2000;89:230–233. doi: 10.1159/000015620. [DOI] [PubMed] [Google Scholar]
- Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CW, Mehmert KK, Kelley SP, McMahon T, Haywood A, Olive MF, Wang D, Sanchez-Perez AM, Messing RO. Supersensitivity to allosteric GABA(A) receptor modulators and alcohol in mice lacking PKCepsilon. Nat Neurosci. 1999;2:997–1002. doi: 10.1038/14795. [DOI] [PubMed] [Google Scholar]
- Joseph RM, Devineni AV, King IF, Heberlein U. Oviposition preference for and positional avoidance of acetic acid provide a model for competing behavioral drives in Drosophila. Proc Natl Acad Sci U S A. 2009;106:11352–11357. doi: 10.1073/pnas.0901419106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun KR, Devineni AV, Heberlein U. Drosophila melanogaster as a model to study drug addiction. Hum Genet. 2012;131:959–975. doi: 10.1007/s00439-012-1146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Lim J, Kliethermes CL, Berger KH, Joslyn G, Brush G, Xue L, Robertson M, Moore MS, Vranizan K, Morris SW, Schuckit MA, White RL, Heberlein U. An evolutionary conserved role for anaplastic lymphoma kinase in behavioral responses to ethanol. PLoS One. 2011;6:e22636. doi: 10.1371/journal.pone.0022636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br J Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder Indra D, Mitra S, Singh RK, Dutta S, Roy A, Mondal RK, Basu PS, Roychoudhury S, Panda CK. Inactivation of CHEK1 and EI24 are associated with the development of invasive cervical carcinoma: Clinical and prognostic implications. Int J Cancer. 2011;129:1859–1871. doi: 10.1002/ijc.25849. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mellert DJ, Knapp JM, Manoli DS, Meissner GW, Baker BS. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development. 2010;137:323–332. doi: 10.1242/dev.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Weafer J, Fillmore MT. Gender differences in alcohol impairment of simulated driving performance and driving-related skills. Alcohol Alcohol. 2009;44:586–593. doi: 10.1093/alcalc/agp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/S0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- Morean ME, Corbin WR. Subjective response to alcohol: a critical review of the literature. Alcohol Clin Exp Res. 2010;34:385–395. doi: 10.1111/j.1530-0277.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- Mork CN, Faller DV, Spanjaard RA. Loss of putative tumor suppressor EI24/PIG8 confers resistance to etoposide. FEBS Lett. 2007;581:5440–5444. doi: 10.1016/j.febslet.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Nässel DR, Winther AM. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol. 2010;92:42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Park D, Veenstra JA, Park JH, Taghert PH. Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS One. 2008;3:e1896. doi: 10.1371/journal.pone.0001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–760. doi: 10.1023/A:1011177318162. [DOI] [PubMed] [Google Scholar]
- Pohorecky LA. Biphasic action of ethanol. Biobehav Rev. 1977;1:231–240. doi: 10.1016/0147-7552(77)90025-0. [DOI] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan AR, Rothenfluh A. The genetics of behavioral alcohol responses in Drosophila. Int Rev Neurobiol. 2010;91:25–51. doi: 10.1016/S0074-7742(10)91002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/S0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LT, Lasek AW, Heberlein U. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127:199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/S0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Terhzaz S, Rosay P, Goodwin SF, Veenstra JA. The neuropeptide SIFamide modulates sexual behavior in Drosophila. Biochem Biophys Res Commun. 2007;352:305–310. doi: 10.1016/j.bbrc.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396:366–369. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Willis B, Stadler J, Reynolds JG, Bernstein IL, McKnight GS. High ethanol consumption and low sensitivity to ethanol-induced sedation in protein kinase A-mutant mice. J Neurosci. 2000;20:RC75. doi: 10.1523/JNEUROSCI.20-10-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, Wang X, Kovács AL, Yu L, Zhang H. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Tsai LT, Bainton RJ, Blau J, Heberlein U. Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol. 2004;2:e408. doi: 10.1371/journal.pbio.0020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila Homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. J Neurosci. 2007;27:4541–4551. doi: 10.1523/JNEUROSCI.0305-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrontou E, Nilsen SP, Demir E, Kravitz EA, Dickson BJ. fruitless regulates aggression and dominance in Drosophila. Nat Neurosci. 2006;9:1469–1471. doi: 10.1038/nn1809. [DOI] [PubMed] [Google Scholar]
- Wells A, Gupta K, Chang P, Swindle S, Glading A, Shiraha H. Epidermal growth factor receptor-mediated motility in fibroblasts. Microsc Res Tech. 1998;43:395–411. doi: 10.1002/(SICI)1097-0029(19981201)43:5<395::AID-JEMT6>3.0.CO%3B2-T. [DOI] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Kanai MI, Demir E, Jefferis GS, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ayer RE, Davis SL, Ames SJ, Florence B, Torchinsky C, Liou JS, Shen L, Spanjaard RA. Apoptosis factor EI24/PIG8 is a novel endoplasmic reticulum-localized Bcl-2-binding protein which is associated with suppression of breast cancer invasiveness. Cancer Res. 2005;65:2125–2129. doi: 10.1158/0008-5472.CAN-04-3377. [DOI] [PubMed] [Google Scholar]
- Zhao YG, Zhao H, Miao L, Wang L, Sun F, Zhang H. The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J Biol Chem. 2012;287:42053–42063. doi: 10.1074/jbc.M112.415968. [DOI] [PMC free article] [PubMed] [Google Scholar]