Abstract
The temporal and spatial patterning involved in the specification, differentiation, and myelination by oligodendroglia is coordinated in part by the activation and repression of various transcriptional programs. Olig2 is a basic helix-loop-helix transcription factor necessary for oligodendroglial development and expressed continuously throughout the lineage. Despite evidence for the critical role of Olig2 in oligodendroglial specification and differentiation, the function for Olig2 during later stages of oligodendroglial development, namely, the transition into mature oligodendrocytes (OLs) and the formation of the myelin sheath, remains unclear. To address the possibility for a stage-specific role, we deleted Olig2 in oligodendrocyte precursor cells (OPCs) under the control of the CNPase-promoter or in immature OLs under the inducible proteolipid protein promoter. As expected, ablation of Olig2 in OPCs significantly inhibits differentiation, resulting in hypomyelination. However, deletion of the Olig2 gene in immature OLs significantly enhances the maturation process and accelerates the kinetics of myelination/remyelination. Underlying the stage-specific roles for Olig2 is the compensatory expression and function of Olig1, a transcription factor that promotes OL maturation and (re)myelination. Olig1 expression is significantly reduced upon Olig2 deletion in OPCs but is dramatically increased by nearly threefold when deleted in immature OLs. By enforcing expression of Olig1 into OPCs in a null Olig2 background, we demonstrate that overexpression of Olig1 is sufficient to rescue the differentiation phenotype and partially compensates for the Olig2 deletion in vitro. Our results suggest a stage-specific regulatory role for Olig2, mediated by Olig1 that conveys opposing functions on the differentiation and maturation of oligodendrocytes.
Introduction
Before myelination in the CNS, the oligodendrocyte precursor cell (OPC) exits from the cell cycle and initiates a transcriptional program resulting in the differentiation into the oligodendrocyte (OL). Newly generated OLs receive cues from their surroundings and concomitantly extend and elaborate processes, ensheath axons, and initiate myelin gene expression (Baumann and Pham-Dinh, 2001; Liu et al., 2003). These processes are orchestrated in part by the activation and repression of various transcriptional programs (Emery, 2010; Fancy et al., 2011). Olig1/2, two close related basic helix-loop-helix transcription factors, have been identified as essential factors in the specification of oligodendroglia (Lu et al., 2000, 2002; Takebayashi et al., 2000; Park et al., 2002) and in the differentiation, maturation and myelination/remyelination by OLs (Arnett et al., 2004; Xin et al., 2005). Olig2-null mice fail to develop cells of the oligodendroglial lineage (Lu et al., 2002; Takebayashi et al., 2002), whereas overexpression of Olig2 in neural stem cells (NSCs) is sufficient to specify OPCs rather than neurons and/or astrocytes, resulting in a dramatic increase in the number of OPCs (Copray et al., 2006; Maire et al., 2009; Kim et al., 2011; Hu et al., 2012). Moreover, conditional deletion of Olig2 at the multipotent progenitor stage in the cerebral cortex results in normal patterning of OPCs but a decrease in myelin gene expression and hypomyelination (Yue et al., 2006), suggesting that Olig2 function may not be required for the specification of cortical progenitors into OPCs but for the differentiation of OPCs into OLs. The findings, combined with the persistent expression of Olig2 throughout oligodendroglial development, suggest that Olig2 may function at multiple stages of the OL lineage. However, transplantation of Olig2-overexpressing NSCs into demyelinating lesions significantly enhances the generation of oligodendrogila, with only modest effects on remyelination (Copray et al., 2006; Maire et al., 2009; Kim et al., 2011; Hu et al., 2012). These findings suggest that overexpression of Olig2 may not facilitate the terminal maturation and myelination by OLs. Is it possible that Olig2 displays stage-specific roles during OL development, conveying opposing functions on differentiation and myelination?
To address these questions, the Olig2 gene is conditionally ablated in OPCs under the control of the CNPase-promoter or in newly generated OLs under the inducible proteolipid protein (PLP) promoter. As expected, ablation of Olig2 in OPCs (CNPase-Cre) significantly inhibits differentiation resulting in hypomyelination. However, deletion of the Olig2 gene in newly generated OLs (PLP-Cre) significantly enhances the maturation process and accelerates the kinetics of myelination/remyelination. To gain insight into the possible mechanisms underlying the stage-specific role of Olig2, we examined the changes in the expression of transcription factors associated with the state-specific deletion of Olig2. We find that Olig1 expression is significantly reduced upon Olig2 deletion in OPCs but is dramatically increased by nearly threefold when deleted in mature OLs. By enforcing expression of Olig1 into OPCs in a null Olig2 background, we demonstrate that overexpression of Olig1 is sufficient to rescue the differentiation phenotype and partially compensates for the Olig2 deletion in vitro. These results suggest a stage-specific regulatory role for Olig2, mediated by Olig1 that conveys opposing functions on the differentiation and maturation of oligodendrocytes.
Materials and Methods
Transgenic mice.
All mice examined in this study were conducted with the approval of the Third Military Medical University Administrative Panel on Laboratory Animal Care. Olig2 flox/flox mice have been described previously (Yue et al., 2006); where the coding region of Olig2 is flanked by LoxP sites. CNPcre/+ mice have been previously described (Cai et al., 2007); where Cre recombinase was knocked into one allele of the CNP gene locus. The PLP-CreER mice, which were also previously described (Doerflinger et al., 2003), CreER(T) expression was placed under the transcriptional control of the regulatory sequences of the myelin proteolipid protein (PLP) gene. For constitutive ablation of Olig2 within the oligodendroglial lineage Olig2flox/flox mice were bred to CNPCre/+ to generate both CNP-Cre; Olig2flox/flox (Olig2cnp-cko) and Olig2 flox/flox (Olig2+/+) littermate offspring. For inducible ablation of Olig2 in immature oligodendrocytes Olig2flox/flox mice were bred to PLP-CreER(T) to generate PLP-CreER(T); Olig2flox/flox (Olig2plp-iko) mice. The offspring were genotyped by PCR for CreER(T), CNPase-Cre, and floxed Olig2 alleles. To induce recombination, the Olig2plp-iko mice were given intraperitoneal injections of tamoxifen (Sigma, 50 mg/kg per day) for 5 d starting from P5.
Cuprizone demyelination.
Demyelination was induced by feeding 6-week-old male mice (Olig2+/+ and Olig2plp-iko) a diet containing 0.2% cuprizone (bis-cyclohexanone oxaldihydrazone; Sigma-Aldrich) mixed into a ground standard rodent chow. Cuprizone feeding was maintained for 6 weeks; thereafter, mice were put on a normal chow for another week with tamoxifen treatment.
OPC purification.
OPCs were purified from P8–P9 mouse cortices by immunopanning as previously described (Chan et al., 2004). Briefly, Petri dishes were incubated with goat anti-mouse IgG + IgM secondary antibody solution (Jackson ImmunoResearch Laboratories) and primary antibody solutions containing Ran-2, GalC, PDGFRα, or O4. Mouse brain cerebral hemispheres were diced and dissociated with papain (Worthington) at 37°C for 75 min. After trituration, cells were resuspended in a panning buffer and then incubated at room temperature sequentially on three immunopanning dishes: Ran-2, GalC, and PDGFRα or O4. PDGFRα-positive mouse OPCs or O4 immature OLs were released from the final panning dish by 0.25% trypsin-EDTA (Invitrogen).
OPC transfection.
Wt mouse Olig1 was subcloned into pEGFP vector. The control plasmid pEGFP and pEGFP-wt-Olig1 were transfected into OPCs (4.5 × 104 cells per well) using Lipofectamine 2000 (Invitrogen) for 48 h, and the cells were cultured in OPC-Differentiation Medium for another 3 d.
Immunofluorescence.
Mice were deeply anesthetized with 1% pentobarbital and transcardially perfused with 4% paraformaldehyde in PBS. Brains and spinal cords were dehydrated in 30% sucrose and sectioned (20 μm) on a cryostat microtome (MS 1900, Leica). OPCs cultured on coverslips were fixed with 4% paraformaldehyde in PBS. Both free-floating sections and coverlsips were blocked with 10% BSA and incubated with primary antibodies overnight at 4°C. Sections were incubated with secondary antibodies for 1 h at room temperature. Primary antibodies included the following: rabbit anti-Olig2 (1:500; Millipore), mouse anti-Olig2 (1:100; Millipore), goat anti-MBP (1:500; Santa Cruz Biotechnology), rabbit anti-Olig1 (1:500; Millipore), mouse anti-Olig1 (1:100; Santa Cruz Biotechnology), rabbit anti-PDGFRα (1:100; Santa Cruz Biotechnology), mouse anti-CC1 (1:200, Millipore), mouse anti-BrdU (1:200, Sigma), and mouse anti-NF200 (1:200, Abcam). Secondary antibodies included the following: AlexaFluor-488-, AlexaFluor-568-, or Cy5-conjugated secondary antibodies to rabbit, mouse, or goat (1:500; Invitrogen).
Image acquisition and analysis.
Fluorescent images were collected on a fluorescence microscope (90i, Nikon) or a laser confocal scanning microscope (PV100, Olympus) with the excitation wavelengths appropriate for AlexaFluor-488 (488 nm), AlexaFluor-568 (568 nm), Cy5 (628 nm), or DAPI (380 nm). Images of fixed areas (the central part of the corpus callosum and the dorsal cerebral cortex) were digitally recorded using a Nikon 90i light microscope (Nikon) equipped with a digital capture system (SPOT, Diagnostic Instruments). Cell quantification was performed using the Image-Pro Plus software (version 5.1, Media Cybernetics). For stereological quantification, serial sections of spinal cord from C2-C6 and brain coronal sections containing the corpus callosum were collected (∼100 sections) and 10 sections were sampled from each animal in a systematic and random manner. For statistical analysis, at least nine representative fields were randomly acquired at 40× magnification from each of the two experiments performed in triplicate. Cell counting for coverslips was conducted on nine randomly chosen fields for each sample (Media Cybernetics). Quantification of MBP immunostaining and cell counting was performed using the Image-Pro Plus software 5.0 (Media Cybernetics).
Western blotting.
Mice (3–5 per group) were anesthetized with sodium pentobarbital and brains and spinal cords were rapidly removed and frozen. Frozen samples were homogenized and proteins were extracted using a RIPA lysis buffer with 1% PMSF solution (Biocolors). After protein determination using Coomassie Brilliant Blue G250, samples containing 50 μg of protein per 10 μl were denatured in gel-loading buffer and separated on 10% SDS-PAGE gels. Protein samples were transferred to polyvinylidene difluoride membranes and visualized by chemiluminescence (ECLplus, GE Healthcare). β-Actin was used as a loading control for the total protein loaded. Quantification of band intensity was analyzed using the Image Pro Plus software. Primary antibodies included the following: mouse anti-Olig2 (1:1000; Millipore), mouse anti-Olig1 (1:1000; Santa Cruz Biotechnology) and β-actin (1:2000; Santa Cruz Biotechnology).
qRT-PCR.
qRT-PCR was performed using the Mx3005P qRT-PCR System (Agilent Technologies). RNA was extracted from spinal cord of wt (Olig2+/+), Olig2plp-iko, and Olig2cnp-cre mice using Trizol (Invitrogen). cDNA were generated using a first-strand cDNA synthesis kit (Takara). qRT-PCR was performed as described previously. Briefly, the fluorescent dye SYBR Green (Takara) was included in each assay to measure DNA concentration during each annealing phase. Triplicate samples were analyzed for each cDNA with test and internal control primers for the house keeping gene, glyceraldehyde-3-phosphatase dehydrogenase (GAPDH). Dilution curves were generated by 10-fold serial dilutions of each target cDNA to estimate copy number of the target cDNA. Fluorescence intensity was plotted against cycle number and standardized using GAPDH to account for sample variability. Primers used for expression analyses are as follows: Olig1F (tcatcctcatcctcatcctcttcc) and Olig1R (gctgctgctgttcctctttgg); Olig2F (ggcggtggcttcaagtcatc) and Olig2R (tcgggctcagtcatctgcttc); Sox10F (ctagccgaccagtaccctcac) and Sox10R (agcctctcagcctcctcaatg); Sox8F (tacccgcatctccataacgc) and Sox8R (tcacactcttccttagccttg); Hes5F (ggagatgctcagtcccaagg) and Hes5R (gctctatgctgctgttgatgc); ID2F (gcagcacgtcatcgtcatcgattaca) and ID2R (ttcagatgcctgcaaggaca); and GAPDHF (aggccggtgctgagtatgtc) and GAPDHR (tgcctgcttcaccaccttct).
Electron microscopy.
Mice were anesthetized with sodium pentobarbital and perfused with PBS followed by 2% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.2. Optic nerves of Olig2plp-iko, Olig2cnp-cko, and wt were removed and fixed in fresh fixative overnight at 4°C. Tissues were rinsed in PBS, postfixed in 1% OsO4 in PBS for 1 h, dehydrated in a graded ethanol series, infiltrated with propylene oxide, and embedded in Epon. Semithin sections were stained with toluidine blue, and thin sections were stained with lead citrate. Measurements were made on electron micrographs from five pairs of mice at either P14 or P21.
BrdU incorporation assay.
To assess proliferation of OPCs, mice were injected with BrdU (Sigma; 5 mg/kg) 2 and 24 h before death. Immunohistochemistry for BrdU was performed as described, with the exception that sections were sequentially incubated in 2N HCl (25 min) and 0.1 m borate buffer (25 min) before the blocking steps.
TUNEL assay.
TUNEL assays were performed using the In Situ Cell Death Detection kit (Roche) according to the manufacturer's instructions. Briefly, the sections were pretreated with 0.2% Triton/PBS for 30 min and subsequently incubated with the TUNEL reaction solution mixture containing terminal deoxynucleotidyltransferase in a humidified 37°C chamber for 45 min.
Statistical analysis.
Statistical analyses were performed using one-way ANOVA followed by Tukey's post hoc test. Comparisons between two experimental groups were made using Student's t test. A p value < 0.05 was considered statistically significant.
Results
Specific deletion of Olig2 from newborn OLs facilitates maturation and myelination
It is well documented that Olig2 plays a crucial role in the specification of NSCs into oligodendroglial lineage cells. However, the persistent expression of Olig2 throughout the oligodendroglial lineage begs the question: does Olig2 display stage-specific roles during OL development? To examine the possible role for Olig2 at later stages of OL development, the PLP-Cre-ERT mice were crossed with the floxed-Olig2 mice (Olig2plp-iko). Conditional knock-out of Olig2 in immature and mature OLs was accomplished by treatment with tamoxifen on 5-day-old pups (P5) for 5 d (Fig. 1A). This time point coincides with oligodendrocyte differentiation and the initiation of myelination in the CNS. Upon observation of both white (corpus callosum) and gray (cortex) matter areas in the Olig2plp-iko mice, MBP expression was increased by ∼35% in the brain of Olig2plp-iko mice compared with wt mice at P14 (Fig. 1B–D). The increase in MBP (green) expression correlated with an approximate 1.4-fold increase in the number of CC1+ OLs (red) (Fig. 1B′,C′,E) and a significant decrease in the number of Olig2+ cells (white) (Fig. 1B′,C′,F). As expected in the Olig2plp-iko, only 10% of the CC1+ cells coexpressed Olig2, whereas >90% of the CC1+ cells coexpressed Olig2 in the wt mice (Fig. 1B″,C″,G). These results demonstrate the efficient and successful deletion of Olig2 in newly generated and mature OLs. Electron microscopic analysis of the optic nerve confirms the increased number of myelinated axons in the Olig2plp-iko mice at P14 (Fig. 2A,B). To exclude the possibility that Olig2 deletion affects neuronal development, NF200 (red) and MBP (green) double labeling was performed on spinal cord sections. Axon numbers were unchanged, whereas MBP intensity was increased in the Olig2plp-iko mice (Fig. 2C,D). As increased cell proliferation of OPCs could result in an enhancement in the number of mature oligodendrocytes and hypermyelination, we analyzed the effect of Olig2 deletion in OPC proliferation. However, BrdU incorporation analysis in the Olig2plp-iko and wt mice spinal cord and corpus callosum did not reveal any significant changes in the number of proliferating cells (Fig. 2E). Additionally, the number of PDGFRα+ OPCs (red) was comparable between Olig2plp-iko (Fig. 2G) and wt mice (Fig. 2F), and continued to coexpress Olig2 (green). These results suggest that Olig2 acts as a negative regulator in the maturation of OLs.
Figure 1.
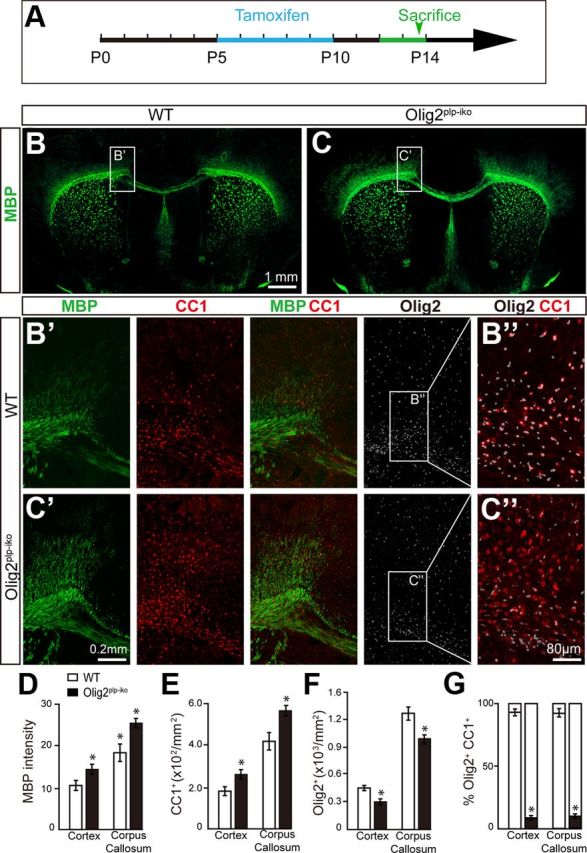
Conditional knock-out of Olig2 at later stages of oligodendroglial development facilitates MBP expression. For inducible ablation of Olig2 in oligodendrocytes, Olig2flox/flox mice were crossed to PLP-CreER(T) to generate Olig2plp-iko mice. Schematic diagram displaying the expression of Cre was induced with tamoxifen injections at P5 for 5 d (A) and MBP expression in the forebrain was analyzed at P14 in wild-type (B) and Olig2plp-iko mice (C). Magnified images of the white matter of Olig2plp-iko mice (C′) and wt mice (B′) immunostained for MBP (red), CC1 (red), and Olig2 (white). Decreased colocalization of Olig2 and CC1 (B″,C″) displays effective deletion of Olig2 specifically in differentiated oligodendrocytes. D–G, Quantification of the immunostaining in the cortex and corpus callosum reveals a 35% increase in MBP expression (D) accompanied with a 1.4-fold increase in CC1+ cell numbers (E). The number of Olig2+ cells decreased by 30% (F) and the number of CC1+/Olig2+ cells was 90% reduced (G) in either cortex or corpus callosum of Olig2plp-iko mice. Scale bars: B, C, 1 mm; B′, C′, 0.2 mm; B″, C″, 80 μm. Data are mean ± SEM (5 animals in each group). *p < 0.05.
Figure 2.
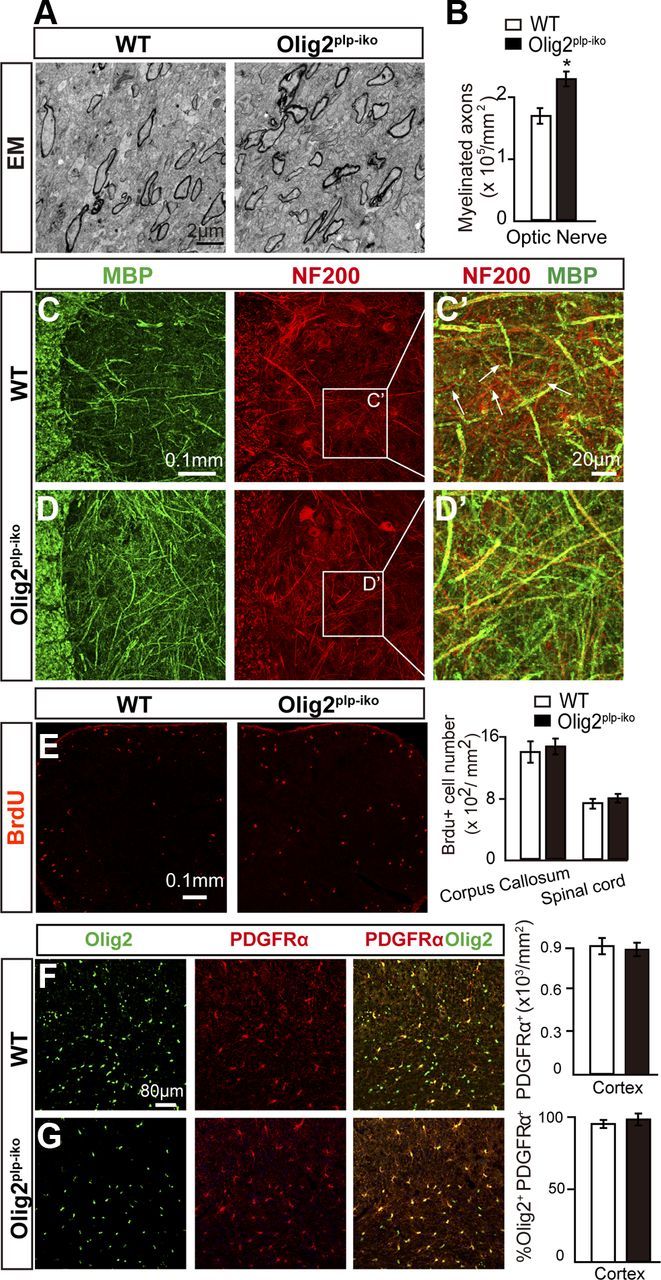
Myelination is enhanced in Olig2plp-iko mice during development. A, Electron microscopic images of optic nerve cross-sections of Olig2plp-iko and wt P14 mice. B, Quantification of electron microscopic images displays a 30% increase in myelinated axons in the absence of Olig2. C, D, Spinal cord sections immunostained for MBP (green) and NF (red) display more myelinated axons in Olig2plp-iko mice but similar axonal staining levels in both genotypes. Unmyelinated axons are more frequently observed in the gray matter of wt mice (C′) than in Olig2plp-iko mice (D′). BrdU (red) incorporation assay displays similar proliferative cell numbers in the spinal cord and corpus callosum (E). Double immunofluorescence labeling of brain sections for PDGFRα (red) and Olig2 (green) in wt (F) and Olig2plp-iko (G) mice demonstrate similar OPC numbers. Scale bars: A, 2 μm; C, D, 0.1 mm; C′, D′, 20 μm; E, 80 μm; F, G, 0.1 mm. Data are mean ± SEM (5 animals in each group). *p < 0.05.
Specific deletion of Olig2 in OPCs inhibits differentiation and myelination
To examine the role of Olig2 in the differentiation of OPCs into OLs, we conditionally deleted the Olig2 gene in OPCs by crossing the CNP-Cre with the floxed-Olig2 mice (Olig2cnp-cko). The number of PDGFRα+ OPCs in the Olig2cnp-cko mice was comparable with wt (Fig. 3B); however, only 3% of the OPCs expressed Olig2 by P0 in the spinal cord (Fig. 3C). Indeed, Olig2 expression was significantly attenuated throughout the CNS, demonstrating efficient recombination (Fig. 3A). As expected from the primary literature, MBP expression was decreased by ∼50% in the brain (Fig. 3E) and the number of CC1+ OLs was decreased by almost 80% (Fig. 3D) in the spinal cord by P14 in the Olig2cnp-cko mice compared with wt. Consistent with these findings, myelinated axons were significantly reduced in the optic nerve of Olig2cnp-cko mice (Fig. 3F), indicating that Olig2 in OPCs functions as a positive regulator of oligodendroglial differentiation. Apoptotic cell numbers in either spinal cord or corpus callosum were not changed between both mouse lines eliminating the possibility that an increase in cell death could be responsible for the observed hypomyelination (Fig. 3G). These findings suggest that Olig2 may convey opposing functions at different stages of the oligodendroglial lineage. Olig2 exhibits a positive role for specification and differentiation of OPCs but may act to repress the maturation and myelination by OLs. What molecular mechanisms underlie the stage-dependent role for Olig2?
Figure 3.
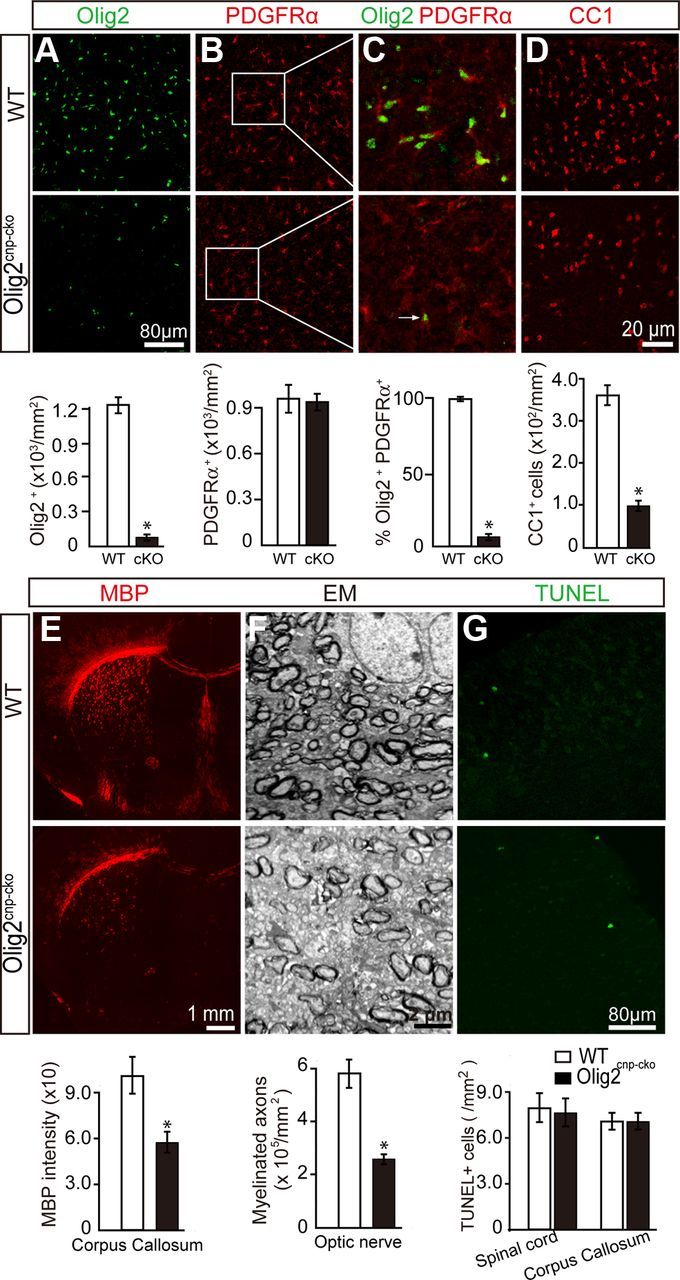
Deletion of Olig2 in OPCs inhibits oligodendroglial differentiation and myelination. Olig2 was specifically deleted in OPCs by crossing Olig2flox/flox mice with CNPcre/+ mice to generate the Olig2cnp-cko mice. Spinal cord sections of wt mice and Olig2cnp-cko mice at P0 were stained for Olig2 (A, green) and PDGFRα (B, red) confirming decreased levels of Olig2+ cells and the deletion of Olig2 in the majority of PDGFRα+ OPCs (C, 3% of PDGFRα+ cells coexpressed Olig2). D, Immunostaining of P14 spinal cord sections shows an 80% decrease in CC1+ cell numbers in the Olig2cnp-cko mice without changing the TUNEL+ apoptotic cell numbers in either spinal cord or corpus callosum (G). E, P14 coronal brain sections of wt and Olig2cnp-cko mice stained for MBP (red) show a 45% reduction of MBP expression in the knock-out mice. F, Optic nerve electron microscopic images of Olig2cnp-cko and wt mice at P21 reveal less myelinated axons in the absence of Olig2 in OPCs. Scale bars: A, B, G, 80 μm; C, D, 20 μm; E, 1 mm; F, 2 μm. Data are mean ± SEM (5 animals in each group). *p < 0.05.
Stage-dependent deletion of Olig2 results in opposing changes to Olig1 expression
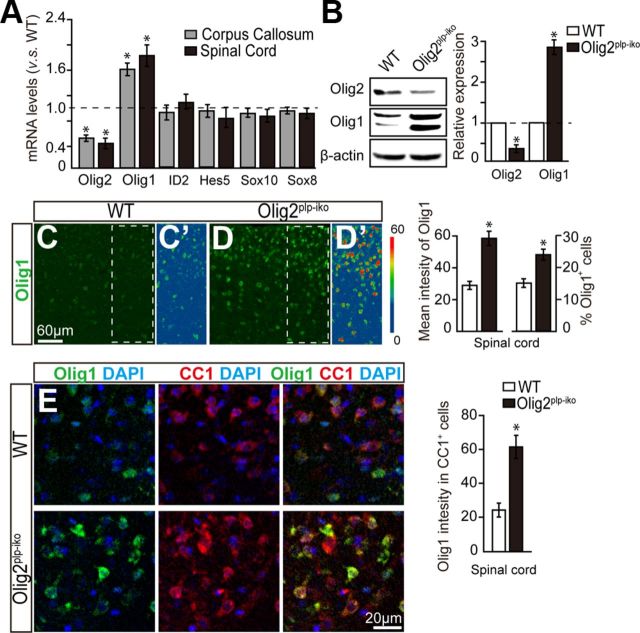
It is well accepted that the temporal and spatial regulation of differentiation and myelination by oligodendroglia is coordinated in part by the activation and repression of various transcriptional programs (Emery, 2010). Therefore, we examined the expression profile of known transcription factors that are important for specification and differentiation of OPCs in the Olig2cnp-cko and wt mice. These factors include Olig1, ID2, Hes5, Sox10, and Sox8. Our results indicate that Olig1 mRNA levels in either spinal cord or corpus callosum of Olig2cnp-cko mice is significantly diminished to less than one-third of wt Olig1 expression by P0 (Fig. 4A), whereas the other transcription factors were unchanged. Western blot analysis confirms a similar decrease of Olig1/2 expression in both brain and spinal cord (Fig. 4B). To examine the relationship between Olig1/2, we find that ∼90% of the Olig1+ cells (red) express Olig2 (green) in the spinal cord of wt mice, whereas only 3% was expressed in the Olig2cnp-cko mice by P14 (Fig. 4C). Concomitantly, we observe a significant decrease in the overall Olig1+ cell numbers (Fig. 4C), suggesting that Olig1 expression is highly coupled to Olig2 expression. It is well established that Olig1 can initiate myelin gene expression and that it plays an important role for OL maturation and (re)myelination (Arnett et al., 2004; Burton, 2005; Xin et al., 2005). To examine Olig1 expression in OLs in the Olig2cnp-cko mice, we analyzed CC1 and Olig1 expression and observed decreased expression levels of Olig1 in the Olig2cnp-cre mice compared with the wt (Fig. 4D). Our results suggest that Olig2 deletion in OPCs may negatively regulate Olig1 expression in oligodendroglial cells and that diminished Olig1 expression may contribute, at least in part, to the decreased differentiation and myelination observed in Olig2cnp-cko mice.
Figure 4.
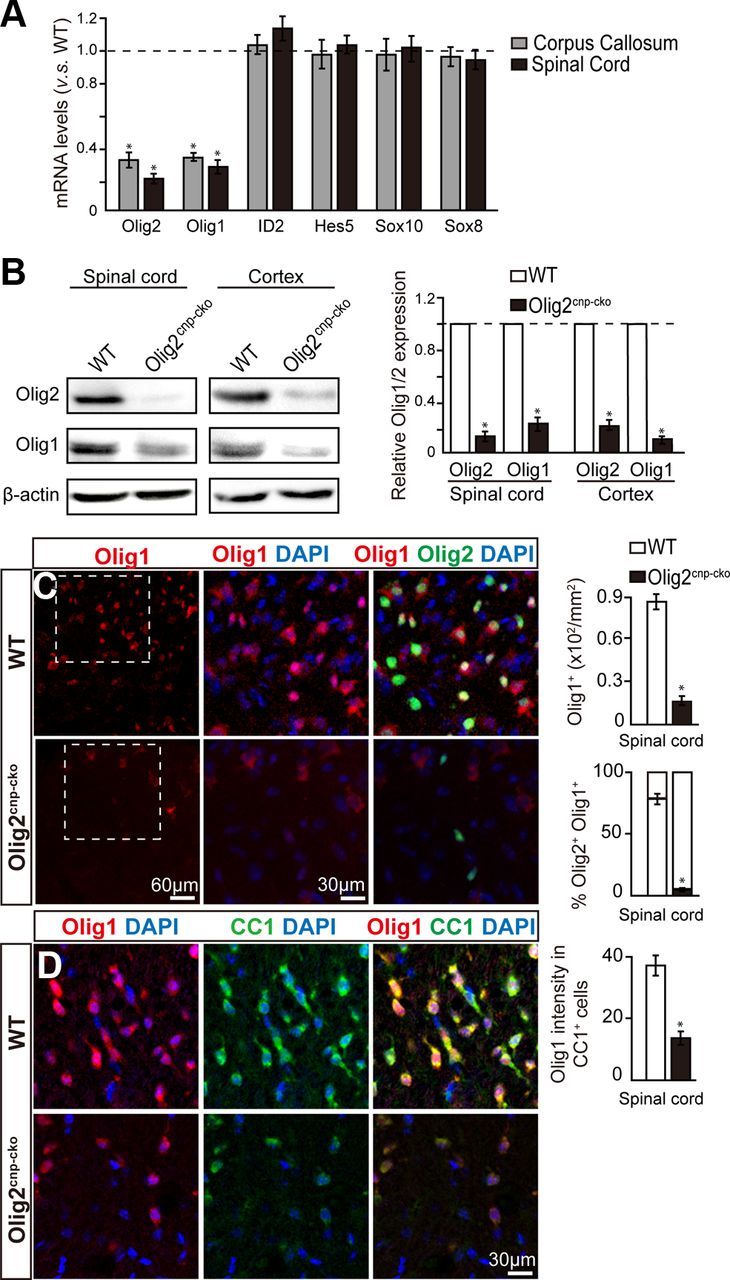
Olig2 deletion downregulates Olig1 expression in oligodendroglial cells. A, mRNA levels of transcription factors from Olig2cnp-cko and wt mice spinal cord at P0 or corpus callosum at P7 were assayed by qPCR. Olig2 and Olig1 mRNA levels, but not ID2, Hes5, Sox10, and Sox8, are significantly diminished in Olig2cnp-cko mice compared with wt mice. B, Western blot analysis illustrates decreased Olig1 and Olig2 expression in both spinal cord and cortex of Olig2cnp-cko mice compared with wt at P0. All results are quantified relative to control levels (dotted line). C, Spinal cord immunostaining for Olig1 (red) and Olig2 (green) in P14 knock-out and wt mice reveals a decrease in Olig1+ cell numbers in parallel with Olig2 deletion. D, Double immunostaining of spinal cord sections with Olig1 (red) and CC1 (green) display decreased expression levels of Olig1 in oligodendroglial cells in the Olig2cnp-cko mice. Scale bars: C, bottom left, 60 μm; C, bottom middle, D, 30 μm. Data are mean ± SEM (4 animals in each group). *p < 0.05.
To further investigate the possible link between Olig1 expression and the stage-dependent deletion of Olig2, we further examined Olig1 expression in the developing spinal cord and corpus callosum of Olig2plp-iko mice by qPCR and Western blot. Our results indicate that Olig1, but not Hes5, Id2, Sox10, or Sox8 mRNA, is significantly enhanced by ∼1.7-fold (Fig. 5A) and Olig1 protein level is increased by threefold (Fig. 5B). Both Olig1-expressing cells and the fluorescence intensity of the immunostaining were significantly increased in the Olig2plp-iko mice (Fig. 5C,D). Moreover, double staining of CC1 and Olig1 showed an increase in Olig1 expression in OLs when Olig2 is deleted (Fig. 5E). These observations demonstrate that Olig1 expression correlates with the differentiation of OLs, suggesting that the stage-specific function for Olig2 on oligodendroglial differentiation may be mediated by Olig1 function.
Figure 5.
Knock-out of Olig2 in immature OLs upregulates Olig1 expression. qPCR and Western blot analysis display decreased Olig2 expression and enhanced Olig1 expression, but not ID2, Hes5, Sox10, and Sox8, in the spinal cord and corpus callosum of Olig2plp-iko mice at P14 by either mRNA (A) or protein levels (B). All results are quantified relative to control levels (dotted line). Both Olig1-expressing cell numbers and Olig1 fluorescence intensity are upregulated in spinal cord of Olig2plp-iko mice (C, C′) compared with the wt mice (D, D′). E, Olig1 (green) immunostaining intensity in CC1+ cells (red) in the spinal cord at P14 is increased in the Olig2plp-iko mice. Scale bars: C, D, 60 μm; E, 20 μm. Data are mean ± SEM (5 animals in each group). *p < 0.05.
Overexpression of Olig1 in Olig2-ablated OPCs rescues differentiation arrest
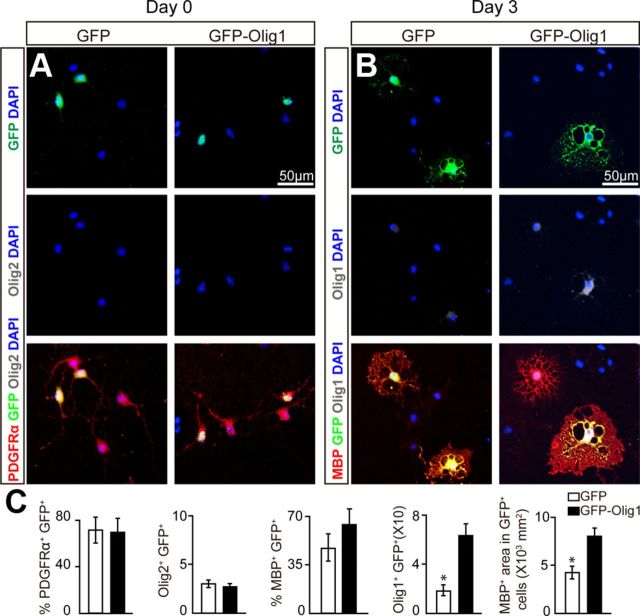
Given that Olig1 is important for OPC differentiation and OL maturation, it is plausible to imagine that forced expression of Olig1 in Olig2-ablated OPCs would rescue the differentiation arrest. To test this possibility, we first characterized OPCs from Olig2cnp-cko mice in vitro. Olig2 expression was deleted in PDGFRα+ OPCs from Olig2cnp-cko mice without altering the number of OPCs compared with wt (Fig. 6A,C). However, upon differentiation for 3 d, the number of MBP+ cells from Olig2cnp-cko mice was decreased compared with the wt, the overall membrane area of the MBP+ cells was diminished, and Olig1 expression was significantly decreased (Fig. 6B,C). The downregulation of Olig1/2 was further confirmed by Western blot analysis of OPCs purified from wt and Olig2cnp-cko mice (Fig. 6D). We next purified immature OLs from Olig2plp-iko mice previously treated with tamoxifen and confirmed the decrease in Olig2 expression by Western blot (Fig. 6E). This decrease was accompanied by an increase in Olig1 expression in immature OLs from Olig2plp-iko mice and was more pronounced after differentiation into mature MBP-expressing OLs. Therefore, Olig1 expression is dependent on the stage-specific deletion of Olig2 and confirms the in vivo data, suggesting that Olig2 may regulate OL differentiation and maturation via Olig1 expression. To test this hypothesis, we examined the role of Olig1 in an Olig2-null background by overexpressing Olig1 into the OPCs from the Olig2cnp-cko mice. The results demonstrate that the number of PDGFRα+ OPCs is unchanged in cultures transfected with or without the Olig1-containing plasmid on the day after transfection (Fig. 7A,C). However, upon 3 d of differentiation, MBP expression and membrane expansion are increased in the OLs expressing the Olig1-containing vector compared with the control vector (Fig. 7B,C). These results suggest that overexpression of Olig1 in Olig2-ablated OPCs can rescue differentiation arrest and compensate for the loss of Olig2.
Figure 6.
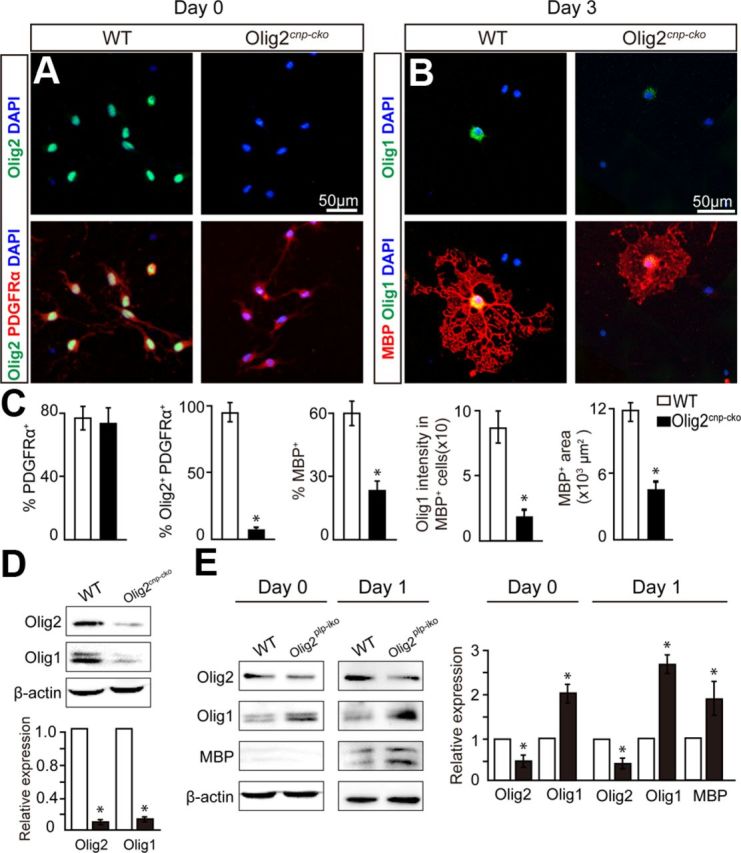
Olig2-null OPCs in vitro display decreased differentiation capacity and Olig1 downregulation, whereas Olig2 deletion in immature OLs displays Olig1 upregulation. A, Purified OPCs from Olig2cnp-cko and wt mice were plated and immunostained (0 d) for Olig2 (green) and PDGFRα (red) demonstrating similar OPC cell numbers upon Olig2 deletion. B, After differentiation for 3 d, Olig2-null OLs have less extensive MBP+ membrane sheets (red) and correlate with a decrease in Olig1 (green) expression compared with wt OLs. C, Quantification of PDGFRα, Olig1/2, and MBP stainings. D, Western blot analysis of Olig1/2 expression in purified OPCs (PDGFRα immunopanning) from wt and Olig2cnp-cko P8 mice. E, Olig2plp-iko and wt mice were treated with tamoxifen for 2 d from purified immature OLs (O4 immunopanning) at P8 and subjected to Western blot analysis. Immature OLs and MBP OLs display decreased Olig2 expression but increased Olig1 expression compared with OLs from wt mice. Scale bar: A, B, 50 μm. Data are mean ± SEM, and all experiments were performed in triplicate. *p < 0.05.
Figure 7.
Overexpression of Olig1 in Olig2-deleted OPCs in vitro rescues differentiation arrest. A, B, OPCs from Olig2cnp-cko mice were purified and transfected with Olig1-GFP or GFP constructs and immunostained the next day after plating (0 d) for Olig1 (white), PDFGRα (red) (A) or cultured in OPC differentiation media for 3 d and stained for Olig1 (white) and MBP (red) (B). C, Quantification of the immunostaining demonstrates that OPCs transfected with the Olig1-GFP plasmid promote the expansion of MBP+ membrane area compared with OPCs transfected with GFP alone without changing the number of PDGFRα+ cells. Scale bar: A, B, 50 μm. Data are mean ± SEM, and all experiments were performed in triplicate. *p < 0.05.
Deletion of Olig2 results in upregulation of Olig1 and facilitates remyelination in cuprizone-induced demyelination
Our findings demonstrate that deletion of Olig2 in immature OLs increases OL maturation and myelination, suggesting that the stage-dependent deletion of Olig2 could represent a potential strategy to promote remyelination. To test this hypothesis, we treated Olig2plp-iko and wt mice with cuprizone for 6 weeks to induce widespread demyelination. After withdrawing the cuprizone, Cre-mediated recombination and Olig2 deletion were induced by treatment with tamoxifen for 7 d (Fig. 8A). MBP expression and remyelination were enhanced in the brains of Olig2plp-iko mice compared with wt controls (Fig. 8B,C). Under higher magnification, more abundant MBP+ myelin segments could be identified in the cortex (Fig. 8B′,C′) and lateral septal regions (Fig. 8B″,C″) in the Olig2plp-iko than that of wt mice. Furthermore, Olig1 expression was upregulated by ∼3-fold in the Olig2plp-iko mice as assayed by Western blot in both cortex and corpus callosum (Fig. 8D). Additionally, immunostaining displayed an increase in the Olig1+/CC1+ OLs in the cortex of the Olig2plp-iko mice compared with wt (Fig. 8E). These observations suggest that deletion of Olig2 in OLs is capable of facilitating maturation and remyelination and may provide a potential therapeutic strategy for myelin repair.
Figure 8.
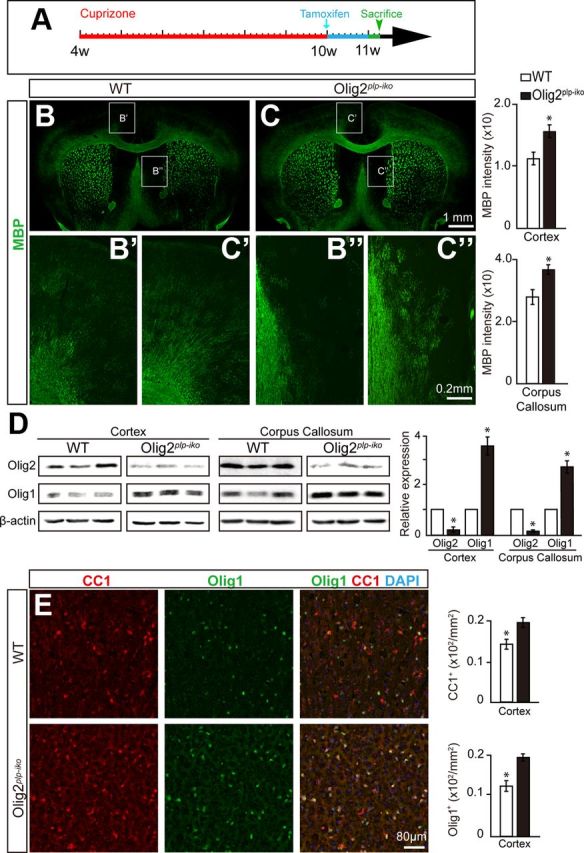
Knock-out of Olig2 in OLs facilitates remyelination and upregulates Olig1 expression in a cuprizone-induced demyelinating model. A, Schematic diagram displaying the time course of cuprizone treatment and induction of Olig2 deletion with tamoxifen treatment in Olig2plp-iko mice. B, C, Brain coronal sections illustrating an increase in MBP expression (green) in the brain of Olig2plp-iko mice (C) compared with wt (B) after cuprizone demyelination. Higher magnification of the cortex and the lateral septal region in the Olig2plp-iko mice (B′,B″) and wt mice (C′,C″) displaying increased remyelination efficiency in the absence of Olig2 in immature OLs. D, Western blot analysis displaying increased Olig1 expression in the brain of Olig2plp-iko mice after cuprizone treatment. The values are normalized to the control, and all experiments were performed in triplicate. E, Both Olig1+ (green) and CC1+ (red) cells are increased during remyelination in the cortex of Olig2plp-iko mice compared with wt. Scale bars: B, C, 1 mm; B′, C′, 0.2 mm; E, 80 μm. Data are mean ± SEM (9 animals in each group). *p < 0.05.
Discussion
Before myelination in the CNS, the OPC exits from the cell cycle and initiates a transcriptional program resulting in the differentiation into the OL. Newly generated OLs receive cues from their surroundings and concomitantly extend and elaborate processes and ensheath axons (Baumann and Pham-Dinh, 2001; Liu et al., 2003). These processes are orchestrated in part by transcriptional regulatory networks (Emery, 2010; Fancy et al., 2011). We hypothesize that the continuous expression of single transcription factor may be responsible for coordinating the transcriptional program at different stages of oligodendroglial development. Two distinct and sequential functions by one molecule could ensure proper temporal differentiation and the spatial regulation for myelination.
Among the transcription factors responsible for oligodendroglia development, Olig1 and Olig2 were identified to play essential roles in determining both the oligodendroglial lineage and in the generation and maturation of oligodendrocytes (Lu et al., 2000; Zhou et al., 2000; Ligon et al., 2006a; Ligon et al., 2006b). Olig2-null mice fail to develop cells of the oligodendroglial lineage (Lu et al., 2002; Takebayashi et al., 2002), whereas overexpression of Olig2 in NSCs is sufficient to specify OPCs rather than neurons and/or astrocytes (Copray et al., 2006; Hu et al., 2012). Moreover, conditional deletion of Olig2 at the multipotential progenitor stage in the cerebral cortex results in normal patterning of OPCs but a decrease in myelin gene expression and hypomyelination (Yue et al., 2006), suggesting that Olig2 function may not be required for the specification of cortical progenitors into OPCs, but for the differentiation of OPCs into OLs. In our study, ablation of Olig2 in OPCs under the control of the CNPase-promoter significantly inhibits differentiation into OLs resulting in hypomyelination, whereas deletion of the Olig2 gene in newly generated OLs under the PLP promoter facilitates myelination (Figs. 1, 2, and 3). For the first time, our study identifies a stage-specific role for Olig2 on oligodendroglial differentiation, namely, Olig2 exhibits a positive role for specification and differentiation of OPCs but may act to repress the maturation and myelination of OLs.
What mechanisms underlie the stage-specific functions of Olig2 during oligodendroglial differentiation? Given that the temporal and spatial regulation of oligodendroglial differentiation and myelination are coordinated in part by the activation and repression of various transcriptional programs (Emery, 2010; Fancy et al., 2011), it is quite possible that Olig2 dynamically modulates transcription of target genes that regulate the differentiation and maturation of oligodendroglia. Upon examination of the expression profile of various transcriptional factors previously identified as essential for specification and differentiation (Olig1, ID2, Hes5, Sox10, and Sox8) (Kondo and Raff, 2000; Lu et al., 2001; Wang et al., 2001; Stolt et al., 2002, 2004; Zhou and Anderson, 2002), we demonstrate that Olig1 expression highly correlates with the stage-specific function of Olig2. We find that Olig1 expression is decreased in Olig2cnp-cko OPCs but is increased in Olig2plp-iko immature OLs (Figs. 4, 5, and 6). Although we do not know whether the changes that we observe in Olig1 expression are caused by the deletion of Olig2 or just a consequence of oligodendroglial cell maturation changes, the fact that the expression of other transcription factors is maintained suggests that Olig2 might directly or indirectly regulate Olig1 expression. As a highly homologous transcription factor to Olig2, Olig1 can facilitate myelin gene expression and is required for (re)myelination (Lu et al., 2001; Zhou and Anderson, 2002; Arnett et al., 2004; Xin et al., 2005; Li et al., 2007). Our recent data indicate that Olig1 enhances membrane expansion and process outgrowth during terminal maturation of OLs (Niu et al., 2012), and enforced expression of Olig1 in Olig2-null OPCs rescues differentiation arrest induced by Olig2 deletion (Figs. 6 and 7). Recently, it has been reported that injection of recombinant retroviruses encoding Olig1 into the injured spinal cord does not increase NG2+ and PDGFRα+ OPCs but increases CC1+ OLs and promotes remyelination, demonstrating that upregulation of Olig1 expression could promote OL maturation and myelination (Kim et al., 2011).
Although cooperation of Olig1/2 with other transcription factors, such as Sox10, Zfp488, and ID2/4, have been reported in oligodendroglial differentiation (Wang et al., 2006; Liu et al., 2007; Guo et al., 2011), the regulatory role of Olig2 on Olig1 gene expression has not been previously reported. Could Olig1 be a target gene that is regulated by Olig2? Indeed, a number of conserved and nonconserved functional enhancer elements of the Olig1 locus have been systematically screened and possess potential binding sites for Olig2 (Friedli et al., 2010). A recent study illustrates that Olig2 directs Sox10 expression by binding to the U2 enhancer of Sox10 gene in oligodendroglial cells (Kuspert et al., 2010). Moreover, it has been suggested that Olig2 functions primarily as a transcriptional repressor in the specification of neurons and astroglia (Lu et al., 2000; Zhou et al., 2000; Sun et al., 2003) while acting synergistically with Olig1 to drive MBP and Sox10 expression in oligodendroglial cells (Li et al., 2007), suggesting that Olig2 functions as either a repressor or activator, depending on interactions with transcription factors (Gokhan et al., 2005). In addition, a recent study demonstrates that phosphorylation of Olig2 can modulate the interaction with other transcription factors and switches neuron-oligodendroglial fate determination (Li et al., 2011). We propose that Olig2 stage-specifically may cooperate with different transcription factors and regulate Olig1 expression in an opposing manner to promote oligodendroglial differentiation and maturation. Further investigation is required to test whether Olig2 regulates Olig1 expression and identify the possible cofactors of Olig2 at different stages of oligodendroglial development.
Differentiation and myelination by oligodendroglia is a graded process, requiring membrane expansion, the initiation of myelin gene expression followed by the ensheathment and wrapping of axons (Liu et al., 2003; Emery, 2010). Coincidentally, myelin gene expression also undergoes a stage-dependent process, namely, that when differentiation is initiated myelin gene expression needs to increase to a maximal level to accommodate active myelination followed by a downregulation in mature oligodendrocytes to adjust for levels that are adequate for myelin maintenance (Kuspert et al., 2010). It seems likely that the stage-specific function of Olig2 on oligodendroglial differentiation and myelination may contribute to these graded changes observed in maturation (Figs. 1, 2, 3, and 4). We speculate that, at early stages of OPC differentiation, Olig2 may act as an activator, interacting with Olig1, Sox10, and other transcription factors to initiate myelin gene expression and drive differentiation. After the appropriate number of OLs is generated, Olig2 may act as a repressor of myelin gene expression in a subset of OLs as they compete for axonal cues and extend and elaborate processes to ensheath axons (Baumann and Pham-Dinh, 2001; Liu et al., 2003).
Currently, differentiation arrest is considered rate-limiting in the failure of remyelination (Fancy et al., 2011; Kotter et al., 2011; Miron et al., 2011). The stage-specific functions of Olig2 may provide an alternative and effective strategy for promoting remyelination. Transplantation of Olig2-overexpressing NSCs into demyelinating lesions significantly enhances the generation of oligodendrogila but with only modest effects on remyelination (Copray et al., 2006; Hwang et al., 2009; Maire et al., 2009; Hu et al., 2012). Overexpression of Olig2 by injection of retroviruses into the injured spinal cord, however, induces glioma (Kim et al., 2011). Furthermore, overexpression of Olig1 and Olig2 simultaneously could increase both OPC generation and OL formation and maturation (Kim et al., 2011), but the possibility of tumorigenesis cannot be excluded. In this study, deletion of Olig2 in newly formed OLs under the PLP promoter upregulates Olig1 expression and facilitates remyelination in cuprizone-induced demyelination (Fig. 8). These findings imply that deletion of Olig2 at the later stages of oligodendroglial development could be a potential strategy in the treatment of demyelinating diseases. We demonstrate a stage-specific regulatory role for Olig2, mediated by Olig1 that conveys opposing functions on the differentiation and maturation of oligodendrocytes. This stage-specific function of Olig2 may provide new insights into the precise regulation of myelination and remyelination.
Footnotes
This work was supported in part by the National Natural Science Foundation of China (NSCF, 81270017), Joint Research Fund for Overseas Chinese Young Scholars (NSCF, 31228011), Natural Science Foundation Project of Chongqing (CSTC2011BA5006), the United States National Multiple Sclerosis Society Harry Weaver Neuroscience Scholar Award JF 2142-A-2, and National Institutes of Health–National Institute of Neurological Disorders and Stroke Grant NS062796-02 to J.R.C. We thank Dr. Richard Lu for the Olig2 mice.
The authors declare no competing financial interests.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Burton A. Olig1 needed for remyelination. Lancet Neurol. 2005;4:80. doi: 10.1016/S1474-4422(05)00978-6. [DOI] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- Chan JR, Watkins TA, Cosgaya JM, Zhang C, Chen L, Reichardt LF, Shooter EM, Barres BA. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron. 2004;43:183–191. doi: 10.1016/j.neuron.2004.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copray S, Balasubramaniyan V, Levenga J, de Bruijn J, Liem R, Boddeke E. Olig2 overexpression induces the in vitro differentiation of neural stem cells into mature oligodendrocytes. Stem Cells. 2006;24:1001–1010. doi: 10.1634/stemcells.2005-0239. [DOI] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Chan JR, Baranzini SE, Franklin RJ, Rowitch DH. Myelin regeneration: a recapitulation of development? Annu Rev Neurosci. 2011;34:21–43. doi: 10.1146/annurev-neuro-061010-113629. [DOI] [PubMed] [Google Scholar]
- Friedli M, Barde I, Arcangeli M, Verp S, Quazzola A, Zakany J, Lin-Marq N, Robyr D, Attanasio C, Spitz F, Duboule D, Trono D, Antonarakis SE. A systematic enhancer screen using lentivector transgenesis identifies conserved and non-conserved functional elements at the Olig1 and Olig2 locus. PLoS One. 2010;5:e15741. doi: 10.1371/journal.pone.0015741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SJ, Hu JG, Zhao BM, Shen L, Wang R, Zhou JS, Lü HZ. Olig1 and ID4 interactions in living cells visualized by bimolecular fluorescence complementation technique. Mol Biol Rep. 2011;38:4637–4642. doi: 10.1007/s11033-010-0597-x. [DOI] [PubMed] [Google Scholar]
- Hu JG, Shen L, Wang R, Wang QY, Zhang C, Xi J, Ma SF, Zhou JS, Lu HZ. Effects of Olig2-overexpressing neural stem cells and myelin basic protein-activated T cells on recovery from spinal cord injury. Neurotherapeutics. 2012;9:442–445. doi: 10.1007/s13311-011-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DH, Kim BG, Kim EJ, Lee SI, Joo IS, Suh-Kim H, Sohn S, Kim SU. Transplantation of human neural stem cells transduced with Olig2 transcription factor improves locomotor recovery and enhances myelination in the white matter of rat spinal cord following contusive injury. BMC Neurosci. 2009;10:117. doi: 10.1186/1471-2202-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Hwang DH, Choi JY, Park CH, Suh-Kim H, Kim SU, Kim BG. Differential and cooperative actions of Olig1 and Olig2 transcription factors on immature proliferating cells after contusive spinal cord injury. Glia. 2011;59:1094–1106. doi: 10.1002/glia.21182. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Kotter MR, Stadelmann C, Hartung HP. Enhancing remyelination in disease: can we wrap it up? Brain. 2011;134:1882–1900. doi: 10.1093/brain/awr014. [DOI] [PubMed] [Google Scholar]
- Küspert M, Hammer A, Bösl MR, Wegner M. Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 2011;39:1280–1293. doi: 10.1093/nar/gkq951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, de Faria JP, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69:918–929. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Fancy SP, Franklin RJ, Rowitch DH. Olig gene function in CNS development and disease. Glia. 2006a;54:1–10. doi: 10.1002/glia.20273. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Kesari S, Kitada M, Sun T, Arnett HA, Alberta JA, Anderson DJ, Stiles CD, Rowitch DH. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci U S A. 2006b;103:7853–7858. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, Casaccia-Bonnefil P. Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia. 2003;44:264–274. doi: 10.1002/glia.10290. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, Qiu M. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/S0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/S0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Maire CL, Buchet D, Kerninon C, Deboux C, Baron-Van Evercooren A, Nait-Oumesmar B. Directing human neural stem/precursor cells into oligodendrocytes by overexpression of Olig2 transcription factor. J Neurosci Res. 2009;87:3438–3446. doi: 10.1002/jnr.22194. [DOI] [PubMed] [Google Scholar]
- Miron VE, Kuhlmann T, Antel JP. Cells of the oligodendroglial lineage, myelination, and remyelination. Biochim Biophys Acta. 2011;1812:184–193. doi: 10.1016/j.bbadis.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Niu J, Mei F, Wang L, Liu S, Tian Y, Mo W, Li H, Lu QR, Xiao L. Phosphorylated olig1 localizes to the cytosol of oligodendrocytes and promotes membrane expansion and maturation. Glia. 2012;60:1427–1436. doi: 10.1002/glia.22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci. 2003;23:9547–9556. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/S0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/S0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/S0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Dulin J, Wu H, Hurlock E, Lee SE, Jansson K, Lu QR. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–3398. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T, Xian K, Hurlock E, Xin M, Kernie SG, Parada LF, Lu QR. A critical role for dorsal progenitors in cortical myelination. J Neurosci. 2006;26:1275–1280. doi: 10.1523/JNEUROSCI.4717-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/S0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/S0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]