The general view of cancer progression is based on a gradual evolutionary process, involving a continuous acquisition of genetic changes in individual cells with successive subclonal selection and expansion.1 However, whole-genome sequencing (WGS) in cancer has led to discover the chromothripsis phenomenon, in which focal regions of the genome undergo extensive chromosome fragmentation and reorganization causing structural genomic complexity in some tumor cells.2 Chromothripsis was first described in a chronic lymphocytic leukemia (CLL) patient with aggressive disease.2 Subsequent WGS has revealed chromothripsis in other tumor types, mostly associated with aggressive clinical courses.2, 3, 4, 5 Breakpoint characteristics suggest that multiple regional lesions occur by a single catastrophic mitotic event acting as a driver of tumor initiation, activating oncogenes and disrupting tumor-suppressor genes randomly.2, 3, 4, 5 However, an evaluation of the genetic evolution of a cancer genome with chromothripsis has not been reported yet, and from a clinical perspective, much remains to be understood. We present here a longitudinal analysis over 11 years of evolution of a CLL case affected by chromothripsis with the goal to answer two main unsolved questions: (a) is chromothripsis an initiating event or is it an episode arising from previous alterations? And (b) does chromothripsis resist successive therapeutic interventions?
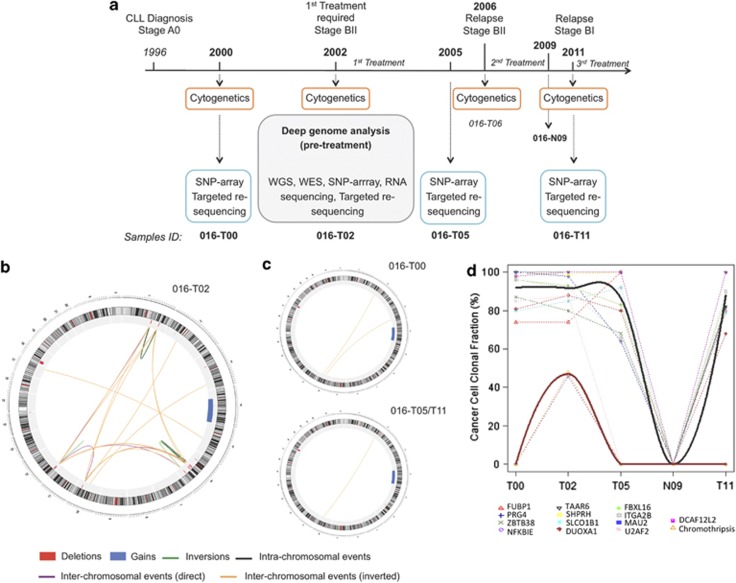
We characterized structural variants (SVs) and single-nucleotide variants (SNVs) by sequencing and molecular approaches in different samples from a 59-year-old IGHV unmutated CLL patient (ICGC-CLL code 016) obtained at four time points over 11 years of clinical evolution. Sample 016-T02 was collected 6 years after diagnosis, just before starting the first treatment due to the increase of CLL aggressiveness, and analyzed by WGS (Figure 1a). WGS results showed a profile of complex SVs with similar characteristics of other previously published cases of chromothripsis.2, 3, 4, 5 Although conventional cytogenetic and fluorescence in situ hybridization analysis showed only a derivative chromosome formed by 4q (duplicated) and 18q (with 18p deletion) (Supplementary Table S1), by single-nucleotide polymorphism (SNP) array and targeted re-sequencing we confirmed 49 SVs detected by WGS (Supplementary Table S3). Most SVs showed chromosome geographical localization, with 80% of breakpoints (involving 35 of 49 SVs) confined to focal points on chromosomes 1, 6, 10 and 12. A significant link between the small regions and an altered copy-number profile alternating two states demonstrated the existence of complex genomic rearrangements consistent with the pattern of chromothripsis2, 6 (Figure 1b, Supplementary Figure S1). The remaining 14/49 SVs affect one locus each (Supplementary Table S3). In addition, RNA sequencing showed that the 1.6-Mb deletion on 6p21 lead to a fusion transcript of UBR2-SPATS1 (Supplementary Figure S2). Furthermore, we confirmed 13 potentially functional SNVs previously detected in this sample by exome sequencing7 and we found one additional somatic de novo SNV (c.G566C:p.R189T) in ATM (Table 1), validated by RNA and Sanger sequencing (Supplementary Figure S3). Interestingly, the SNV that generates a premature stop codon (c.G853T:p.E285X) in NFKBIE, located in 6p21, should lead to the total absence of a functional gene due to the 1.6-Mb deletion of the other allele.
Figure 1.
Longitudinal analysis of SVs of case 016 over a period of 11 years of disease evolution. (a) A 59-year-old male was diagnosed with CLL, IGHV unmutated subtype at stage A0 in 1996. He did not require treatment until 2002, when CLL progressed to stage BII, because of the development of massive lymph nodes, splenomegaly and constitutional symptoms. He received fludarabine-cyclophosphamide-mitoxantrone (six cycles), with complete response and negative detection of minimal residual disease (MRD). In 2006, the patient relapsed (BII), and was retreated with six cycles of fludarabine-cyclophosphamide-mitoxantrone-rituximab, obtaining a complete response, MRD negative. He was free of disease in 2009 when normal mononuclear cells were collected. After a second relapse (BI) in 2011 with an axillar bulky lymphadenopathy, the patient received six cycles of rituximab-bendamustine, with excellent response, MRD negative, which is maintained until December 2012. Somatic SV characterization was done by WGS of samples 016-T02 and 016-N09 (Supplementary Table S2). Mosaicism analysis of CNAs of sample 016-T02 was done by Illumina HumanOmni1-Quad array (Illumina, Inc., San Diego, CA, USA). Somatic point mutations analysis of sample 016-T02 was performed as described by exome sequencing7 and using an additional in-house pipeline. Validation of somatic SVs and point mutations in all samples was done by a targeted re-sequencing approach (Agilent SureSelect Target Enrichment; Agilent Technologies, Santa Clara, CA, USA) and by Affymetrix 6.0 SNP array (Affymetrix 6.0; Affymetrix, Santa Clara, CA, USA). Detection of fusion transcripts of RNA in sample 016-T02 were performed by RNA sequencing.7 Further detailed information is provided in Supplementary Methods. (b) Circos representing the SVs detected in sample 016-T02, before starting treatment, in consistence with the chromothripsis pattern. (c) Circos representing the SVs detected in pre-treatment (016-T00) and post-treatment (016-T05 and 016-T11) samples. (d) Evolution of cancer cell CF over time for SNVs and chromothripsis rearrangements. Smoothed solid curves represent the average profile of the time evolution of cancer cell percentages of the mutations detected in all time points (black) and only in sample 016-T02 (red).
Table 1. Somatic point mutations in all time-point samples.
| Chromosome | Position | Reference | Observed | Gene | Exonic function | AA change | 016-T00 | 016-T02 | 016-T05 | 016-T11 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 78425887 | G | A | FUBP1 | Stopgain SNV | NM_003902:c.C1558T:p.Q520X | + | + | + | + |
| 1 | 186277552 | A | G | PRG4 | Nonsynonymous SNV | NM_001127710:c.A2299G:p.T767A | + | + | + | + |
| 3 | 141163154 | C | T | ZBTB38 | Nonsynonymous SNV | NM_001080412:c.C1924T:p.P642S | + | + | + | + |
| 6 | 44230329 | C | A | NFKBIE | Stopgain SNV | NM_004556:c.G853T:p.E285X | − | + | − | − |
| 6 | 132892234 | A | C | TAAR6 | Nonsynonymous SNV | NM_175067:c.A774C:p.K258N | + | + | + | + |
| 6 | 146264389 | T | C | SHPRH | Nonsynonymous SNV | NM_001042683:c.A2128G:p.M710V | + | + | + | + |
| 11 | 108114749 | G | C | ATM | Nonsynonymous SNV | NM_000051:c.G566C:p.R189T | + | + | + | + |
| 12 | 21358921 | C | A | SLCO1B1 | Nonsynonymous SNV | NM_006446:c.C1451A:p.P484H | + | + | + | + |
| 15 | 45409873 | A | G | DUOXA1 | Nonsynonymous SNV | NM_144565:c.T1292C:p.V431A | + | + | + | + |
| 16 | 745751 | T | C | FBXL16 | Nonsynonymous SNV | NM_153350:c.A806G:p.N269S | + | + | + | + |
| 17 | 42463002 | G | A | ITGA2B | Nonsynonymous SNV | NM_000419:c.C491T:p.A164V | + | + | + | + |
| 19 | 19431945 | G | A | MAU2 | Splicing site | NA | + | + | + | + |
| 19 | 56173950 | A | T | U2AF2 | Nonsynonymous SNV | NM_001012478:c.A569T:p.Q190L | + | + | − | + |
| X | 125299321 | C | A | DCAF12L2 | Nonsynonymous SNV | NM_001013628:c.G587T:p.W196L | + | + | + | + |
+, present; −, absent.
To determine if chromothripsis was involved in the tumorigenesis and survived therapeutic intervention, we analyzed from the same patient three samples obtained at different time points (016-T00, 016-T05 and 016-T11) (Figure 1a). These samples were collected in pre-treatment and two post-treatment phases. Cytogenetics and fluorescence in situ hybridization analyses, performed in each stage, showed the same profile of sample 016-T02 (Supplementary Table S1). The 49 additional SVs were analyzed by SNP array and targeted re-sequencing (Figure 1a). We also validated the 13 potential deleterious somatic SNVs and the new mutation in ATM. Only seven non-complex SVs and twelve SNVs were detected in all samples (Figure 1c, Table 1, Supplementary Table S3), indicating that they remained during disease progression and suggesting their potential role in the CLL initiation. We did not detect the pattern of complex rearrangements in the additional samples (Figure 1c, Supplementary Table S3, Supplementary Figure S4), demonstrating that chromothripsis was reversible by treatment and it did not participate in the tumor development and subsequent relapses. Three additional deletions (on 2p16, 3p14 and 6p22), a tandem duplication (1q44) and the NFKBIE mutation were also exclusive of 016-T02, suggesting that they progressed and disappeared together with the chromothripsis. SNP array did not identify any additional somatic copy-number alteration (CNA) at any other time point (Supplementary Figure S5), revealing no other subclonal populations carrying different CNA.
To evaluate whether the SVs exclusive of sample 016-T02 occurred in the same subclonal population, we analyzed the clonal fraction (CF) of all of them. We found all in a CF of 40–50%, strongly suggesting that occurred at the same subpopulation. In contrast, those SVs common in all time points presented a CF of ∼90% in 016-T02, indicating that they were present in most tumor cells at chromothripsis appearance (Supplementary Table S4, Supplementary Figure S6). This suggests that a subclonal competing population carrying chromothripsis emerged from a major population of tumor cells. These results were supported by the SNV frequencies in sample 016-T02, revealing that the tumor was composed by two main subpopulations corresponding to regions of CF of ∼50% and ∼90%, respectively (Supplementary Figure S9). Interestingly, the NFKBIE mutation was found in a CF of 48% and the remaining SNVs of ∼90% (Supplementary Table S5). This supports that NFKBIE mutation appeared in the same subpopulation as chromothripsis, whereas the remaining SNVs were in almost all tumor cells.
Targeted re-sequencing and SNP array performed in all samples did not allow calculating the CF for the shared CNA. However, CNA parameters obtained by SNP array for these regions suggested similar CF along the disease progression (Supplementary Figure S10). Conversely, targeted re-sequencing enabled a good estimate of the CF for SNVs in sample 016-T02 (Supplementary Table S6). Thus, we observed highly similar CF values for SNVs in all samples and no signs of mutated alleles in sample 016-N09 (Figure 1c, Supplementary Table S6).
The predominant population of tumor cells observed in sample 016-T02 has been prevailing along disease evolution of patient 016, reappearing even after two lines of treatment and a short disease-free period, in contrast of previous findings relating chemotherapy-treated CLL with different clonal expansions.8 Therefore, this preponderant clone might entail driver mutations for the CLL development. Appealing driver alterations could be the 18p deletion, recently observed in 3% of CLL cases,9 the 6p21 deletion, entailing the formation of fusion transcript UBR2-SPATS1, and the ATM mutation (R189T), because ATM mutations occur in 12% of CLL cases10 and deletions are associated with high-risk CLL.9, 11, 12 The ATM mutation could also explain the rise of chromothripsis because of its role in regulating DNA-damage response through p53 signaling. Cells bearing defects in this pathway should be predisposed to chromothripsis.4 Finally, chromothripsis has been detected in 5% of CLL patients, most with unmutated IGHV, high-risk CLL genomic aberrations (11q22 and 17p13 deletions) and/or TP53 mutations.9 Hence, although patient 016 had no TP53 alterations, the high CF of cells with the ATM mutation supports its potential functional implications both in CLL and chromothripsis.
Our study clearly shows that chromothripsis was not the tumor-initiating event but it participated in the increase of CLL aggressiveness observed in 016-T02, as subclones carrying complex SVs were in expansion and competing with a predominant tumor population. Within the chromothripsis rearrangements, we found a 2-Mb deletion affecting the 6q21 locus, involved in 6% of CLL and linked to shorter progression-free survival.9, 11, 13, 14 In addition, we detected a 10q24 deletion involving NFKB2, also detected in a small percentage of CLL patients.9 The E285X mutation in NFKBIE was potentially important for chromothripsis, as the deletion of the other allele in affected subclones led to the absence of a functional NFKBIE allele. Interestingly, Nfkbie-knockout mice exhibit increased B-cell proliferation and survival,15 supporting its implication in CLL evolution.
We confirmed the single catastrophic model for chromothripsis, but we showed that it appeared years after the CLL diagnosis, rejecting a hypothetic role in the tumor initiation. Our data suggest that chromothripsis might have been the consequence of previous alterations expanded from an early leukemia cell, which would have led to chromosome instability associated with defects in DNA repair. The complex rearrangements of chromothripsis added on a previous tumor cell carrying expanded alterations might have conferred a selective advantage to the subclonal population increasing its malignancy, providing a more aggressive CLL phenotype.
Our findings also revealed that chromothripsis subclones did not survive chemotherapy and that it has not reappeared in a period of 10 years, having therefore no apparent implications for patient prognosis. This differs from previous data strongly associating chromothripsis with chemotherapy surviving and/or poor clinical outcome.2, 3, 4, 5 Chemotherapeutic regimens followed by our case achieved continuously excellent responses, but a subset of non-chromothripsis alterations led to the relapse of the CLL phenotype. This indicates that an undetectable percentage of cells carrying these alterations persisted after therapy and subsequently expanded.
In summary, our study demonstrates the importance of analyzing cancer samples at different time points to elucidate clonal genotypes that could be therapy resistant, which might help in therapeutic decisions along the CLL clinical course.16 This is the first description of the rise and fall of chromothripsis, demonstrating that it was a secondary effect of the genomic instability typical of tumor cells. However, there is the need of other cases to be analyzed to better understand the dynamics of chromothripsis and to determine the exceptionality of our case.
Acknowledgments
We thank the Spanish Plan Nacional funding SAF2008-00357 (NOVADIS), the Generalitat de Catalunya funding AGAUR 2009 SGR-1502, the Ministry of Economy and Competitiveness funding to CLL-Consortium project and the ESGI–European Commission Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 262055. JMCT was supported by two fellowships from Xunta de Galícia (Spain) and FSE European funds, and a grant from Xunta de Galícia (10PXIB918057PR). LZ is supported by a ‘La Caixa' pre-doctoral fellowship. SNP genotyping services were provided by the Spanish ‘Centro Nacional de Genotipado (CEGEN-ISCIII)' (www.cegen.org)
Author contributions
LB designed research, analyzed the data and wrote the manuscript. SB designed research, collected data and helped with the data analysis. GE designed research and performed the computational analysis for SV detection using sequencing data. CT carried out computational steps for the sequencing data analysis. IS collected data and helped with the data analysis. LZ carried out the analysis of subclonal mutations by whole-genome, whole-exome and targeted re-sequencing. OD and SO performed the computational analysis of somatic point mutations by whole-exome and targeted re-sequencing. PGF carried out the analysis of fusion transcripts by RNA sequencing. BR-S performed the detection of clonal fractions by Illumina SNP array platform. JMCT participated in the data analysis. AN and DMG carried out the PCR analysis. CL performed the cytogenetic analysis. AM-T and AL-G provided and revised the clinical history of the patient. MG coordinated sequencing experiments. CL-O and EC revised carefully the manuscript and coordinated the CLL consortium. XE designed research and revised carefully the manuscript. All the authors read and approved the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrangeas F, Avet-Loiseau H, Munshi NC, Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118:675–678. doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS, et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann J, Holzmann K, Miller F, Winkler D, Buhler A, Zenz T, et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;120:4783–4794. doi: 10.1182/blood-2012-04-423517. [DOI] [PubMed] [Google Scholar]
- Guarini A, Marinelli M, Tavolaro S, Bellacchio E, Magliozzi M, Chiaretti S, et al. ATM gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica. 2012;97:47–55. doi: 10.3324/haematol.2011.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- Finn WG, Kay NE, Kroft SH, Church S, Peterson LC. Secondary abnormalities of chromosome 6q in B-cell chronic lymphocytic leukemia: a sequential study of karyotypic instability in 51 patients. Am J Hematol. 1998;59:223–229. doi: 10.1002/(sici)1096-8652(199811)59:3<223::aid-ajh7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Gunnarsson R, Mansouri L, Isaksson A, Goransson H, Cahill N, Jansson M, et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica. 2011;96:1161–1169. doi: 10.3324/haematol.2010.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JM, Aleksiyadis K, Martin A, McNamee K, Tharmalingam T, Williams RO, et al. Inhibitor of kappa B epsilon (IkappaBepsilon) is a non-redundant regulator of c-Rel-dependent gene expression in murine T and B cells. PLoS One. 2011;6:e24504. doi: 10.1371/journal.pone.0024504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Lopez-Otin C. The evolutionary biography of chronic lymphocytic leukemia. Nat Genet. 2013;45:229–231. doi: 10.1038/ng.2556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.