In order to maintain cellular homeostasis, excess or misfolded proteins must be removed and/or recycled.1 In myeloma, plasma cells produce large quantities of immunoglobulin, and excess unfolded immunoglobulin results in high levels of endoplasmic reticulum (ER) stress. These proteins are removed either via the proteasome2 or via autophagy,3 suggesting that these pathways may be specifically exploited therapeutically. Although proteasome inhibition has been extensively explored as a therapeutic option in myeloma, little work has been performed on targeting the pathways leading to autophagy. We have previously demonstrated the importance of the unfolded protein response (UPR) in dealing with ER stress in myeloma cells.2 Importantly, activation of the UPR, elimination of proteins by the proteasome and autophagy are interlinked. Proteasome inhibition leads to the compensatory upregulation of autophagy,4 whereas inhibition of autophagy reduces the clearance of ubiquitinated proteins.5, 6
Autophagy has an important protective role in sustaining homeostasis in normal cells.1 Due to high levels of excess protein in myeloma cells, basal levels of autophagy are high and are upregulated in response to ER stress and proteasome inhibition, protecting against apoptosis.3 However, there are conflicting reports on the impact of the combination of proteasome and autophagy inhibition on myeloma cells. Synergistic cytotoxicity was seen in vitro with Bortezomib and the autophagy inhibitor, Bafilomycin A1 (Bfa), whereas cell survival was increased with Bortezomib and a different autophagy inhibitor, 3-methyladenine.3, 7 If we are to optimally manipulate these pathways, we need to fully understand how they interact, are differentially regulated and the signals that initiate one pathway if the other is compromised.
It is a well-established observation that autophagy is negatively regulated by the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway.8, 9 Consequently inhibitors of PI3K, AKT and mTOR can be used as investigational tools to examine therapeutic interactions with autophagy. Targeting the PI3K/AKT/mTOR pathway is known to inhibit myeloma cell growth,10 and this activity is enhanced by the use of dual PI3K/mTOR inhibitors that block the negative feedback loop that is activated when single-agent PI3K inhibitors are used.1, 11 However, clinical trials of these inhibitors in other cancers have been disappointing;11 potentially this could be due to the undesired side-effect of autophagy induction. Therefore, usingPI-103, an ATP-competitive dual inhibitor of PI3K and mTOR, we have examined the interactions of the proteasome, UPR and autophagy.
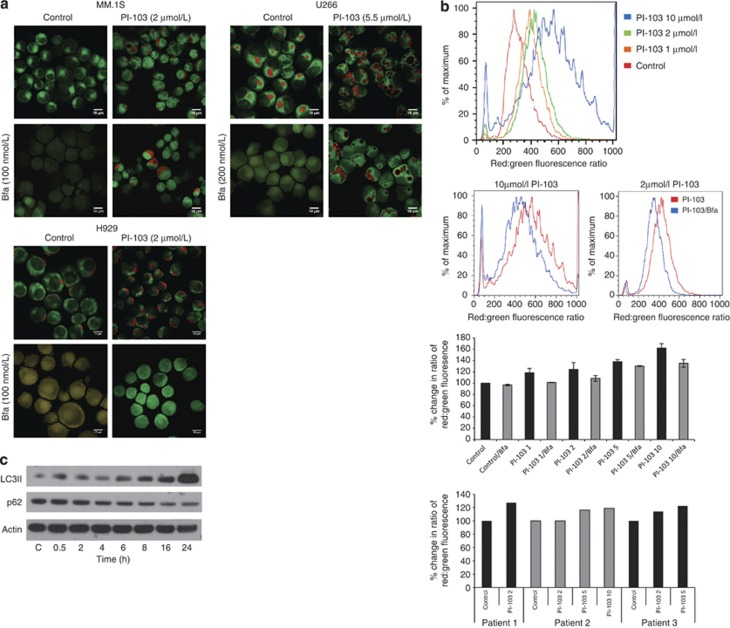
We have been able to confirm that inhibition of thePI3K pathway induces autophagy in myeloma cells and primary patient samples by treating cells with PI-103, staining them with acridine orange and examining cleavage of the autophagosome marker, microtubule-associated protein 1 light chain 3 (LC3).An increase in acidic vesicles in response to PI-103 is seen. These then disappear or are greatly reduced upon exposure to Bfa (Figures 1a and b). Immunoblotting for LC3 shows an increase in membrane bound LC3II by 6 h following PI-103 treatment while, over the same time course, we demonstrate a decrease in p62, a protein degraded via autophagy (Figure 1c). These results demonstrate that PI-103 induces autophagy, with normal autophagic flux, in myeloma cells.
Figure 1.
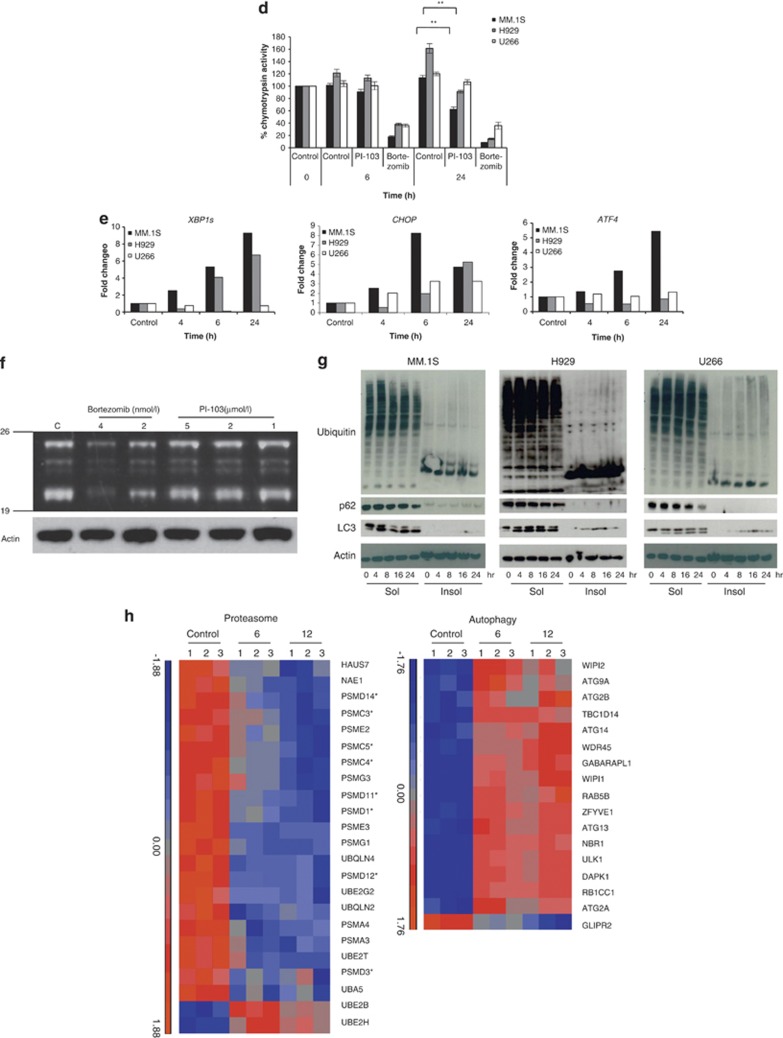
Autophagy clears ubiquitinated proteins when proteasome stability is compromised by dual PI3K/mTOR inhibition. In order to demonstrate that PI-103 activated autophagy, (a) we stained cells with acridine orange and examined the development of acidic vesicles in response to PI-103, after 24 h treatment, by confocal microscopy at 63x magnification (top panels). Acridine orange fluoresces red in an acidic milieu such as autophagosomes but fluoresces green when bound to DNA. Treatment with the autophagy inhibitor, Bfa, for the final 1 h of treatment reduced the formation of these vesicles (bottom panels). Scale bars represent 10 μm. (b) Results obtained by confocal microscopy were confirmed in triplicate by flow cytometry in MM.1S and primary patient samples. Treatment with increasing concentrations of PI-103 for 24 h resulted in an increase in the ratio of red:green fluorescence (1st and 3rd panels), while 1h treatment with 25 nmol/l Bfa blocked this increase at all tested concentrations (2nd and 3rd panels). The same pattern was observed in patient cells (bottom panel). (c) Further evidence for autophagy induction is shown by western blotting in MM.1S for the autophagosome marker, LC3II, and p62 over a 24 h timecourse with PI-103. (d) Activation of autophagy by PI-103 was accompanied by inhibition of the chymotrypsin-like catalytic activity of the proteasome by 24 h as shown in MM.1S and H929. The decrease observed in U266 was not significant. **P<0.01. Results represent mean of three separate experiments. Bortezomib was used as a positive control. (e) Inhibition of the proteasome can lead to an increase in the UPR in order to cope with excess proteins in the cell. Treatment of MM.1S, H929 and U266 with PI-103 for 24 h lead to an increase in UPR markers, XBP1s, CHOP and ATF4, as assessed by reverse transcription-PCR (RT-PCR). Data were normalized using actin. Representative plots are shown. (f) Competitive binding assay with a fluorescently labeled proteasome activity probe (Me4BodipyFL-Ahx3Leu3VS) showed that proteasome inhibition by PI-103 was not due to inhibition of the catalytic sites, as no change was observed in any of the proteasome β subunits in response to PI-103 over 24 h. Bortezomib was used as a positive control and actin as a loading control. (g) To demonstrate proteasome inhibition visually, we performed western blotting for ubiquitin in both the soluble and insoluble cell fractions. In MM.1S, H929 and U266, ubiquitin decreased over 24 h in response to PI-103. As can be seen in the insoluble fractions, ubiquitin was not accumulating as insoluble aggregates either. Blotting for p62 and LC3 showed a decrease and increase, respectively, in response to PI-103 indicative of autophagy activation. (h) Relative expression of proteasome and autophagy genes was examined in MM.1S using Affymetrix Human Gene ST 2.0 arrays at 6 and 12 h after PI-103 treatment. The indicated genes had at least a 1.5-fold change over the timecourse. Asterisks denote components of the proteasome 19S lid and base. Drug concentrations are as follows unless otherwise stated. PI-103: MM.1S and H929, 2 μmol/l; U266, 5.5 μmol/l. Bfa: MM.1S and H929, 100 nmol/l; U266, 200 nmol/l. Bortezomib: MM.1S, H929 and U266, 8 nmol/l.
Next we sought to determine the effects of autophagy induction by PI-103 on proteasome activity. We examined the effect of PI-103 on the chymotrypsin-like catalytic activity of the proteasome and showed that by 24 h, PI-103 significantly inhibited the proteasome (P<0.01) in MM.1S and H929 by 50% and 70%, respectively, compared to control values. In contrast, PI-103 did not affect the proteasome in U266 (Figure 1d). Proteasome inhibition has previously been shown to trigger ER stress via the UPR. In order to assess whether proteasome inhibition by PI-103 had the same physiological consequences as if it were inhibited by other mechanisms, we examined the levels of critical UPR effector molecules following treatment with PI-103. Levels of X-box binding protein1 (XBP1s), C/EBP homologous protein (CHOP) and activating transcription factor 4 (ATF4) were increased to varying degrees, indicating the induction of ER stress in response to PI-103 (Figure 1e). Thus, PI-103 not only inhibits the proteasome, it also upregulates the UPR.
In order to determine whether proteasome inhibition by PI-103 was a direct or indirect mechanism, we performed competition experiments with PI-103 and a proteasome activity probe (Me4BodipyFL-Ahx3Leu3VS), which binds to all the catalytic proteasome subunits. No decrease in any β catalytic subunits was observed, indicating that the mechanism of action of PI-103 on the proteasome is not via active site inhibition (Figure 1f). Despite proteasome inhibition, western blotting for ubiquitinated proteins following exposure to PI-103 indicated that the levels of these proteins decreased over 24 h, as did p62, whereas LC3 increased (Figure 1g). This observation suggests that when the proteasome is inhibited by PI-103, ubiquitinated proteins are degraded by autophagy, highlighting an important link between these pathways. To examine this further, we performed gene expression profiling on MM.1S cells treated for 6 and 12 h withPI-103 (Figure 1h, Supplementary Tables 1 and 2). Following PI-103 treatment, 21 out of 23 proteasome genes were downregulated, and 16 out of 17 autophagy genes were upregulated. Of the proteasome genes that were downregulated, 8 comprise the 19S complex lid and base, and no effect was observed on the catalytic β subunits, confirming that PI-103 does not inhibit the active site. Of particular interest is the downregulation of PSMD14, the gene responsible for removing ubiquitin on target proteins,12 and which also influences proteasome stability. PSMG1, the gene responsible for the formation and maturation of the 20S proteasome,13 was also downregulated. Downregulation of these genes, together with components of the 19S complex, would therefore interfere with proper assembly of the 26S proteasome, reducing degradation of ubiquitinated proteins, and thereby act as a cue to trigger autophagy. To this end, autophagy genes that are upregulated include members of the Unc-51-like kinase 1 (ULK1) complex that are negatively regulated by mTOR, and which initiate autophagy and genes that are important for the formation and elongation of the isolation membrane, both early events in the autophagy pathway.9
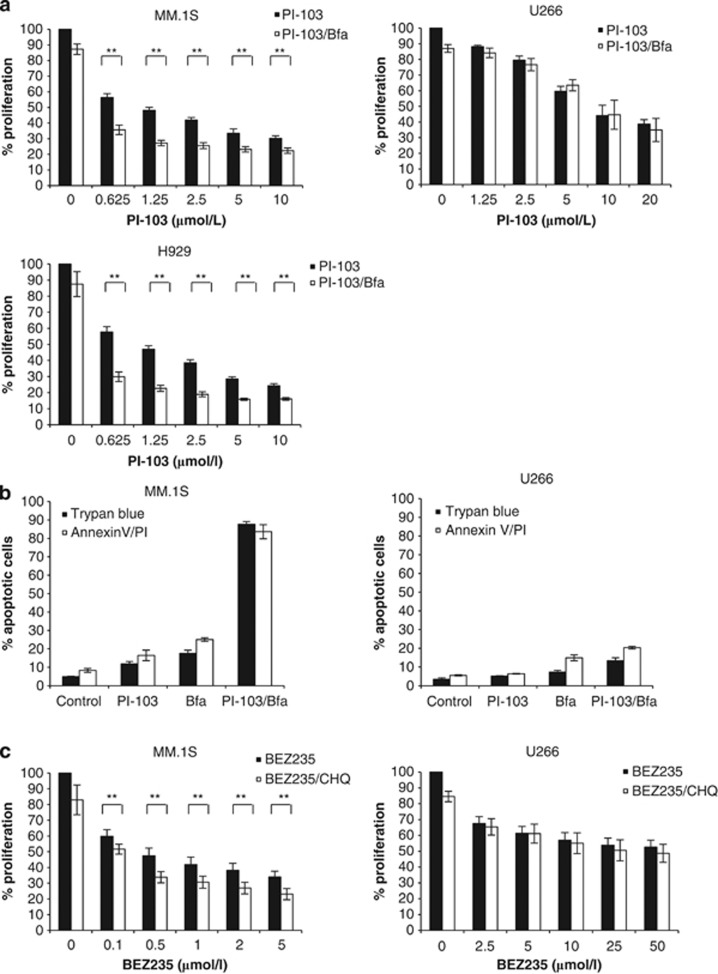
As PI-103 induces autophagy and autophagy is a known tumor survival response which may mitigate the proapoptotic effects of PI-103, we hypothesized that blocking autophagy in the context of PI-103 may initiate apoptosis. In order to address this, we performed proliferation and apoptosis assays using PI-103 in combination with the known autophagy inhibitor, Bfa. We clearly demonstrate that in MM.1S and H929, inhibition of autophagy results in enhanced apoptosis, but the same effect on proliferation and apoptosis was not seen in U266, the cell line in which PI-103 did not inhibit the proteasome (Figures 2a and b). To confirm the specificity of the observed effect, we treated cells with a second dual PI3K/mTOR inhibitor, BEZ235, in combination with a second autophagy inhibitor, chlororquine (CHQ). Again we observed that in MM.1S but not U266, the combination of BEZ235 and CHQ significantly reduced proliferation, and enhanced apoptosis (Figures 2c and d). In order to determine which kinase, either PI3K or mTOR, was more important for the observed effect, we treated cells with Rapamycin, an exclusive mTOR inhibitor, in combination with Bfa. This indicated that in order to enhance apoptosis following exposure to an autophagy inhibitor, inhibition of both PI3K and mTOR is required (Figure 2e).
Figure 2.
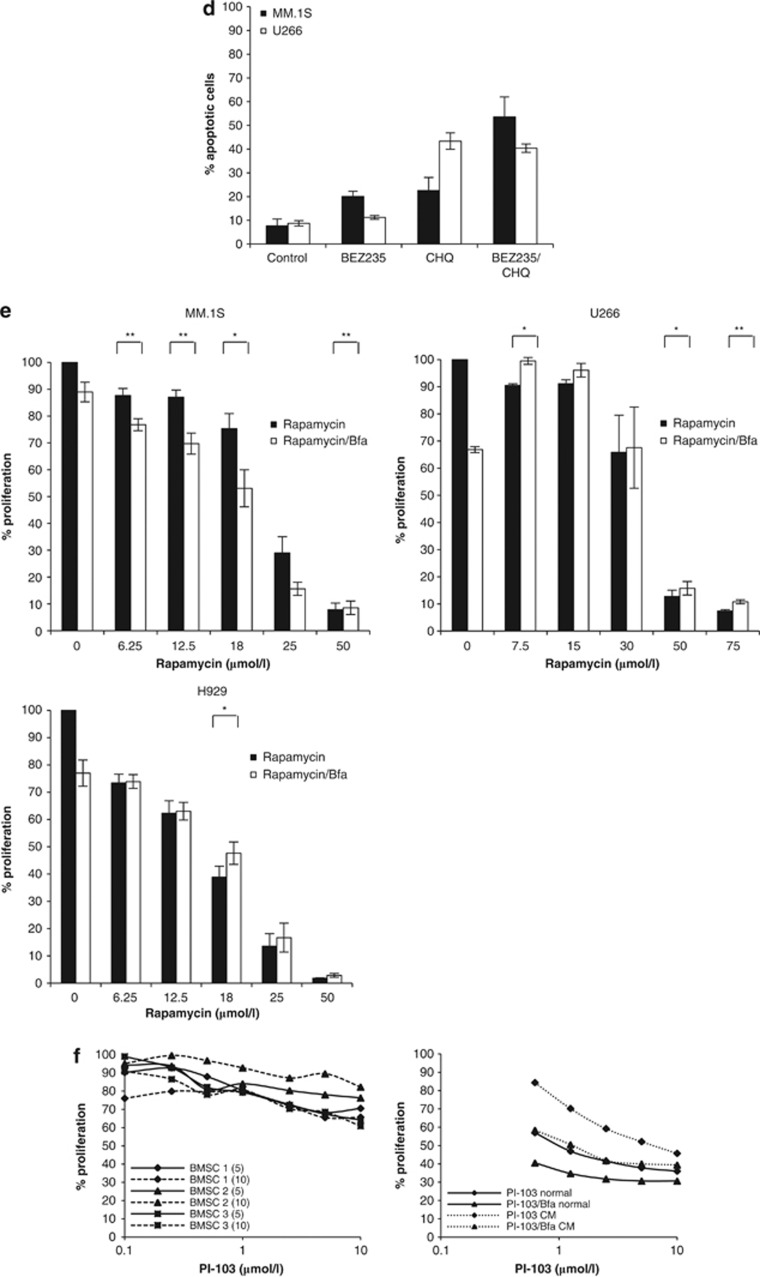
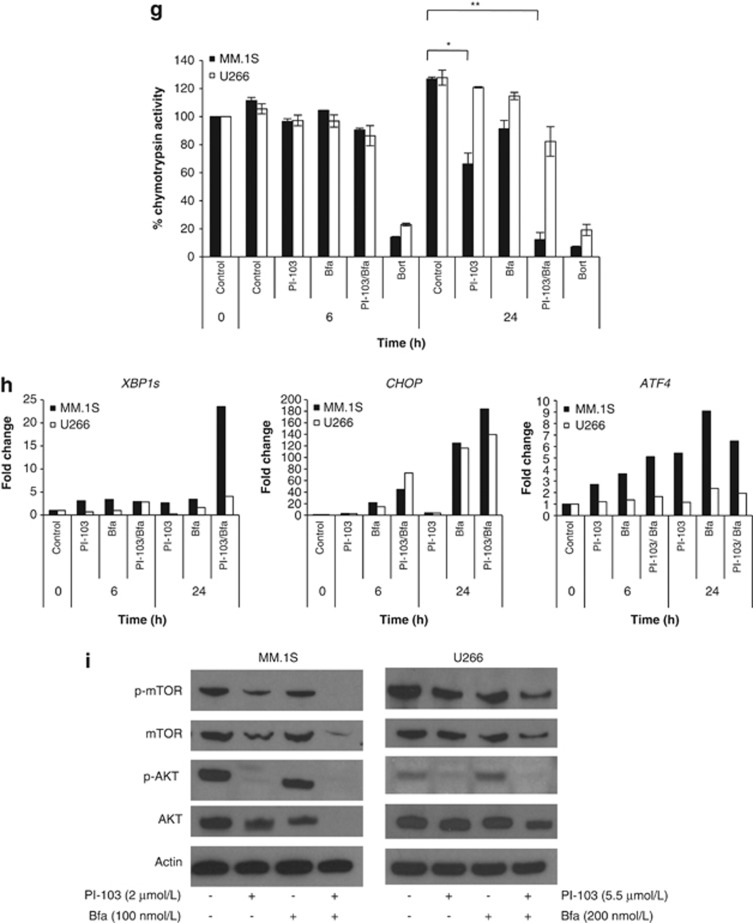
Dual PI3K/mTOR inhibition, combined with autophagy inhibition, enhances apoptosis of myeloma cells. Given that PI-103 induces autophagy in myeloma cells, we were interested to understand the effect on cells when autophagy was blocked. (a) The combination ofPI-103, with a constant subtoxic concentration of Bfa, over 24 h effectively inhibited the proliferation of MM.1S and H929 (left panels) at all PI-103 concentrations tested, as determined by triplicate MTT assays. **P<0.01. The combination was no more effective in U266 than PI-103 alone (right panel). (b) Initiation of apoptosis in MM.1S (left) and U266 (right) was confirmed by trypan blue exclusion and Annexin V/PI (propidium iodide) staining over a 24 h period with PI-103 and Bfa. Results represent the average of three independent experiments. (c) Specificity of the observed effect with PI-103 and Bfa was demonstrated by triplicate MTT assay by treating cells for 24 h with a second PI3K/mTOR inhibitor, BEZ235, in combination with another autophagy inhibitor, CHQ. When CHQ was applied at a constant subtoxic concentration, it significantly inhibited proliferation of MM.1S, but not U266, at all tested concentrations of BEZ235. (d) This was confirmed, in triplicate, by Annexin V/PI staining, indicating that apoptosis is enhanced by combination treatment in MM.1S but not U266. (e) In order to delineate whether it was the inhibition of PI3K or mTOR that was more important for the observed effect, we performed triplicate MTT assays with the mTOR inhibitor, Rapamycin, in combination with Bfa for 24 h. The combination significantly enhanced apoptosis at most of the tested Rapamycin concentrations in MM.1S but not in H929 or U266. (f) Left, PI-103 had minimal effect on patient-derived BMSCs (BMSC 1-3) seeded at two different densities (5 × 104 (5) and 10 × 104 (10) cells per well) as assessed by 24 h Wst1 assays. Right, Using conditioned medium from patient BMSCs, 24 h MTT assays were performed to determine the efficacy of the combination in MM.1S in the presence of bone marrow cytokines. Representative results from one of the three experiments are shown. CM, conditioned medium; normal, normal growth medium. (g) The chymotrypsin-like activity of the proteasome was determined in triplicate in MM.1S and U266 following 6 and 24 h treatment with PI-103, Bfa or both. Bortezomib (Bort) was used as a positive control. *P<0.05; **P<0.01. (h) Detection of UPR markers, XBP1s, CHOP and ATF4, by RT-PCR in MM.1S and U266 following 6 and 24 h treatment with PI-103, Bfa or both. Data were normalized using actin. Representative plots are shown.(i) Western blot analysis was carried out at 24 h in MM.1S (left) and U266 (right) treated with PI-103, Bfa or both. Drug concentrations were as follows. PI-103: MM.1S, 2 μmol/l; U266, 5.5 μmol/l. Bfa: MM.1S and H929, 100 nmol/l; U266, 200 nmol/l. BEZ235: MM.1S, 800 nmol/l; U266, 25 μmol/l. CHQ: MM.1S and U266, 500 nmol/l. Bortezomib was used at 8 nmol/l for both cell lines.
The bone marrow microenvironment has an important role in protecting myeloma cells from apoptosis. Therefore, we tested whether PI-103 affected the proliferation of patient-derived bone marrow stromal cells (BMSCs) and whether the combination of PI-103 and Bfa was still effective in the presence of bone marrow cytokines by using conditioned medium from BMSCs. PI-103 had minimal effect on patient's BMSCs (Figure 2f, left panel), but the conditioned medium did offer some protection to the cells, ranging from 10 to 30%. However, the combination ofPI-103 and Bfa was still able to effectively inhibit myeloma cell proliferation across all tested PI-103 concentrations (Figure 2f, right panel). Taken together, these results suggest that blocking the protective effects of autophagy induced by dual PI3K and mTOR inhibition has the ability to elicit a strong apoptotic response.
As our results demonstrated that PI-103 results in proteasome inhibition and UPR induction, we next sought to determine whether these effects were still present in the presence of an autophagy inhibitor. In MM.1S, following exposure to PI-103/Bfa, the combination significantly reduced the catalytic activity of the proteasome reaching 12% of the control value (P<0.01); levels that are similar to those found after exposure to Bortezomib. Again, this effect was not observed in U266 (Figure 2g). In addition, the effect of the combination on UPR markers was also enhanced (Figure 2h). Taken together these findings suggest that the apoptotic response elicited by PI-103/Bfa is due to a significant effect on proteasome inhibition which, given its magnitude, may be clinically relevant. In order to understand what may be responsible for the increased apoptotic effect seen when PI-103 is combined with an autophagy inhibitor, we performed blotting for PI3K pathway members. We demonstrate that in addition to the loss of the phosphorylated forms of AKT and mTOR, there is a complete loss of both of these proteins (Figure 2i). This result could be explained as being a consequence of either proteasome14 or AKT inhibition.8 We suggest that due to the central nature of the PI3K pathway in normal cellular proliferation, complete loss of these critical signaling molecules is responsible for the increased apoptosis.
The induction of pro-survival autophagy may limit the efficacy of PI3K inhibitors. Given the intense interest in this class of inhibitors and the promise they hold for a multitude of cancer types,11 it is important to fully understand the consequences of autophagy induction. In this work, we demonstrate that PI-103 induces autophagy and has the potentially beneficial effect of inhibiting the proteasome in myeloma cells but despite this, apoptosis induction is low. It is likely that this is due to the fact that autophagy compensates by removing ubiquitinated proteins that accumulate when the proteasome is blocked, relieving the potentially hazardous accumulation of these proteins that would lead to apoptosis.4 We demonstrate the reciprocal interaction between these pathways and propose that the decrease in subunits of the 19S lid and base provide an important link between these pathways. Finally, we exploit the upregulation of autophagy by combining PI-103 with autophagy inhibitors. The result is a strong apoptotic response with enhanced proteasome inhibition and increased ER stress, an effect that may be clinically relevant. In conclusion, PI3K inhibitors hold great promise for the treatment of myeloma in the combination setting, however, in order to achieve maximum clinical benefit, the interactions between autophagy and the proteasome need to be carefully considered.
Acknowledgments
Research grants and financial support were received from Cancer Research UK, Myeloma UK and The Kay Kendall Leukemia Fund. We acknowledge NHS funding to the NIHR Biomedical Research centre at the Royal Marsden Hospital. FED is a Cancer Research UK Senior Cancer Research Fellow (Grant number C20826/A1210). We are grateful to Ian Titley and Gowri Vijaykrishnan for assistance with flow cytometry, Louise Howell for assistance with confocal microscopy and Brian Walker for assistance with the microarrays.
Author Contributions
LIA, ELD and FED designed the research, analyzed data. LIA, ELD and FM performed research and analyzed data. LIA, GJM and FED wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Aronson LI, Davies FE. DangER: protein ovERload. Targeting protein degradation to treat myeloma. Haematologica. 2012;97:1119–1130. doi: 10.3324/haematol.2012.064923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport EL, Moore HE, Dunlop AS, Sharp SY, Workman P, Morgan GJ, et al. Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood. 2007;110:2641–2649. doi: 10.1182/blood-2006-11-053728. [DOI] [PubMed] [Google Scholar]
- Hoang B, Benavides A, Shi Y, Frost P, Lichtenstein A. Effect of autophagy on multiple myeloma cell viability. Mol Cancer Ther. 2009;8:1974–1984. doi: 10.1158/1535-7163.MCT-08-1177. [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Miyazawa K, Moriya S, Ohtomo T, Che XF, Naito M, et al. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effectand induces endoplasmic reticulum stress in U266 myeloma cells: crosstalk among proteasome, autophagy-lysosome and ER stress. Int J Oncol. 2011;38:643–654. doi: 10.3892/ijo.2010.882. [DOI] [PubMed] [Google Scholar]
- Fu L, Kim YA, Wang X, Wu X, Yue P, Lonial S, et al. Perifosine inhibits mammalian target of rapamycin signaling through facilitating degradation of majorcomponents in the mTOR axis and induces autophagy. Cancer Res. 2009;69:8967–8976. doi: 10.1158/0008-5472.CAN-09-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment andthe identification of new targets for myeloma therapy. Leukemia. 2009;23:10–24. doi: 10.1038/leu.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz J-E, Ray-Coquard I. PI3 kinase inhibitors in the clinic: an update. Anticancer Res. 2012;32:2463–2470. [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hendil KB, Yashiroda H, Iemura S-i, Nagane R, Hioki Y, et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- Hutter G, Zimmermann Y, Rieken M, Hartmann E, Rosenwald A, Hiddemann W, et al. Proteasome inhibition leads to dephosphorylation and downregulation of protein expression of members of the Akt/mTOR pathway in MCL. Leukemia. 2012;26:2442–2444. doi: 10.1038/leu.2012.118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.