Abstract
Small molecules that can induce and stabilize G-quadruplex DNA structures represent a novel approach for anti-cancer and anti-parasitic therapy and extensive efforts have been directed towards discovering lead compounds that are capable of stabilizing quadruplexes. The purpose of this study is to explore conformational modifications in a series of heterocyclic dications to discover structural motifs that can selectively bind and stabilize specific G-quadruplexes, such as those present in the human telomere. The G-quadruplex has various potential recognition sites for small molecules; however, the primary interaction site of most of these ligands is the terminal tetrads. Similar to duplex-DNA groove recognition, quadruplex groove recognition by small molecules offers the potential for enhanced selectivity that can be developed into a viable therapeutic strategy. The compounds investigated were selected based on preliminary studies with DB832, a bifuryl-phenyl diamidine with a unique telomere interaction. This compound provides a paradigm that can help in understanding the optimum compound-DNA interactions that lead to quadruplex groove recognition. DNA recognition by the DB832 derivatives was investigated by biophysical experiments such as thermal melting, circular dichroism, mass spectrometry and NMR. Biological studies were also performed to complement the biophysical data. The results suggest a complex binding mechanism which involves the recognition of grooves for some ligands as well as stacking at the terminal tetrads of the human telomeric G-quadruplex for most of the ligands. These molecules represent an excellent starting point for further SAR analysis for diverse modes of quadruplex recognition and subsequent structure optimization for drug development.
Keywords: Heterocyclic dications, Telomere, Telomerase, G-quadruplex, Circular dichroism, Nuclear magnetic resonance, Groove binding, Dimer
INTRODUCTION
DNA is an important biological target for cells ranging from cancer to eukaryotic parasites. The remarkable conformational plasticity exhibited by linear strands of DNA to fold into non-canonical structures has been demonstrated in sequences that can form G-quadruplexes and in more subtle microstructural differences in the local conformations of the double helix [1–3]. These structural differences provide variations for drug targeting in addition to sequence differences. G-quadruplexes, for example, have become a particularly attractive target for drug development due to their numerous biological functions, ability to act as genetic switches and relevance to cancer [4–9]. Quadruplex DNAs consist of a series of coplanar arrays of stacked, H-bonded guanine tetrads and are stabilized by aromatic stacking interactions of the G tetrads and by coordination with cations [10, 11]. Guanine-rich sequences, which are capable of forming quadruplex structures, are present in many biologically significant regions of genomic DNA, such as telomeres and many oncogene promoters [11–13]. Because of the critically essential roles of telomere DNA structures in cancer cells, the telomere is a particularly attractive target for drug design. As a cell replicates, DNA polymerase is unable to copy the extreme end of the 3′ lagging strand. This “end replication problem” results in shortening of the telomere in each round of cell division. While the enzyme is inactive in most normal somatic cells, in over 85% of cancer cells telomerase is activated and provides a potentially specific therapeutic target for cancer [14–16]. In order to maintain the telomere the primer sequence must be accessible to the enzyme and formation of ordered structures in the primer, such as quadruplexes, inhibits the enzyme from binding to the telomere DNA.
The unique geometry of G-quadruplexes is predicted to allow specific recognition by small molecules through various binding modes that would make the DNA inaccessible to telomerase in cancer cells. Various classes of quadruplex-interactive ligands have been identified in an intense research effort and most of these molecules have or can assume relatively planar aromatic scaffolds. Such compounds are highly favored to interact with quadruplexes by stacking on either one or both ends of the terminal G-tetrads [17–24]. Heterocyclic diamidines constitute an important class of DNA minor-groove targeting agents that have demonstrated effective cell uptake and excellent biological activity in animals as well as humans [25–27]. Diamidine compounds with an appropriate curved shape have a general preference for AT-rich duplex sequences [28–30]. Development of compounds for targeting quadruplex grooves, as versus end-stacking, is a very attractive strategy for increasing selectivity of recognition of quadruplexes over duplexes as well as among different quadruplex DNAs. In fact, quadruplex grooves are very different from double helix grooves in shape and dimensions. Moreover, due to the high polymorphism of the structures, quadruplex grooves can vary significantly according to the type of sequence, the nature of the complexed cation and the syn/anti conformation of guanine residues [31–33]. Targeting of the grooves can allow the recognition of different quadruplex DNAs with a high degree of selectivity. A possible strategy for the discovery of quadruplex groove-binding agents would be to use heterocyclic cations, similar to those that usually bind well to the duplex minor groove, which do not form strong complexes with duplexes [34]. If such compounds can bind to quadruplexes, they would have poor duplex interactions and high potential for quadruplex groove complex formation.
Compound Design
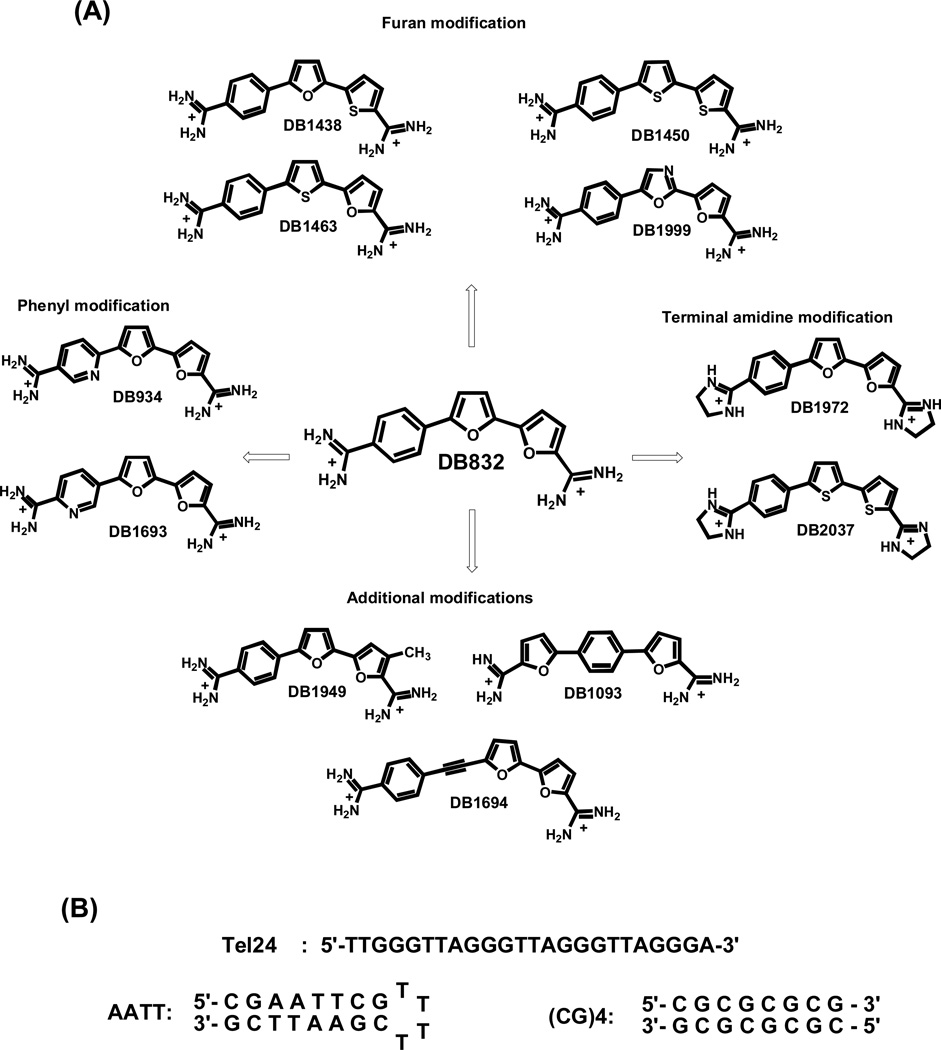
Our strategy for design of compounds that can selectively recognize quadruplexes over duplexes is to use compounds that are too curved to fit the topology of the DNA minor groove [34]. The discovery of such compounds that could then bind well to quadruplexes would yield the desired selectivity. A first step in the search for compounds of this type focused on compounds such as those shown in Fig. (1A), DB832, a bifuryl-phenyl diamidine, binds relatively weakly to duplexes but binds more strongly to quadruplexes. A very important and interesting feature of DB832 is its interaction with the human telomeric quadruplex sequence with a unique CD exciton-type splitting pattern. This is an exciting finding that suggests that DB832 can form a stacked molecular complex and is bound in the quadruplex grooves [35]. This is clearly a significant step forward in quadruplex recognition and it is important to gain a better understanding of all the key molecular features of DB832 contributing towards quadruplex binding. To do this, a series of structurally similar aromatic diamidines related to DB832 Fig. (1A), were synthesized and evaluated for telomere binding. Particular emphasis was directed towards compounds that maintained the core structure of DB832 with the 5-5-6 heterocyclic system. Systematic atom-wise and group-wise modifications and substitutions were undertaken to discover more favorable motifs for quadruplex recognition. The compounds were divided into the following groups according to the modifications of the different molecular units of DB832 - the two furans, the phenyl and the two terminal amidine groups:
Furan modifications: The modifications in this group consisted of the replacement of either one or both furans with either thiophene (DB1438, DB1450, and DB1463) or oxazole (DB1999) systems. The relatively larger size and lower electronegativity of sulfur than oxygen changes the compound curvature and the hydrogen bonding potential respectively, and can affect interactions in the grooves of the quadruplex. The additional nitrogen of the oxazole can act as a potential hydrogen-bond acceptor.
Phenyl modifications: The modifications in this group mainly consist of the replacement of the terminal phenyl with pyridine where the pyridine nitrogen is either ortho (DB934) or meta (DB1693) to the central furan. The additional lone-pair of nitrogen can act as a potential hydrogen-bond acceptor from the guanine –NH2 groups of the quadruplex grooves.
Terminal amidine modifications: The terminal amidines of DB832 were replaced with imidazolines in this group (DB1972 and DB2037). The imidazolines have a relatively planar conformation for the molecules which can affect the stacking ability with the quadruplex while maintaining the high pKa values of the amidines.
Additional modifications: These compounds consisted of miscellaneous modifications that would affect the compound stacking ability with the DNA. Switching the order of the two terminal furan and phenyl groups (DB1093) renders the molecule relatively linear compared to DB832. The addition of an alkyne linker between the phenyl and furans (DB1694) also decreases the curvature of the ligand. The additional –CH3 group on the terminal furan (DB1949) should also affect the stacking ability of the ligand in the complex.
Fig. (1).
(A) Chemical structures of potential quadruplex-targeting agents, derived from DB832, classified based on the modifications. (B) The telomeric quadruplex (Tel24) and control duplex DNA sequences (AATT and (CG)4) used in this study.
An array of biophysical and biological techniques was used to comprehensively characterize the binding of these ligands to the telomeric quadruplex-DNA and their selectivity for duplex-DNA. The results clearly show that, with a combination of systematic design principles, focused synthesis, and careful analysis, it is possible to develop compounds with enhanced selectivity and affinity. We have identified a new series of quadruplex-interactive agents, derived from duplex groove binders, which can be used as paradigms to develop improved compounds.
MATERIALS AND METHODS
DNA and compound synthesis
DNA oligonucleotides Fig. (1B) were purchased from Integrated DNA Technologies (Coralville, IA) with HPLC purification and mass spectrometry characterization. NMR and mass spectral analysis performed on these sequences further confirmed the purity. The concentration of oligonucleotides was determined from absorbance at 260 nm with extinction coefficients calculated by the nearest-neighbor method [36]. The syntheses of ligands DB832, DB934, DB1450, DB1463, DB1438, DB1693 and DB1694 have been published [37]. The syntheses of ligands DB1093, DB1949, DB1972, DB1999 and DB2037 will be reported elsewhere. Distamycin, a commercially available drug, was purchased from Sigma-Aldrich Chemicals. RHPS4 ligand was kindly provided by Dr. Malcolm Stevens, University of Nottingham. Appropriate stock solutions of each compound were prepared in double deionized water or in deuterated water according to the experimental needs. Experiments were performed in 10 mM K2HPO4 buffer containing 80 mM KCl and 3 mM EDTA adjusted to pH 7.3 using 1N HCl.
UV-Thermal melting studies
Thermal denaturation studies were conducted on a Cary 300 BIO UV-visible spectrophotometer in quartz cuvettes of 1 cm pathlength. DNA solutions were prepared in buffer at single-strand concentrations in the range of 2–3 µM. The absorbance of the oligonucleotides was monitored at the recommended wavelength of 295 nm for Tel24 and 260 nm for duplex DNA sequences as a function of temperature. Samples of compound to DNA ratios ranging from 0:1 to 6:1 were prepared. Cuvettes were mounted in a thermal block, and the solution temperatures were monitored by a thermistor in a reference cuvette with computer-controlled heating and cooling rates of 0.5 °C/min. Data were analyzed and plotted using Kaleidagraph 4.0 software.
Circular dichroism (CD)
CD spectra were recorded using a Jasco J-810 spectrophotometer with a 1-cm pathlength quartz cuvette at a scan speed of 50 nm/min and response time of 1 second over the wavelength range of 220 nm to 550 nm. Quadruplex-DNA solutions (4–5 µM) were annealed overnight prior to the collection of spectra. Appropriate amount of ligands were sequentially titrated from the stock solution into the DNA solution in the cuvette until the desired mole ratios of compound to quadruplex were obtained. The spectra were averaged over four scans. A buffer baseline scan was collected in the same cuvette and subtracted from the average scan of each ratio. Data were processed and plotted using Kaleidagraph 4.0 software.
Nuclear magnetic resonance
Tel24 DNA samples were prepared in degassed phosphate buffer and reconstituted in 90% H2O:10% D2O with 0.01 mM DSS as an internal reference. Tel24 concentrations were in the range of 0.1 mM to 0.3 mM unless otherwise mentioned. The final DNA samples were adjusted to pH 7.3 using 1N HCl solution. The final NMR samples were heated past their transition temperature and annealed to room temperature before collecting the spectra. Experiments were performed on a Varian Unity 600 spectrometer. Temperature-dependent 1H-spectra were recorded from 15 °C to 45 °C using jump-return and WATERGATE methods for solvent suppression [38, 39]. Ligands were titrated into the quadruplex DNA with compound to DNA ratios varying from 1 to 4. All NMR data were processed and analyzed with a combination of VNMR (Varian Inc.) and MNova (Mestrelab Research) softwares.
Mass spectrometry
The mass spectrometry analysis was performed on a Waters Q-TOF micromass spectrometer equipped with electrospray ionization source (ESI) in negative mode (Waters Corporate, Milford, MA). The mass range was from 500–3000Da. Since mass spectrometry generally requires DNA sequences dissolved in a volatile salt solution, all the sequences tested with mass spectrometry were dialyzed several times with a 1000 Da cut-off membrane (Spectrum Laboratories Inc., Rancho Dominguez, CA, USA) in 80 mM ammonium acetate buffer at pH 7.2 in order to remove any low molecular weight impurities from DNA. The final concentrations of the dialyzed DNA were determined as above. Samples containing 25–30 µM Tel24 sequence and appropriate amounts of ligands in the desired molar ratios to DNA were prepared in 80 mM ammonium acetate buffer with the final volume of 100 µL. The samples were introduced into the ion source through direct infusion at 5 µl/min flow rate. The instrument parameters were as follows: capillary voltage of 2200V, sample cone voltage of 30V, extraction cone voltage of 2.5V, desolvation temperature of 70 °C and source temperature of 100 °C. Nitrogen was used as nebulizing and drying gas. Spectra were collected for 8 minutes and the last two minutes of the scan were used for data analysis. Data analysis and interpretation were performed using MassLynx 4.1 software.
Taq Polymerase Assay
PCR reactions were performed to amplify the 906–1064 sequence of pBR322 (2.5 ng) in the presence/absence of increasing concentrations of tested derivatives. The reaction was carried out in an Eppendorf thermocycler performing 25 cycles of: 30 s at 94 °C, 30 s at 65 °C and 30 s at 72 °C. The reaction products were resolved on a 2% agarose gel in TBE 1X (89 mM TRIS base, 89 mM boric acid, 2 mM Na2EDTA) and stained by ethidium bromide.
Telomerase Activity Assay
An aliquot of 5×106 HeLa cells (human cervical carcinoma) in exponential phase of growth was pelleted and lysed for 30 min on ice using 100 µl of 0.5% CHAPS, 1 mM EGTA, 25% 2-mercaptoethanol, 1.74% PMSF and 10% w/v glycerol. The lysate was centrifuged at 13000 rpm for 30 min at 4 °C and the supernatant collected, stored at –80 °C. Telomerase activity was assayed using the TRAPeze Telomerase Detection Kit (Millipore) including 1 µg of protein extract as telomerase source. The reaction products were loaded onto a 10% polyacrylamide gel (19:1) in TBE 0.5X. Gels were stained with SybrGreen I (Sigma) and visualized on a Geliance apparatus (Perkin Elmer). For selected compounds, before the amplification step, elongation reaction products were purified by a QIA quick nucleotide purification kit (Qiagen) to remove the ligand (TRAP-lig).
Cell cultures and toxicity assays
HeLa cell lines were maintained in DMEM medium supplemented with 10% heat-inactivated fetal calf serum, 50 U/ml of penicillin G and 50 µg/ml of streptomycin, at 37 °C in humidified atmosphere and 5% of CO2. MTT assays were performed by plating 96 well plates at 10,000 cells/well. After overnight incubation, compounds were added in triplicate and the plates were incubated in presence of the drug for 72 hours. At the end of this period, MTT was added to a final concentration of 0.8 mg/ml. After two additional hours of incubation, the medium was removed and 150 µl/well of DMSO were added. Soluble formazan salts were homogenate by manual pipetting and absorbance at 540 nm was read. The curves consisted of 8 serial dilutions in triplicate in each case, and results were analyzed as sigmoidal dose-response curves.
RESULTS
Selectivity for Quadruplex DNA: UV-Thermal Melting
To determine the selectivity of ligands for human telomeric quadruplex sequence over duplex DNA sequences, thermal melting (Tm) studies were performed with a modified telomeric quadruplex DNA, Tel24, and two different duplex sequences: AATT and (CG)4 Fig. (1B). NMR solution structural studies on Tel24 shows the formation of a highly stable hybrid-1 type quadruplex structure by this sequence [40], which is also predicted to be the predominant structure formed by the wild-type human telomeric sequence. An AT-rich hairpin DNA sequence was chosen since small molecules of the type shown in Fig. (1A) typically exhibit a preferential binding towards double helices with a narrow minor groove, and a GC-rich duplex DNA sequence was used to duplicate the wider grooves of quadruplex DNA. This would eliminate bias towards any particular groove width for the tested ligands.
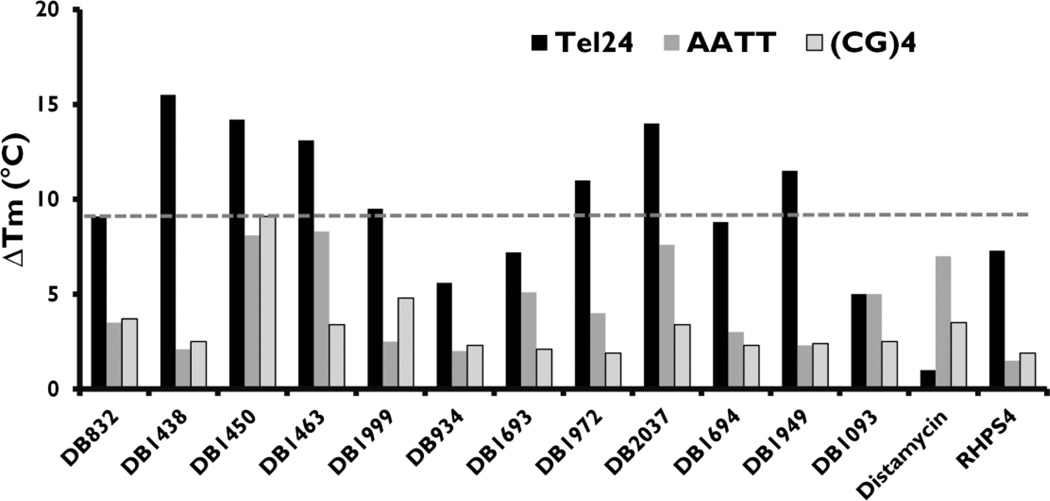
Melting curves were obtained for Tel24 up to a ratio of 6:1 ligands per quadruplex and 4:1 ligands per duplex sequences. This high ratio of ligands per quadruplex was used since the recently solved NMR structure of distamycin bound to an intermolecular quadruplex structure shows that up to four distamycin molecules are bound in the opposite grooves of the d[TG4T]4 intermolecular quadruplex [41]. The ∆Tm values for Tel24 (4:1 ratio) and duplex sequences (2:1 ratio) are represented by bar graphs in Fig. (2). Ratios higher than those listed in Fig. (2) resulted in significant complex aggregation and produced inconsistent melting curves for some compounds.
Fig. (2).
UV-Thermal melting studies of the ligands in Fig. (1) with Tel24 (black bars), AATT duplex sequence (grey bars) and (CG)4 duplex sequence (line bars). Full DNA sequences are listed in Fig. (1). The Tm values plotted are for the drug:DNA ratio of 4:1 with Tel24 and 2:1 with hairpin-DNA. The Tm of ligand-free Tel24, AATT and (CG)4 are 67, 69 and 63 °C respectively. Distamycin and RHPS4 (structures not shown) are employed as reference compounds. The thermal melting values reported are reproducible within ±1 °C.
The melting temperature of Tel24, AATT and (CG)4 sequences in 80 mM KCl are 67, 69 and 63 °C respectively. DB832, the lead compound, showed a ∆Tm of 9.1 °C at 4:1 ratios indicating significant stabilization of the quadruplex. Also, with the two duplex sequences, AATT and (CG)4, DB832 shows very little increase in the Tm (3.5 °C and 3.7 °C respectively) suggesting selectivity over duplex DNA. Interestingly, DB1093, a linear structural isomer of DB832, showed a small increase in Tm at 4:1 ratio (5 °C) with quadruplex DNA, highlighting the importance of the neighboring furans for effective recognition of quadruplex architecture. DB1093 also exhibited very little change in ∆Tm for duplex sequences suggesting that the compound is too linear to interact with the duplex DNA minor groove. DB1999, the oxazole derivative of DB832, and DB1949, with methyl-furan substitution, had slightly better quadruplex stabilization potential than DB832 (9.5 and 11.5 °C respectively) and with lower affinity for duplex sequences. DB1694, with an additional acetylene linker between the phenyl and furan, did not change the thermal stability of either quadruplex or duplex sequences.
Modification of the furans with thiophenes showed the greatest change in the stability of the Tel24 sequence. Replacing the terminal furan with a thiophene, DB1438, significantly increased the stability of Tel24 sequence by 15.5 °C and with very low affinity for duplex sequences. Similar quadruplex stabilization was observed when the central furan or both the furan systems were replaced by thiophenes, DB1463 (13.1 °C) and DB1450 (14.2 °C) respectively, but the latter two modifications also resulted in an increased stability for the AATT sequence. However, DB1463 showed much lower affinity for the (CG)4 sequence as compared to DB1450, suggesting the effect of compound curvature on recognizing different duplex structures.
Nitrogen substitution on the phenyl ring of DB832 decreased quadruplex stabilization when placed in the meta position to the central furan (DB1693, 7.2 °C) and an even further decreased affinity when it is ortho (DB934, 5.6 °C). The conversion of terminal amidine groups to imidazolines (DB1972 and DB2037) yields a more planar arrangement while still maintaining a dicationic charge system. These two compounds exhibited an increase in the thermal stability of Tel24 sequence but also bound relatively stronger to duplex sequences.
Two well-characterized compounds, distamycin and RHPS4 (structures not shown) were also evaluated as reference. Distamycin, a groove binder with the four-stranded [d(TG4T)]4 intermolecular quadruplex, did not show any significant increase in the thermal stability of Tel24 sequence, but it is a strong binder to AT-rich sequences in duplex DNA. Distamycin is the only compound of this group that has reverse selectivity for duplex over quadruplex. RHPS4, a well-known quadruplex stabilizing compound known to interact at the terminal tetrads of human telomeric sequence [42], exhibited very similar melting values as DB832 and with very little affinity for duplex sequences.
The thermal stabilities in Fig. (2) clearly indicate that some ligands are effectively stabilizing the quadruplex conformation without simultaneously improving duplex interaction. The results also show that it is possible to design out the strong binding to duplex sequences that are characteristic of classical compounds such as distamycin. Therefore, these compounds make ideal candidates for further study as highly selective quadruplex interacting agents.
Binding Mode Analysis: Circular Dichroism Pattern Recognition
Circular dichroism (CD) has emerged as an important non-invasive technique for conformation analysis of biomolecules and for providing insights about the binding modes of small molecules with DNA using pattern recognition [43]. When an achiral ligand, which has no CD by itself in solution, binds to a chiral macromolecule such as DNA, an induced CD (ICD) signal can be observed in the wavelength region corresponding to the bound achiral ligand [44, 45]. The ICD signal can be used to obtain information on the binding mode of the ligand. The ICD signal can be a small positive or negative signal, as in the case of duplex intercalators, or can be a large positive signal, as in the case of duplex groove binders [30, 34, 46–49]. A combination of both positive and negative CD signals, coupled with different ligand ICD signal shapes generally indicates formation of stacked complexes, as was previously shown with DODC complexes with quadruplex systems [50].
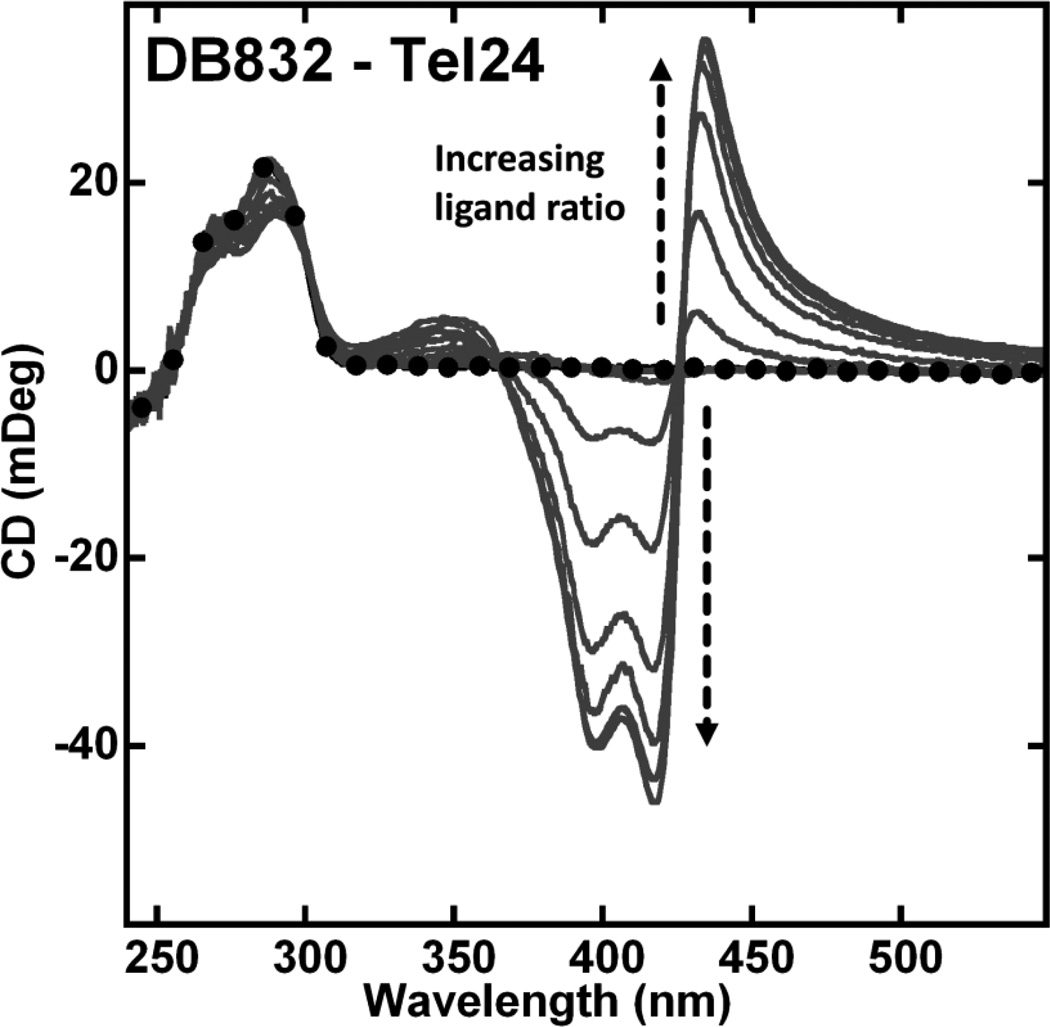
Circular dichroism studies were performed to evaluate how modifications on the DB832 system influence ligand interaction modes and the DNA conformation in the complex. CD studies with DB832 binding to the human telomeric quadruplex DNA are characterized by a large positive (435 nm) and two negative (390 and 418 nm) ICD signals in the wavelength range corresponding to the absorbance of bound DB832 Fig. (3) in agreement with previous studies [35]. As the added ratio of DB832 is increased, increasing ICD signals occur until saturation of quadruplex binding sites. The ICD pattern observed with this sequence is very similar to the previously reported pattern with the wild-type telomeric sequence suggesting the formation of a similar stacked complex. Small CD changes in the DNA absorbance region upon complex formation indicate the pre-formed hybrid conformation as the preferred quadruplex conformation by the compound. Using the ICD pattern exhibited by DB832 as a model for quadruplex multiple-site recognition, CD studies were conducted for the compounds in Fig. (1A) with Tel24 to investigate if a similar pattern exists.
Fig. (3).
CD spectra of DB832 with Tel24 quadruplex sequence at 25 °C. Single strand quadruplex concentration is 5 µM. Filled circles represent free Tel24. The arrow indicates increasing ratio (0:1 to 8:1) of ligand to Tel24 until no further change in ICD is observed.
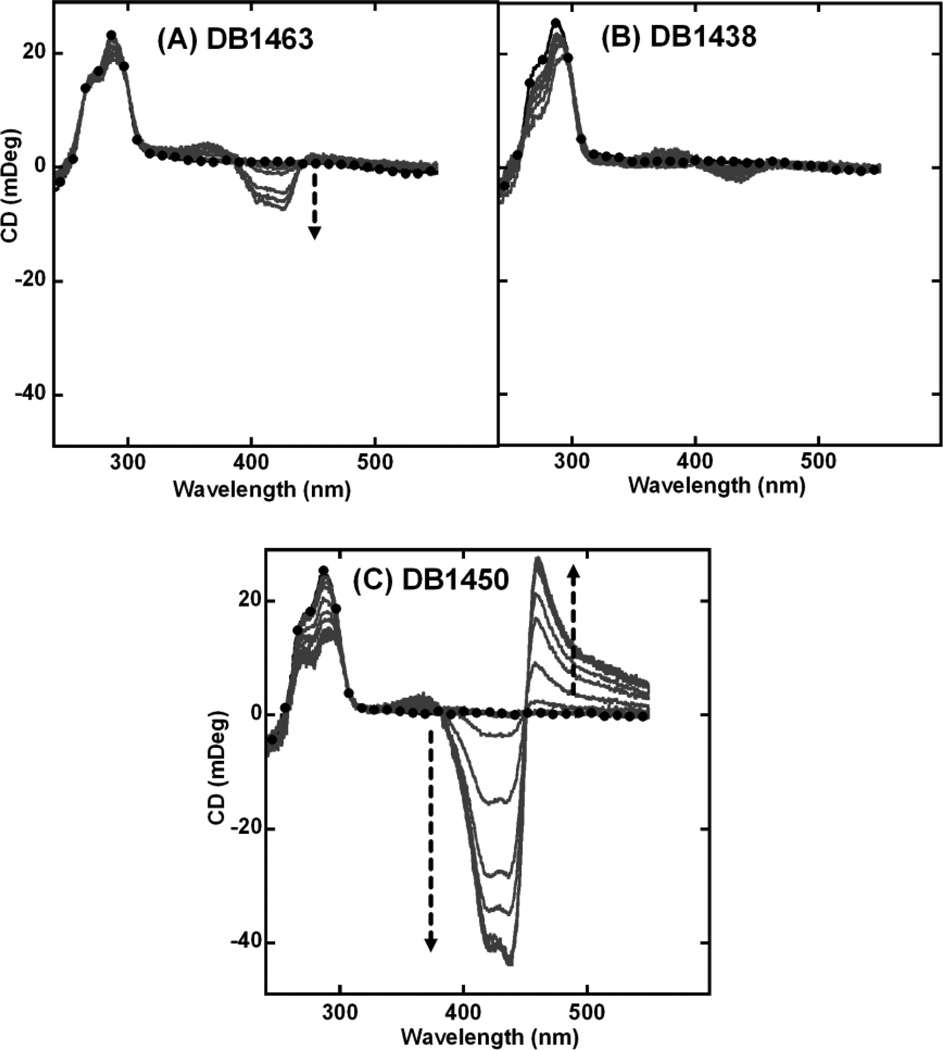
Thermal melting studies of thiophene containing diamidines showed that these compounds have significant quadruplex stabilization potential with varying degrees of duplex selectivity. The two monosubstituted thiophene analogs, DB1463 and DB1438, exhibit a smaller magnitude of ICD signals (Figs. (4A and 4B), respectively) as compared to DB832. However, the disubstituted DB1450 has a very similar exciton-type splitting as DB832 but of relatively lower magnitude Fig. (4C).
Fig. (4).
CD spectra of thiophene containing DB compounds (A) DB1463, (B) DB1438, and (C) DB1450 with Tel24 quadruplex sequence. Single strand quadruplex concentration is 4–5 µM. Filled circles represent free Tel24. The arrow indicates increasing ratio (0:1 to 10:1) of ligand to Tel24 until no further change in ICD is observed.
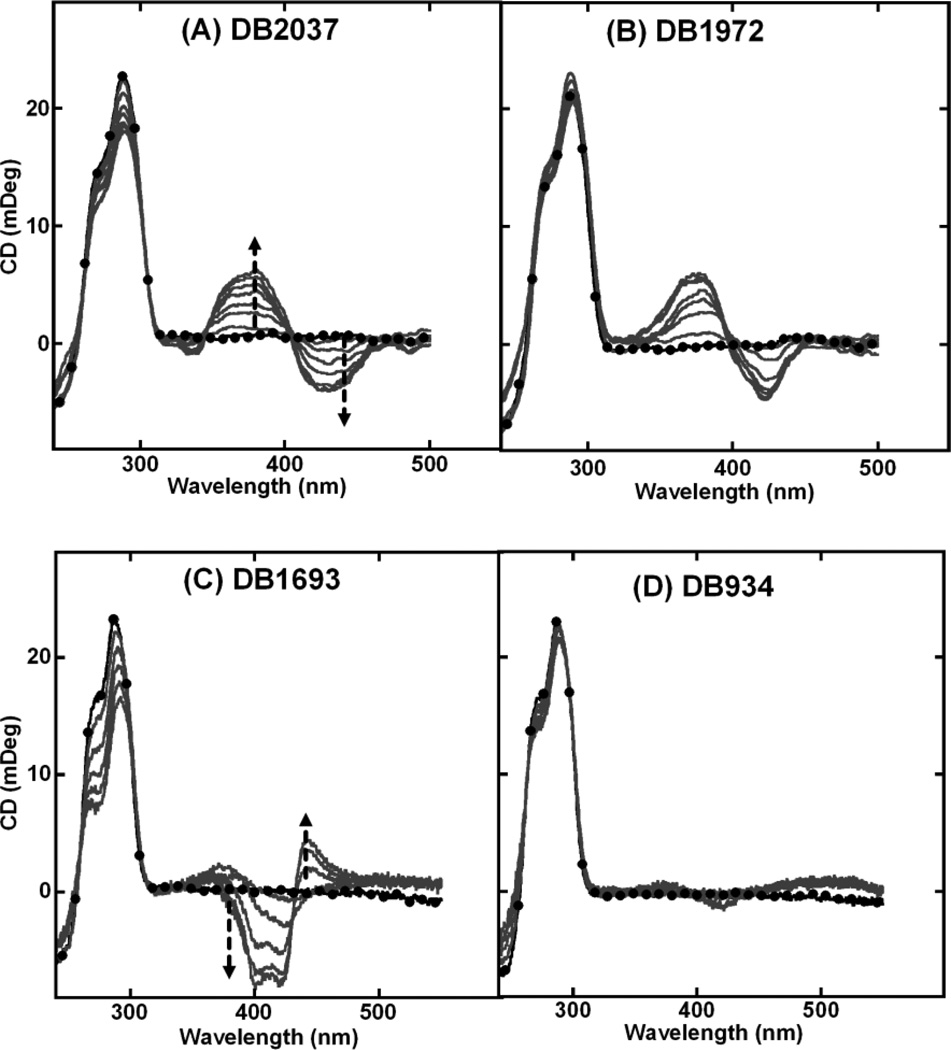
The imidazolines of DB2037 and DB1972 affect the equilibrium structure of the compound because the imidazoline relative to an amidine reduces the torsional angle with the neighbor ring system resulting in a more planar shaped molecule. The magnitudes of the ICD observed for these two ligands are very small with small positive and negative peaks Figs. (5A and 5B) suggesting a transition to a primary end-stacking mode. DB1693, with meta nitrogen substitution on the phenyl ring, exhibited a weak exciton-type splitting Fig. (5C), and DB934, with ortho nitrogen substitution, exhibited virtually no ICD even at very high ratios Fig. (5D).
Fig. (5).
CD spectra of (A) DB2037, (B) DB1972, (C) DB1693 and (D) DB934 with Tel24 sequence. Single strand quadruplex concentration is 4–5 µM. Filled circles represent free Tel24. The arrow indicates increasing ratio (0:1 to 10:1) of ligand to Tel24 until no further change in ICD is observed.
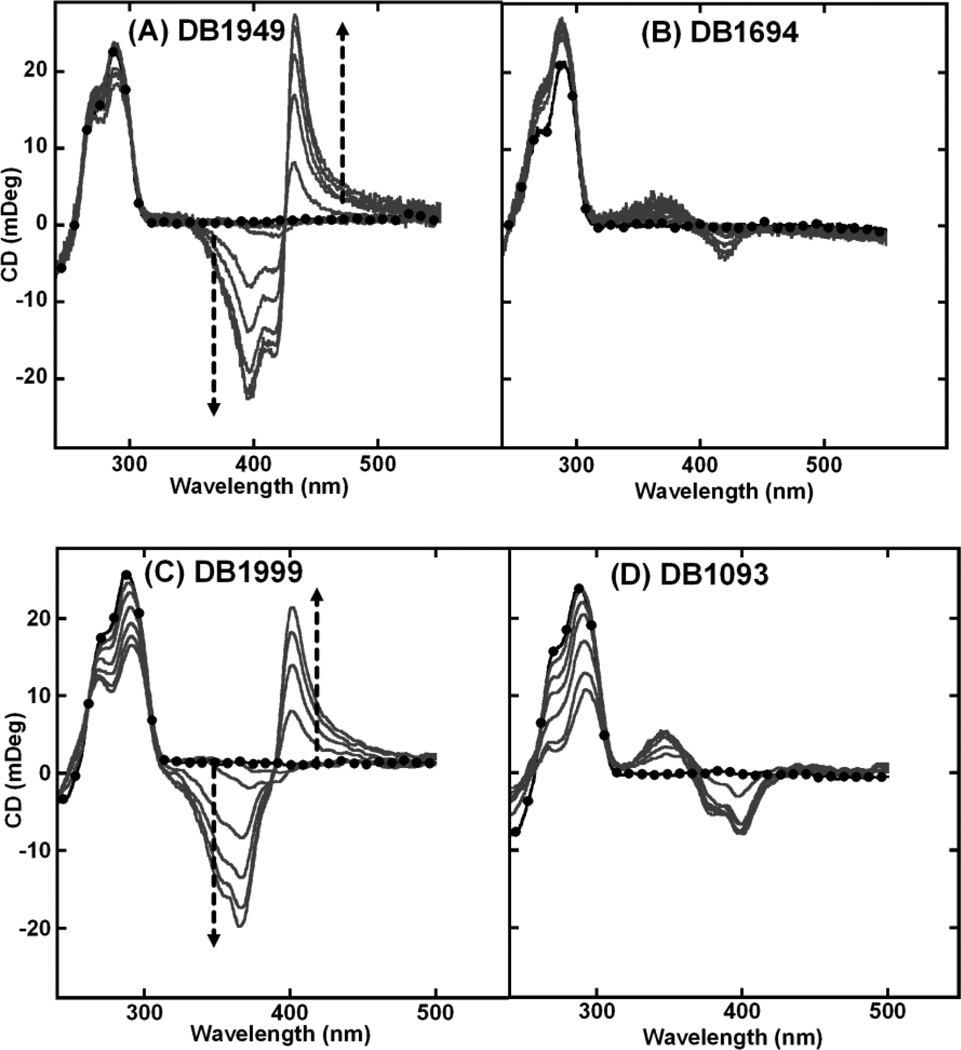
The addition of a methyl group on the terminal furan (DB1949) and the substitution of a central furan with an oxazole (DB1999) yielded ICD patterns similar to DB832 (Figs. (6A and 6C) respectively). These modifications apparently did not significantly affect the interaction mode with Tel24. The only difference that emerges by comparing the spectra is the magnitude of the signal at the ICD signal saturation ratio. DB1694, with an additional triple bond, and DB1093, a structural isomer of DB832, exhibit similar ICD patterns but of different magnitudes with Tel24 sequence (Figs. (6B and 6D) respectively).
Fig. (6).
CD spectra of (A) DB1949, (B) DB1694, (C) DB1999 and (D) DB1093 with Tel24 sequence. Single strand quadruplex concentration is 4–5 µM. Filled circles represent free Tel24. The arrow indicates increasing ratio (0:1 to 12:1) of ligand to Tel24 until no further change in ICD is observed.
Multiple-site recognition: Nuclear magnetic resonance studies
Telomeric quadruplex sequence exists as a mixture of conformations under physiological conditions in K+, with the hybrid conformation as a predominant structure [40, 51–53]. The NMR structure of Tel24 shows that this sequence primarily folds into a single, mixed (3+1) parallel/antiparallel hybrid-1 type structure with three accessible grooves of varying dimensions [40, 54]. To test the binding modes exhibited by some of the compounds, 1H-NMR titration studies were conducted with Tel24 with some of the ligands in the series. Imino proton signals of guanines are excellent probes to determine if a G-rich sequence folds into a quadruplex conformation [55]. These imino protons resonate between 10.0 to 12.5 ppm in a quadruplex structure and are readily observable in water sample. Moreover, the possible presence of multiple quadruplex conformations can also be readily addressed by monitoring the number of imino protons. Binding of a small molecule to a quadruplex structure affects the chemical environment of imino protons which can also be identified from simple titration experiments.
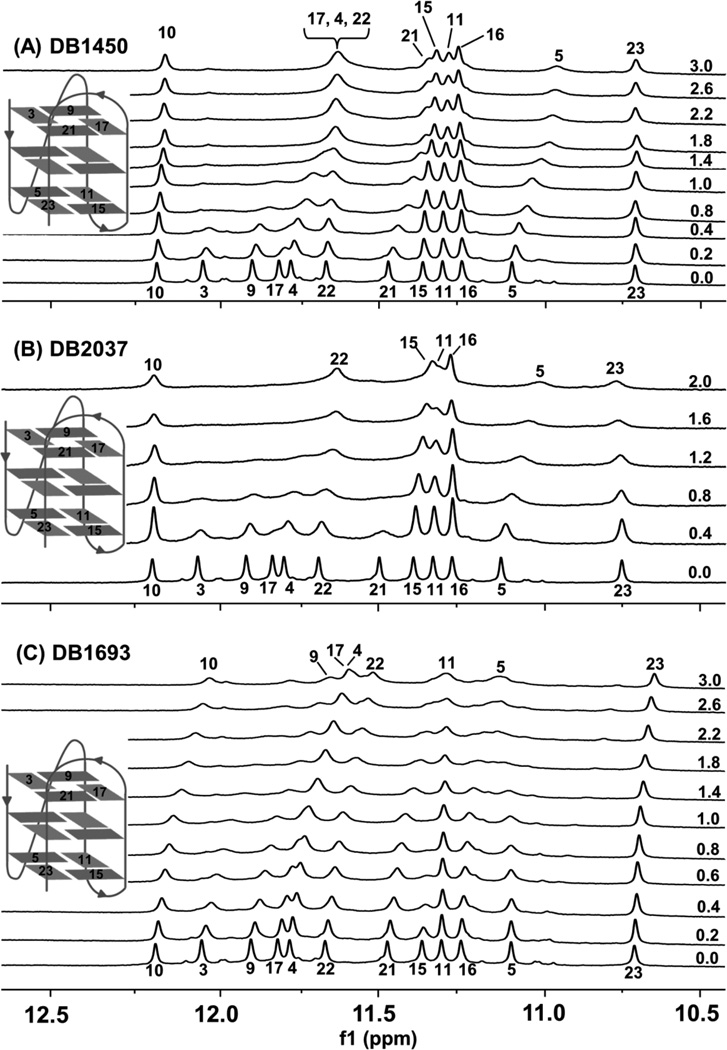
Figs. (7A–7C) show the guanine imino proton NMR spectra of selected ligands with the Tel24 sequence at 25 °C. The solubility of the compound and complex become an issue at compound to quadruplex ratios higher than 3:1 at NMR concentrations. Considerable aggregation is observed at high compound concentrations resulting in significant line broadening and rendering it difficult to obtain spectra for analysis. In the absence of any ligands (Fig. (7), 0:1 ratios), the spectra show distinct number of signals for imino protons corresponding to the total number of guanines in the Tel24 sequence. Titration of the compounds with Tel24 resulted in significant changes in the imino proton spectra and spectral broadening. Changes in the chemical shifts reveal that the ligands were in fast-exchange on the NMR timescale. It is quite informative that a number of imino resonances broaden in a site-specific fashion, consistent with the local chemical environment of various residues being perturbed differentially by ligand binding.
Fig. (7).
Imino proton spectra of Tel24 with (A) DB1450 and (B) DB2037 and (C) 1693 at 25 °C. Inset shows the hybrid-1fold of Tel24 reported in Luu et al [40]. The drug/quadruplex molar ratios are shown along the side of the spectra. Proton assignments of Tel24 were kindly provided by Dr. Anh T. Phan, Nanyang Technological University.
DB1450 recognizes multiple sites of the quadruplex sequence based on CD results. The imino protons of G3, G9, G17 and G21, that constitute the 5’-terminal tetrad of the quadruplex, are the most perturbed at the lowest DB1450 to quadruplex ratio Fig. (7A). They exhibit significant upfield changes accompanied by complete broadening and this suggests that the primary binding site of DB1450 is the 5’-terminal tetrad. Upon further addition of the ligand distinct site-specific chemical shift changes are observed. Imino protons of G4 and G22 that correspond to the guanines of the middle tetrad and G5 of the bottom tetrad exhibit site-specific perturbations with small upfield shifts accompanied by broadening. Imino protons of G10 and G11 encompassed by the G9, G11, G15 and G17 plane of Tel24 exhibit very little change suggesting that the dimensions of this quadruplex groove are not optimal for the initial interaction of DB1450.
Fig. (7B) shows the imino proton spectra of Tel24 sequence with DB2037. At low drug-DNA ratios site-specific changes are observed in the imino protons of terminal tetrads. The 5’ G-tetrad imino protons broaden considerably and disappear completely at high ratios. The high perturbation of the imino protons at the top end suggests the primary stacking of the ligand on the 5’ G-quartet. The imino protons of the bottom tetrad comprised of G5, G11, G15 and G23 exhibit small perturbations in chemical shifts. The G5 imino proton, in particular, exhibits downfield shifts followed by complete broadening; whereas, G11, G15 and G23 exhibit broadening in a similar way. Therefore, the 3’ terminal tetrad might be the secondary binding site for DB2037. G11 and G15 exhibit broadening to some extent; this is probably due to the presence of a stable A24·T13 base pair capping at the 3’ terminal (as observed in NMR [40]), offering protection of the imino protons from solvent exchange.
Imino proton spectra of DB1693 with Tel24 Fig. (7C) reveals some interesting binding characteristics. Based on the CD results, this compound was predicted to exhibit a mixed binding mode with an initial stacking at the terminal tetrad followed by binding in the grooves. The imino protons of the 5’ tetrad exhibit very similar changes (downfield shifts and broadening) confirming the initial stacking at the terminal tetrad. Interestingly, G15 and G16 located in the “narrow” groove of the quadruplex exhibits complete broadening consistent with a site-specific broadening due to the local perturbation upon complex formation. This narrow groove might have the optimal width to accommodate the DB1693 molecule resulting in the observed changes. It must be emphasized, however, that the imino proton signals broaden at relatively low ratios and contain little information at the drug-DNA ratios where the largest ICD signal is observed.
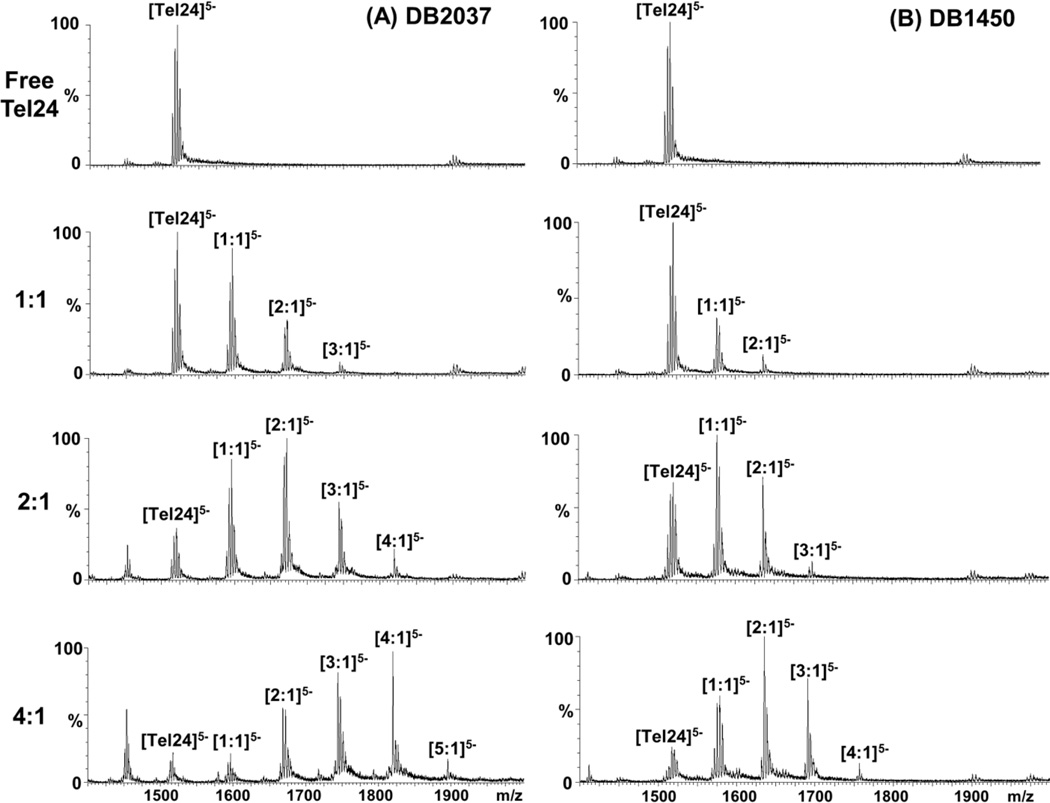
Stoichiometry evaluation: Mass spectrometry
Mass spectrometry has developed into a powerful analytical tool to determine important features of biomolecular interactions such as binding kinetics, conformational changes and stoichiometry [56, 57]. To evaluate the binding stoichiometries with Tel24 sequence, ESI-mass spectra were analyzed with DB2037 and DB1450 (Figs. (8A and 8B) respectively). The mass spectra of free DNA and the ligand-DNA complexes at different ratios were detected at three different charge states (−Z=3, 4, 5) with the −5 charge state being the dominant species. The peaks are labeled only for the charge state of −5 for data interpretation. The Tel24 sequence with DB2037 at 1:1 ratio showed the formation of the 1:1 complex as the predominant species and with a relatively smaller percentage of 2:1 species. At a ratio of 2:1, DB2037 shows the formation of a predominant 2:1 complex and a decreased 1:1 complex. Interestingly, at this ratio, the formation of high stoichiometry complexes (3:1 and 4:1 peaks) is also observed. The relative abundance of free DNA is reduced by a significant proportion at this ratio. Upon further titration of the ligand (at 4:1 DB2037 to Tel24), the most abundant peak observed was for the 4:1 complex and a smaller percentages of 5:1, 3:1, 2:1 and 1:1 peaks. The relative abundance of free DNA at this ratio is negligible.
Fig. (8).
ESI mass spectra of Tel24 with (A) DB2037 and (B) DB1450 at different ratios. Tel24 concentration is 25 µM in 80 mM ammonium acetate buffer adjusted to pH 7.2. The peaks at different ligand to DNA ratios are labeled only for the predominant charge state of −5.
Fig. (8B) shows the ESI-mass spectra of Tel24 with increasing ratios of DB1450. At a ratio of 1:1, DB1450 shows the formation of a 1:1 complex and a weaker 2:1 complex. As the ratio is increased (at 2:1 ratio), there is a significant increase in the relative abundance of the 1:1 and 2:1 complexes and a small amount of 3:1 complex is observed. Upon further titration of DB1450 (4:1 ratio), the formation of the 2:1 complex is dramatically increased with a relative reduction of 1:1 complex and the free DNA. The relative abundance of 3:1 complex is also significantly increased as compared to the 2:1 ratio and a small amount of 4:1 complex is observed. The mass spectra results, thus, clearly support the CD finding that these compounds can exhibit complexes with multiple stoichiometries and binding modes.
Biological studies
All derivatives were assayed for their ability to interfere with enzyme-mediated DNA polymerization and telomere extension. In the first instance, we used TAQ polymerase as target protein. Generally, all test compounds impaired the amplification reaction with IC50 values in the 10–40 µM concentration range. The two most efficient derivatives turned out to be DB1450 and DB2037 (IC50 of 1.8 and 3 µM, respectively). This actually correlates with the higher stabilization of different DNA arrangements (like an AATT sequence) promoted by these derivatives. None of the test compounds showed a significant level of selective inhibition of the telomerase elongation process. As expected from first generation compounds, further optimization in the selective recognition of G-rich sequence will be required based on the SAR from the compounds.
Finally, preliminary assays on the cytotoxic properties of these compounds were performed. MTT assays carried out after a 72h drug-treatment on HeLa cells indicate that the diamidine derivatives are poorly cytotoxic after this short term treatment. Distinctly, DB1972 and DB2037, which contain the two terminal imidazolines, showed IC50 values < 1 µM. For these two compounds an end-stacking DNA binding mode is suggested which can possibly describe their cytotoxic properties [58]. Additionally, a theoretical evaluation of the distribution coefficient confirmed that the diamidine-substitution with the imidazolines causes a reduction of the LogD7.5 which can result in an altered drug cell uptake. Thus, it will be worthwhile to further investigate if this result is related to a different cellular distribution or to a modulation of DNA recognition.
DISCUSSION
With the evidence for in vivo existence of G-quadruplexes becoming more compelling and the abundance of information available regarding their potential role in cancer-specific and other biological activities, G-quadruplex structures represent a new class of DNA-based therapeutic targets. The design and development of small molecules that specifically bind to these unique DNA secondary structures can allow specific targeting and significantly decrease the side effects associated with current chemotherapeutic regimens. The extensive variation of quadruplex structures throughout the genomic DNA also allows the design of compounds that can target particular quadruplex structures with high selectivity.
The single-stranded G-rich sequences of telomeric DNA provide an optimal platform for anticancer therapy since small molecules can successfully induce and stabilize these G-quadruplex structures, a template not recognized by the telomerase enzyme which is active in many types of malignant tumor cells. Small molecules that can selectively bind to the various human telomeric quadruplex conformations or induce a conformational switch from one conformer to the other can have a significant therapeutic impact. The dynamic structural variations exhibited by telomeric G-rich DNA have resulted in several classes of small molecules that can effectively bind to or induce various G-quadruplex conformations, and thereby inhibit telomerase activity. The porphyrin and acridine core, for example, have been excellent starting points to develop several important telomerase inhibitors such as TMPyP4, BRACO19 and RHPS4 [22, 23, 59–61]. Most of the quadruplex-interactive small molecules previously discovered have relatively planar, highly aromatic scaffolds that are conformationally constrained and, as a result, primarily interact with quadruplex units by stacking on either one or both ends of the terminal G-tetrads. Moreover, the relatively exposed terminal tetrads of different quadruplex architectures offer very little diversity and therefore successfully designing ligands to recognize subtle differences at the terminal sites of G-tetrad is difficult.
Screening for compounds able to interact with alternative sites of quadruplex DNA is an attractive strategy because of the potential to increase specificity for quadruplex binding. Because of the different geometries of quadruplex and duplex grooves as well as different patterns of hydrogen-bond donors and acceptors along the grooves, compounds that can bind in quadruplex grooves can have excellent structure-specific recognition potential. Duplex DNA minor groove binders such as distamycin and its derivatives demonstrate weak interactions in various grooves of simple quadruplex systems with high stoichiometry [41, 62]. As can be seen in Fig. (2), these compounds bind much more strongly to duplex than quadruplex DNA. A cyanine molecule, DODC (3, 3’-diethyloxadicarbocyanine iodide), exhibits exciton-type ICD signals in the compound absorbance region upon complex formation that can be attributed to interactions in the quadruplex grooves [50]. These results clearly demonstrate that targeting grooves of quadruplexes is a viable strategy that needs further investigation.
Heterocyclic diamidines represent an important class of duplex-DNA minor groove targeting agents with excellent cell uptake and biological activity in animals and humans [25–27]. Compounds in this group that have relatively weak DNA duplex interactions can act as initial model systems to screen for novel structural motifs for selective G-quadruplex grooves recognition. DB832 was shown to interact with the human telomeric quadruplex with a unique induced CD pattern suggestive of stacking of the ligands in the grooves. This finding is important since small molecules that can bind in the grooves of biologically relevant G-quadruplex motifs such as telomeric DNA have not been reported so far. To understand all the molecular features of DB832 that contribute towards quadruplex groove recognition, a series of structurally similar heterocyclic diamidines were prepared and evaluated with a telomeric quadruplex model system as well as with duplex DNAs. Several well-established techniques such as thermal melting, circular dichroism, nuclear magnetic resonance and mass spectrometry were performed to determine the selectivity, binding mode and stoichiometry of the ligands upon complex formation.
UV-thermal melting measurements provide a very effective screening tool to rank the relative affinities of related compounds for nucleic acid sequences and structures [63, 64]. Thermal melting studies with DB832 and related analogs showed varying degrees of telomere affinity with different functional group modifications suggesting that subtle structural changes in the compounds affect their thermal stabilization of quadruplexes as well as duplexes. The ligand-induced thermal stabilization of quadruplexes can also be used as a screening device to select compounds for more detailed studies. In this case the ∆Tm value of the Tel24 quadruplex sequence with DB832 was used as the cutoff point and several compounds in Fig. (2) have ∆Tm values of greater or approximately equal value to DB832. The thiophene derivatives (DB1438, DB1450 and DB1463, Fig. (1A)) clearly constitute the majority of the ligands with high affinity for the Tel24 sequence. Two of the thiophenes have quite high ∆Tm values for the duplexes and are of less development interest. They are, however, quite useful for SAR studies to distinguish and understand quadruplex and duplex recognition features. DB1438, with a central furan and a terminal thiophene, is clearly a compound of interest due to a slightly lower affinity than DB832 for duplex sequences. DB1949 has a terminal methyl-substituted furan which adds affinity to the quadruplex interaction while slightly reducing the duplex binding. The acetylene linkage in DB1694 does not significantly change the quadruplex ∆Tm but it slightly lowers the duplex value, probably due to an unfavorable shape for minor groove recognition. The two imidazoline derivatives have improved quadruplex ∆Tm values but also bind more strongly to the duplex. It is worthwhile, however, to consider other future additions to the amidines that could enhance quadruplex stability as well as additional methyl groups on the aromatic systems. In summary, the ∆Tm studies have identified several compounds that have impressive telomere binding and selectivity over duplex interactions.
Circular dichroism pattern recognition has been validated for determination of ligand binding modes with duplex DNA [43]. Intercalating compounds, for example, usually show very small negative or positive induced CD signals. Minor groove binders are generally characterized by the presence of a large positive induced CD signal upon complex formation with DNA duplexes [43, 65]. The magnitude of ICD can be a qualitative indication about the affinity of the compounds to the DNA, if they interact with the same binding mode [65]. CD patterns attributable to specific recognition modes for quadruplex DNA have not been definitively established since most of the quadruplex-interactive ligands reported so far primarily recognize the terminal G-tetrads and are characterized by weak induced CD changes upon complex formation. Induced CD patterns obtained with molecules such as DODC and DB832 can be used as a reference to screen for compounds interacting in quadruplex grooves.
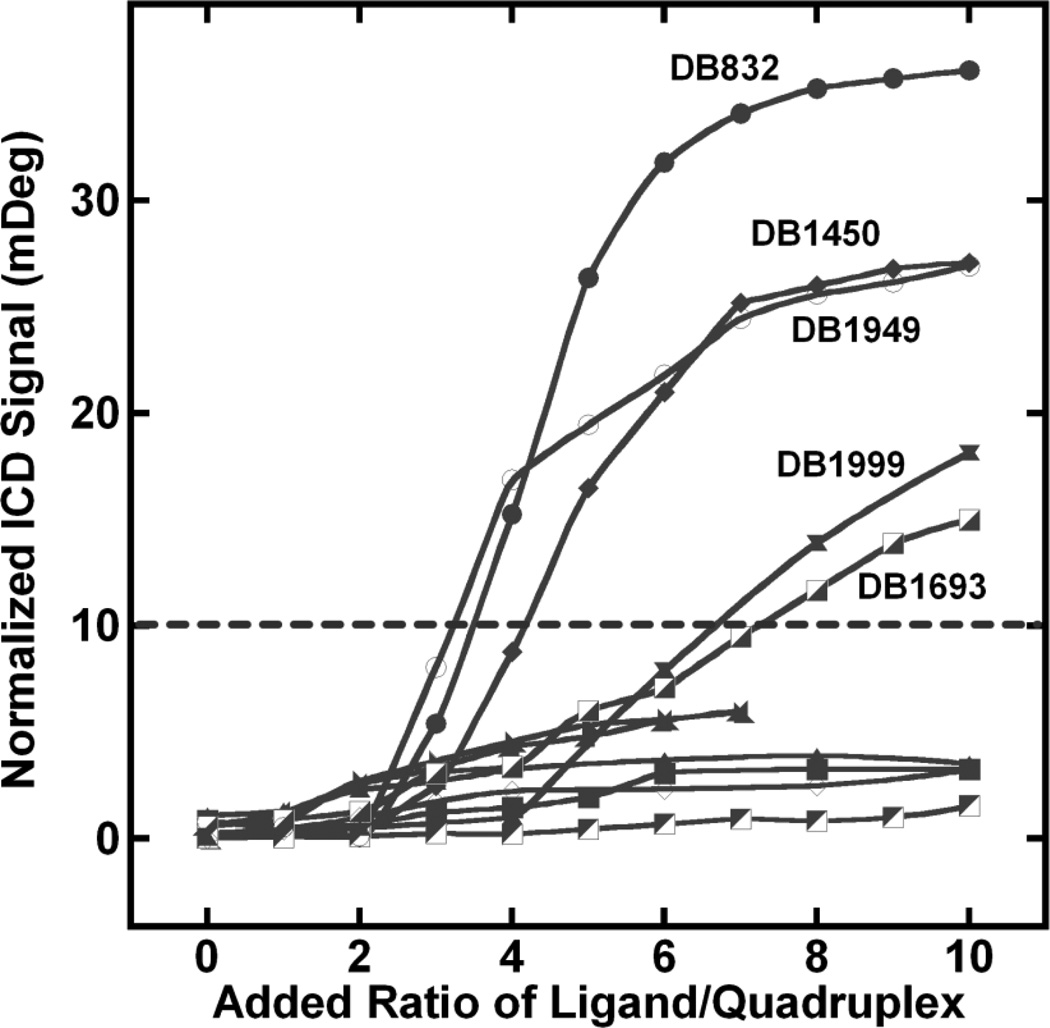
Analysis of the ICD signal exhibited by many of the compounds reveals a unique pattern emerging upon complex formation. Fig. (9) shows the plot of ICD signal at the wavelength corresponding to the maximum positive CD signal of different compounds versus the added ratio of the ligand. A very weak ICD signal is observed for all of the compounds until the added ratio of 2:1 and is followed by a dramatic increase in ICD above ratios 2:1 for some ligands. A weak induced CD signal with duplex DNA is generally observed for intercalation-type mechanisms [49]. However, with quadruplexes, intercalation is not commonly observed due to the high energetic cost required to replace the coordinating cations and tetrad stacking interactions along the quadruplex helical axis. Therefore, intercalation has been ruled out as the possible binding mode for the compounds. The small, initial ICD signals thus suggest a primary end-stacking mode with the Tel24 structure. The aromatic groups of these ligands can favorably stack on the terminal G-tetrads to form optimal interactions.
Fig. (9).
Plot of added ligand ratio versus the ICD signal at wavelength corresponding to the maximum absorbance of the bound ligand with Tel24.
As the ratio of ligand to quadruplex raised above 2:1, a new CD pattern and binding mode is observed in some cases. The compounds can be divided into two groups based on the ICD changes: one group shows an exciton-type spectrum that is relatively large while a second group shows much smaller or no ICD. At ratios higher than 2:1, DB1450, DB1949, DB1999 and DB1693 show ICD signals very similar to DB832. DB1450 (with two thiophenes) has a very similar ICD pattern to DB832, suggesting analogous interactions in the quadruplex grooves. Interestingly, either of the single thiophene substitutions (DB1438 or DB1463) removes the large ICD signal. The reason for the different ICD patterns for the thiophene analogs is not clear at present. Possible reasons for the differences are that sulfur is larger than oxygen, has lower electronegativity, can significantly affect the curvature of the compound, the stacking ability, and has weaker hydrogen bonding potential.
DB1949, the methyl-substituted furan analog, also shows ICD patterns very similar to DB832, but of lower magnitude. Tm studies of this compound showed an increase in the thermal stability with Tel24 and a relative decrease in Tm for duplex sequences. The additional methyl group on the furan of DB1949 can affect the torsional angle with the neighboring amidine group and also can improve the ligand stacking energetics. DB1999, the oxazole derivative of DB832, and DB1693, with the meta-pyridine, show weaker but similar ICD signals suggestive of a possible reduced recognition of the quadruplex grooves.
The different patterns of ICD suggest formation of different stacked complexes within the quadruplex grooves. Most of the compounds in this study have a very small effect on the quadruplex conformation as can be seen from the CD signals corresponding to the DNA region Figs. (3–6). The very small effect on DNA conformation upon ligand binding indicates that the ligands are preferentially binding to an already preformed hybrid structure. The fact that substitution of a single atom makes a large change in compound ICD signal shows that recognition of the quadruplex by a stacked species is extremely sensitive to the compound structure. The four compounds (DB1450, DB1949, DB1999 and DB1693) clearly show that systematic modifications can greatly influence the recognition of alternate sites on a quadruplex unit.
Other compounds in Fig. (1) show weak ICD signals suggesting an end-stacking binding mode with the quadruplex. Two compounds of particular interest are the imidazoline analogs, DB1972 and DB2037 Fig. (1A). The planar conformation of these two ligands suggests a more favorable end-stacking interaction with the external tetrads of the quadruplex. As predicted, these ligands have ICD patterns that are suggestive of pure end-stacking interactions. The slightly higher quadruplex ∆Tm observed with these ligands might be due to the increase in the stacking energetics of the planar imidazolines with the terminal tetrads [58].
1H-NMR studies of some of the DB832 analogs with the Tel24 sequence confirms multiple-site recognition by the ligands Figs. (7A–7C). Significant perturbations in the chemical shifts (downfield shifts and broadening) for some guanine imino protons of the 5’-terminal tetrads (G3, G9, G17 and G21) are observed suggesting the 5’-site as the primary binding site for the ligands. Titration of DB1450 also exhibits site-specific chemical shift changes of G4, G5 and G22 bases upon complex formation Fig. (7A). G4, G5 and G22 are located in an accessible groove that has been classified as a “medium-sized” groove in the Tel24 structure. Therefore, after the initial end-stacking mode at the 5’-terminal, there is a very likely possibility that stacked species of DB1450 are interacting with the “medium-sized” groove encompassed by the G3, G5, G23 and G21 plane. Imino protons of G15 and G16 do not exhibit any chemical shift changes upon titration of the ligands. This groove is classified as the “narrow” groove and therefore might not have the optimal width to facilitate stacking of the ligands. The terminal imidazolines of DB2037 render a more planar conformation which is suitable for stacking at the terminal tetrads, as also observed in CD studies. Titration of DB2037 with Tel24 also exhibits significant perturbations of the 3’-terminal guanine imino protons suggesting the 3’-terminal tetrad as a secondary binding site for DB2037 Fig. (7B). A crystal structure of an acridine derivative with telomeric RNA quadruplex shows two molecules stacked in the same plane next to each other on the terminal tetrad of the quadruplex [66]. We hypothesize a similar interaction of DB2037 with Tel24 sequence where two molecules each are bound side-by-side on the top and bottom end of the tetrads.
NMR studies of distamycin, Hoechst-33258, and ethidium with different quadruplex forming sequences has revealed similar perturbations of the imino protons of the top tetrad of the quadruplex indicating stacking on the top end of the quadruplex [55, 67]. Small molecules, which have excellent duplex-DNA groove binding properties, such as Hoechst-33258 and distamycin, thus generally bind as weak end-stackers with natural quadruplex systems. Therefore, it is probable for the compounds of Fig. (1) to exhibit similar end-stacking as the primary mode of binding to the telomere. This also highlights the severe limitations in designing ligands that have pure groove-binding characteristics.
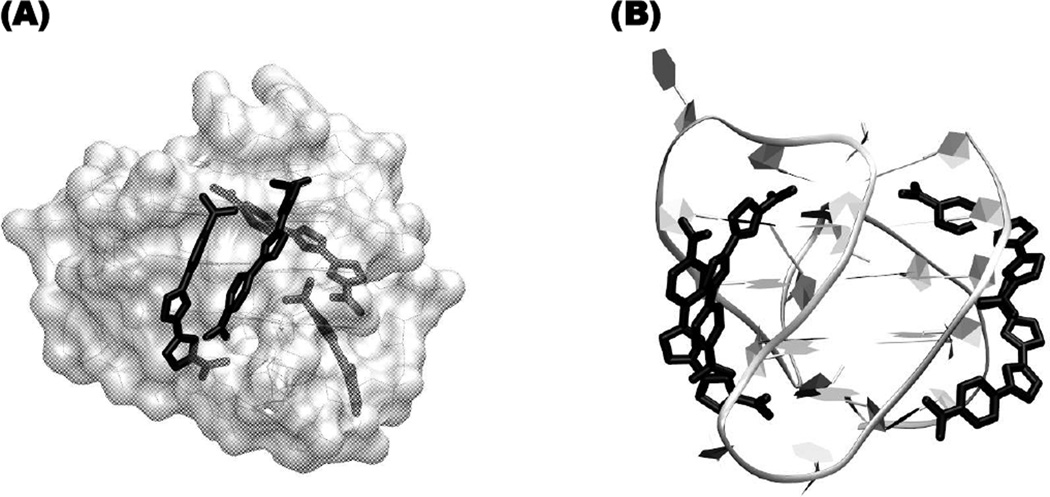
In order to provide some visual concept of how some of the currently studied ligands might stack in the grooves of the human telomeric sequence, DB1450 was docked into the two wider grooves of Tel24 sequence (PDB ID: 2GKU, [40]). The orientation of DB1450 stacked complex shown in Fig. (10) was selected because it fits well into the quadruplex grooves and makes extensive favorable contacts. We also believe that the compound is stacked on the terminal tetrads, a more common binding mechanism, but these bound DB1450 molecules are not shown to help with visualization of the less common groove binding complex. The DNA structure was held rigid in the complex and the compounds were optimized to give favorable dimer stacking in the grooves. All the compounds that show strong ICD signals must bind in some similar stacked orientation. The four molecules shown in the groove-binding mode along with the end-stacking compounds (not shown in the Figure), give a stoichiometry that reasonably agrees with our predicted values.
Fig. (10).
The proposed model shown in (A) is a space-filling representation for Tel24 with the stacked dimer, DB1450, as black tubes in the opposite grooves and (B) represents the view through the narrower groove of Tel24 sequence (ribbons) with DB1450 (tubes) stacked in the two opposite wider grooves.
The current studies suggest that the binding mode of the heterocyclic amidines is very sensitive to subtle changes in compound structural features and chemical groups. Here, we have identified a subset of diamidines that can recognize quadruplex DNA conformations with high selectivity over duplex DNA. More importantly, it has been shown that the recognition of telomeric quadruplex DNA occurs by two different binding mechanisms. End stacking was found to be the most common binding mode for these compounds but the second binding event, formation of stacked complexes in the grooves of the quadruplex, offers enhanced potential for specific recognition among quadruplexes. Studies with duplex DNA show that compounds which bind as stacked dimers or covalent hairpins have increased binding affinity and selectivity over similar compounds that bind as monomers [68, 69]. Similar stacking in the grooves of quadruplex DNA structures is an attractive method to enhance quadruplex binding affinity and specificity of the heterocyclic compounds. Potentially, the stacked monomer units could be covalently linked to additionally reduce the duplex binding while enhancing quadruplex groove affinity. The molecules of Fig. (1) thus form an exciting first generation of human telomere groove binding compounds and provide structure binding information to help in the design of next generation ligands.
ACKNOWLEDGEMENTS
Support from NIH NIAID grant AI064200 and AIRC (Associazione Italiana per la Ricerca sul Cancro), is gratefully acknowledged. We thank Marianna Toniolo, Silvia Benedetti and Davide Sabbadin for their technical support.
LIST OF ABBREVIATIONS
- DNA
Deoxyribonucleic Acid
- DSS
4,4-Dimethyl-4-silapentane-1-sulfonic acid
- DODC
(3, 3’-diethyloxadicarbocyanine iodide)
- EDTA
Ethylene diamine tetraethanoic acid
- G4
G-Quadruplex
- HPLC
High-performance Liquid Chromatography
- ICD
Induced Circular Dichroism
- K2HPO4
Potassium phosphate
- KCl
Potassium chloride
- mdeg
millidegrees
- NMR
Nuclear Magnetic Resonance
- nm
nanometers
- ppm
parts per million
- RHPS4
3,11-difluoro-6,8,13-trimethyl- 8H-quino[4,3,2-kl] acridinium methosulfate
- SAR
Structure Activity Relationship
- Tm
Thermal Melting
Footnotes
CONFLICT OF INTEREST
None declared.
REFERENCES
- 1.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J Biol Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 2.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: From G to Z. Curr Opin Struct Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 4.Neidle S, Parkinson GN. Quadruplex DNA crystal structures and drug design. Biochimie. 2008;90:1184–1196. doi: 10.1016/j.biochi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Neidle S, Thurston DE. Chemical approaches to the discovery and development of cancer therapies. Nat Rev Cancer. 2005;5:285–296. doi: 10.1038/nrc1587. [DOI] [PubMed] [Google Scholar]
- 6.Han H, Hurley L. G-quadruplex DNA: A potential target for anti-cancer drug design. Trends Pharmacol Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 7.Hurley L, Wheelhouse R, Sun D, et al. G-quadruplexes as targets for drug design. Pharmacol Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 8.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 9.Rezler EM, Bearss DJ, Hurley LH. Telomeres and telomerases as drug targets. Curr Opin Pharm. 2002;2:415–423. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 10.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucl Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neidle S, Balasubramanian S. Quadruplex nucleic acids. Cambridge: RSC Pub; 2006. [Google Scholar]
- 12.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rankin S, Reszka AP, Huppert J, et al. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H, Ju Z, Rudolph K. Telomere shortening and ageing. Z Gerontol Geriatr. 2007;40:314–324. doi: 10.1007/s00391-007-0480-0. [DOI] [PubMed] [Google Scholar]
- 15.Colgin LM, Reddel RR. Telomere maintenance mechanisms and cellular immortalization. Curr Opin Genet Dev. 1999;9:97–103. doi: 10.1016/s0959-437x(99)80014-8. [DOI] [PubMed] [Google Scholar]
- 16.Allsopp RC, Chang E, Kashefi-Aazam M, et al. Telomere shortening is associated with cell division in vitro and in vivo. Exp Cell Res. 1995;220:194–200. doi: 10.1006/excr.1995.1306. [DOI] [PubMed] [Google Scholar]
- 17.Monchaud D. A hitchhiker's guide to G-quadruplex ligands. Org Biomol Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 18.Monchaud D, Granzhan A, Saettel N, et al. "One ring to bind them all"- part I: The efficiency of the macrocyclic scaffold for G-quadruplex DNA recognition. J Nucleic Acids. 2010;2010:1–19. doi: 10.4061/2010/525862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertrand H, Bombard S, Monchaud D, Teulade-Fichou M-P. New platinum(II) complexes targeting the loops of the human telomeric G-quadruplex. Nucleic Acids Symp Ser (Oxf) 2008;52:163–164. doi: 10.1093/nass/nrn083. [DOI] [PubMed] [Google Scholar]
- 20.Boulikas T, Vougiouka M. Cisplatin and platinum drugs at the molecular level. (review) Oncol Rep. 2003;10:1663–1682. [PubMed] [Google Scholar]
- 21.Burger AM, Dai F, Schultes CM, et al. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 22.Gowan SM, Heald R, Stevens MF, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 23.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J Am Chem Soc. 1999;121:3561–3570. [Google Scholar]
- 24.Leonetti C, Amodei S, D'Angelo C, et al. Biological activity of the g-quadruplex ligand RHPS4 (3,11-difluoro-6,8,13-trimethyl-8h-quino[4,3,2-kl]acridinium methosulfate) is associated with telomere capping alteration. Mol Pharmacol. 2004;66:1138–1146. doi: 10.1124/mol.104.001537. [DOI] [PubMed] [Google Scholar]
- 25.Wenzler T, Boykin DW, Ismail MA, et al. New treatment option for second-stage african sleeping sickness: In vitro and in vivo efficacy of aza analogs of DB289. Antimicrob Agents Chemother. 2009;53:4185–4192. doi: 10.1128/AAC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purfield A, Tidwell R, Meshnick S. The diamidine DB75 targets the nucleus of plasmodium falciparum. Malar J. 2009;8:1–9. doi: 10.1186/1475-2875-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lansiaux A, Tanious F, Mishal Z, et al. Distribution of furamidine analogues in tumor cells. Cancer Res. 2002;62:7219–7229. [PubMed] [Google Scholar]
- 28.Mallena S, Lee MP, Bailly C, et al. Thiophene-based diamidine forms a “super” AT binding minor groove agent. J Am Chem Soc. 2004;126:13659–13669. doi: 10.1021/ja048175m. [DOI] [PubMed] [Google Scholar]
- 29.Wilson WD, Tanious FA, Mathis A, et al. Antiparasitic compounds that target DNA. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao Y, Lee MP, Parkinson GN, et al. Out-of-shape DNA minor groove binders:Induced fit interactions of heterocyclic dications with the DNA minor groove. Biochemistry. 2005;44:14701–14708. doi: 10.1021/bi051791q. [DOI] [PubMed] [Google Scholar]
- 31.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan AT. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 33.Bloomfield V, Crothers D, Tinoco I. Nucleic acids: Structures, properties, and functions. Sausalito: University Science Books; 2000. [Google Scholar]
- 34.Nguyen B, Tardy C, Bailly C, et al. Influence of compound structure on affinity, sequence selectivity, and mode of binding to DNA for unfused aromatic dications related to furamidine. Biopolymers. 2002;63:281–297. doi: 10.1002/bip.10073. [DOI] [PubMed] [Google Scholar]
- 35.White EW, Tanious F, Ismail MA, et al. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys Chem. 2007;126:140–153. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Fasman GD. Handbook of biochemistry and molecular biology, nucleic acids. Cleveland: CRC Press; 1975. [Google Scholar]
- 37.Ismail MA, Bialy SA, Brun R, et al. Dicationic phenyl-2,2'-bichalcophenes and analogues as antiprotozoal agents. Bioorg Med Chem. 2011;19:978–984. doi: 10.1016/j.bmc.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee GS, Wilson MA, Young BR. The application of the "WATERGATE" suppression technique for analyzing humic substances by nuclear magnetic resonance. Org Geochem. 1998;28:549–559. [Google Scholar]
- 39.Liu M, Mao X-a, Ye C, et al. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J Magn Reson. 1998;132:125–129. [Google Scholar]
- 40.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J Am Chem Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martino L, Virno A, Pagano B, et al. Structural and thermodynamic studies of the interaction of Distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J Am Chem Soc. 2007;129:16048–16056. doi: 10.1021/ja075710k. [DOI] [PubMed] [Google Scholar]
- 42.Gavathiotis E, Heald RA, Stevens MF, Searle MS. Recognition and stabilization of quadruplex DNA by a potent new telomerase inhibitor: NMR studies of the 2:1 complex of a pentacyclic methylacridinium cation with d(TTAGGGT)4. Angew Chem Int Ed Engl. 2001;40:4749–4751. doi: 10.1002/1521-3773(20011217)40:24<4749::aid-anie4749>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 43.Rodger A, Norden B. Circular dichroism and linear dichroism. New York: Oxford University Press; 1997. [Google Scholar]
- 44.Nordén B. Applications of linear dichroism spectroscopy. Applied Spectroscopy Reviews. 1978;14:157–248. [Google Scholar]
- 45.Norden B, Kubista M, Kurucsev T. Linear dichroism spectroscopy of nucleic acids. Quarterly Reviews of Biophysics. 1992;25:51–170. doi: 10.1017/s0033583500004728. [DOI] [PubMed] [Google Scholar]
- 46.Wilson WD, Nguyen B, Tanious FA, et al. Dications that target the DNA minor groove: Compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr Med Chem Anticancer Agents. 2005;5:389–408. doi: 10.2174/1568011054222319. [DOI] [PubMed] [Google Scholar]
- 47.Nishimura T, Okobira T, Kelly AM, et al. DNA binding of tilorone 1H NMR and calorimetric studies of the intercalation. Biochemistry. 2007;46:8156–8163. doi: 10.1021/bi602402m. [DOI] [PubMed] [Google Scholar]
- 48.Munde M, Ismail MA, Arafa R, et al. Design of DNA minor groove binding diamidines that recognize GC base pair sequences: A dimeric-hinge interaction motif. J Am Chem Soc. 2007;129:13732–13743. doi: 10.1021/ja074560a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee T, Mukhopadhyay R. Structural effects of nogalamycin, an antibiotic antitumour agent, on DNA. Biochem Biophys Res Commun. 2008;374:264–268. doi: 10.1016/j.bbrc.2008.07.053. [DOI] [PubMed] [Google Scholar]
- 50.Chen Q, Kuntz ID, Shafer RH. Spectroscopic recognition of guanine dimeric hairpin quadruplexes by a carbocyanine dye. Proc Natl Acad Sci U S A. 1996;93:2635–2639. doi: 10.1073/pnas.93.7.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg Med Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 53.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 54.Ambrus A, Chen D, Dai J, et al. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat Chem Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofstadler SA, Griffey RH. Analysis of noncovalent complexes of DNA and RNA by mass spectrometry. Chem Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 57.Rosu F, De Pauw E, Gabelica V. Electrospray mass spectrometry to study drug-nucleic acids interactions. Biochimie. 2008;90:1074–1087. doi: 10.1016/j.biochi.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Bailly C, Dassonneville L, Carrasco C, et al. Relationship between topoisomerase II inhibition, sequence-specificity and DNA binding mode of dicationic diphenylfuran derivatives. Anticancer Drug Des. 1999;14:47–60. [PubMed] [Google Scholar]
- 59.Cookson JC, Dai F, Smith V, et al. Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-difluoro-6,8,13-trimethyl-8h-quino[4,3,2-kl]acridinium methosulfate (RHPS4) in vitro: Activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol Pharmacol. 2005;68:1551–1558. doi: 10.1124/mol.105.013300. [DOI] [PubMed] [Google Scholar]
- 60.Fu Y-T, Keppler BR, Soares J, Jarstfer MB. BRACO19 analog dimers with improved inhibition of telomerase and hPot 1. Bioorg Med Chem. 2009;17:2030–2037. doi: 10.1016/j.bmc.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 61.Taetz S, Baldes C, Mürdter T, et al. Biopharmaceutical characterization of the telomerase inhibitor BRACO19. Pharm Res. 2006;23:1031–1137. doi: 10.1007/s11095-006-0026-y. [DOI] [PubMed] [Google Scholar]
- 62.Cosconati S, Marinelli L, Trotta R, et al. Structural and conformational requisites in DNA quadruplex groove binding: Another piece to the puzzle. J Am Chem Soc. 2010;132:6425–6433. doi: 10.1021/ja1003872. [DOI] [PubMed] [Google Scholar]
- 63.Mergny J-L, Lacroix L. Current protocols in nucleic acid chemistry. John Wiley & Sons, Inc.; 2001. [Google Scholar]
- 64.Rachwal PA, Fox KR. Quadruplex melting. Methods. 2007;43:291–301. doi: 10.1016/j.ymeth.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 65.Munde M, Kumar A, Nhili R, et al. DNA minor groove induced dimerization of heterocyclic cations: Compound structure, binding affinity, and specificity for a TTAA site. J Mol Biol. 2010;402:847–864. doi: 10.1016/j.jmb.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collie GW, Sparapani S, Parkinson GN, Neidle S. Structural basis of telomeric RNA quadruplex–acridine ligand recognition. J Am Chem Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]
- 67.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 68.Bailly C, Tardy C, Wang L, et al. Recognition of atga sequences by the unfused aromatic dication DB293 forming stacked dimers in the DNA minor groove. Biochemistry. 2001;40:9770–9779. doi: 10.1021/bi0108453. [DOI] [PubMed] [Google Scholar]
- 69.Wang L, Carrasco C, Kumar A, et al. Evaluation of the influence of compound structure on stacked-dimer formation in the DNA minor groove. Biochemistry. 2001;40:2511–2521. doi: 10.1021/bi002301r. [DOI] [PubMed] [Google Scholar]