Abstract
Background: Previous studies focused on the association of the telomerase reverse transcriptase (TERT) gene polymorphism rs2736100 with lung cancer did not reach the same conclusion. In the present study, we performed a meta-analysis to systematically summarize the possible association between TERT polymorphism rs2736100 and the risk for lung cancer. Method: We conducted a search of case–control studies on the association of TERT with susceptibility to lung cancer in PubMed, EMBASE, ISI Web of Science, Wanfang database in China, and Chinese National Knowledge Infrastructure (CNKI) databases. Data from eligible studies were extracted for meta-analysis. Lung cancer risk associated with rs2736100 was estimated by pooled odds ratios (ORs) and 95% confidence intervals (95% CIs). Results: Six independent case–control studies on rs2736100 were included in our meta-analysis. Our results showed that rs2736100 was associated with the risk of lung cancer not only in an additive model (OR=1.19, 95% CI: 1.04–1.35; p=0.01), but also in a dominant model (OR=1.14, 95% CI: 1.01–1.28; p=0.03). Conclusions: This meta-analysis suggests that rs2736100 is associated with the risk of lung cancer.
Introduction
Lung cancer is the most frequent cause of cancer-related deaths throughout the world. And China bears a rising burden of lung cancer incidence and mortality. The major risk factor for lung cancer includes several environmental factors such as cigarette smoking and air pollution. However, there is considerable variation in the susceptibility to lung cancer (Shields and Harris, 2000; Kiyohara et al., 2002). Therefore, identification of genetic determinants associated with lung cancer risk has tremendous importance from a public health perspective.
Recently, a number of case–control studies have suggested that rs2736100 in the TERK gene is associated with the risk of lung cancer (Chen et al., 2010, 2012; Miki et al., 2010; Wang et al., 2010; Bae et al., 2012; Brenner et al., 2013; Myneni et al., 2103). The telomerase reverse transcriptase (TERT) gene is located on chromosome 5 (locus 5p15.33) and synthesizes the TTAGGG DNA sequences onto the ends of chromosomes in cooperation with other proteins of the core telomerase complex (e.g., telomerase RNA component [TERC] and dyskerin [DKC1]). With its activity, telomerase helps maintain the integrity of the genome in embryonic stem cells and in proliferating progenitor cells derived from quiescent normal stem cells (Bianchi and Shore, 2008; Osterhage and Friedman, 2009). Telomerase is silent in the vast majority of human tissues and is only expressed in a small number of normal cell types such as dividing male germline spermatocytes and a subset of proliferating somatic adult stem cells (Osterhage and Friedman, 2009).
In light of the already abundant evidence linking telomerase activity to the development of many tumor types, many researchers are testing the hypothesis that variability of the TERT gene sequence might be a general mechanism affecting individual cancer predisposition (Baird, 2010). Regarding the latter field of investigation, tens of thousands of patients affected with different cancer histotypes have been so far enrolled in molecular epidemiology studies, and some TERT polymorphisms have been reported to be associated with cancer risk, although findings are not always concordant (Baird, 2010). Therefore, we systematically reviewed the data published to date on the relationship between TERT polymorphisms and lung cancer risk and quantitatively summarized the available evidence by performing a formal meta-analysis.
Materials and Methods
Search strategy, eligibility criteria, and data extraction
A systematic review of original articles, reviews, and meta-analyses analyzing the association between TERT locus polymorphisms and lung cancer risk was performed by searching PubMed, EMBASE, ISI Web of Knowledge, Wanfang database in China, and Chinese National Knowledge Infrastructure (CNKI) databases with the following search terms: “TERT or rs2736100” AND “Lung cancer” or “Lung tumor,” by two independent investigators. Disagreements were resolved by iteration, discussion, and consensus. To unravel potential systematic biases, a third investigator performed a concordance study by independently reviewing all eligible studies; complete concordance (100%) was reached for all variables assessed.
In addition, we performed the following to retrieve other potentially relevant data: (1) cited references from selected articles were reviewed; (2) publicly available databases dedicated to associations between genotype and phenotype (e.g., Database of Genotypes and Phenotypes [dbGaP], www.ncbi.nlm.nih.gov/gap) were searched; (3) authors were contacted whenever unreported data were potentially useful for the systematic review or to rule out overlapping data reported in different publications.
Publication date and publication language were not restricted in our search. Reference lists were examined manually to further identify potentially relevant studies. The following data were extracted from eligible studies: the first author's last name, year of publication, country of origin, ethnicity, the numbers of genotyped cases and controls, and genotyping methods. All studies matching the inclusion criteria were retrieved for further examination and data extraction. All of the investigators have received training in literature search, statistics, and evidence-based medicine.
Statistical analysis
Meta-analysis was performed by using RevMan 5.0 software provided by the Cochrane Collaboration. We directly used the Q-test and I2 test to examine the heterogeneity between each study. By the heterogeneity test, if p>0.05, we select the Fixed Effect Mode1, and if p<0.05, we select the Random Effect Mode1 to merge the odds ratio (OR). p<0.05 was considered as a significant difference. Analysis of sensitivity includes the difference of point estimation and confidence intervals (CIs) of the combined effects value at a different model, to observe whether it changes the result. To test the publication bias, we used the RevMan 5.0 statistical software to make the funnel plot. The SIPA1 was tested for the associations with breast cancer susceptibility based on different genetic models. The meta-analysis examined the overall association of the SIPA1 with the risk of breast cancer measured by ORs at 95% CIs. The statistical significance of the pooled OR was determined with the Z-test, and a p value of<0.05 was considered significant.
Results
Study characteristics
A total of 246 articles were retrieved after first search in the databases above. After our selection, six case–control studies fulfilled the inclusion criteria. The qualities of the studies were considered acceptable for our meta-analysis. Characteristics of included studies are summarized in Table 1. A total of six studies involving 3100 cases and 11,392 controls were ultimately analyzed in our meta-analysis. There was one study carried out in the Europeans, whereas the remaining studies were performed in the Asians. Four genotyping methods were employed in the studies, including fluorescence-labeled hybridization probes method, TaqMan, Illumina Human550 BeadChips, and sequencing. The genotype distribution in the controls was in agreement with Hardy–Weinberg equilibrium (HWE) in all of the included studies.
Table 1.
Characteristics of Studies Included in the Meta-Analysis
| |
|
|
|
|
|
Genotypes case (%) |
Genotypes control (%) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Country | Ethnicity | Genotyping methods | No. (cases/controls) | TT | GT | GG | TT | GT | GG |
| Myneni | 2013 | China | Asian | Sequenom platform | 399/466 | 122 (34.6) | 141 (40.1) | 89 (25.3) | 157 (35.1) | 212 (47.4) | 78 (17.5) |
| Wang | 2010 | UK | European | Illumina Human550 BeadChips | 239/553 | 42 (17.6) | 115 (48.1) | 82 (34.3) | 136 (24.6) | 259 (46.8) | 158 (28.6) |
| Chen | 2012 | China | Asian | TaqMan | 196/229 | 50 (25.5) | 101 (51.5) | 45 (23.0) | 48 (21.0) | 112 (48.9) | 29 (30.1) |
| Miki | 2010 | Japan | Asian | TaqMan | 525/7676 | 157 (29.9) | 273 (52.0) | 95 (18.1) | 2830 (36.9) | 3664 (47.7) | 1182 (15.4) |
| Miki | 2010 | Korea | Asian | TaqMan | 557/1458 | 174 (31.2) | 277 (49.7) | 106 (19.1) | 567 (38.9) | 692 (47.5) | 199 (13.6) |
| BAE | 2012 | Korea | Asian | Fluorescence-labeled hybridization probes | 1094/1100 | 402 (36.7) | 501 (45.8) | 191 (17.5) | 422 (38.4) | 522 (47.5) | 156 (14.2) |
Meta-analysis
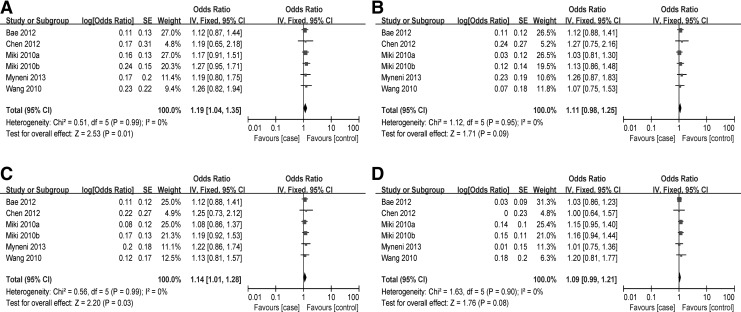
The associations between rs2736100 and susceptibility to lung cancer were analyzed in six independent studies with 3,100 cases and 11,392 controls. Results of the meta-analysis are shown in Figure 1. The Q-test in all of the genetic models showed no significant heterogeneity. Therefore, the fixed-effects model was used to analyze the association. In our analysis, significant differences were observed for the comparison of GG versus TT (OR=1.19, 95% CI: 1.04–1.35; p=0.01; Fig 1A) and GG+GT versus TT (OR=1.14, 95% CI: 1.01–1.28; p=0.03; Fig 1C), no difference was observed for the comparison of GG versus GT (OR=1.11, 95% CI: 0.98–1.25; p=0.09; Fig 1B) and GG versus GT+TT (OR=1.09, 95% CI: 0.99–1.21; p=0.08; Fig 1D).
FIG. 1.
Forest plot of lung cancer risk associated with rs2736100. (A) GG versus TT; (B) GG versus GT; (C) GG+GT versus TT; (D) GG versus GT+TT. The squares and horizontal lines correspond to the study-specific odds ratio (OR) and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95% CI. In this analysis, the fixed-effects model was used. CI, confidence interval.
Publication bias
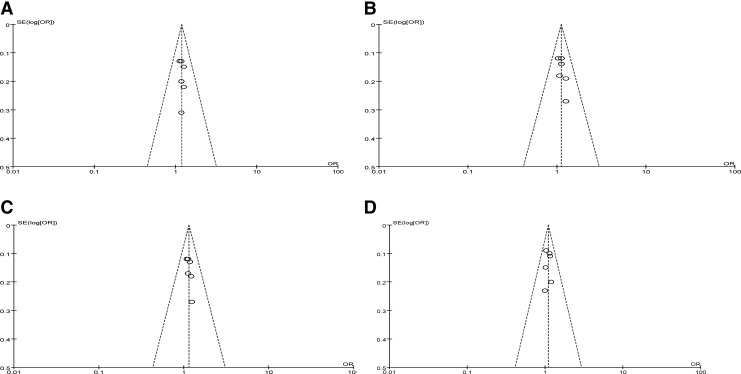
The funnel plot and Egger's test were performed to assess the publication bias of the literature. Symmetrical funnel plots were obtained in the single-nucleotide polymorphism (SNP) tested in all of the models. The Egger's test further confirmed the absence of publication bias in this meta-analysis (p>0.05) (Fig. 2).
FIG. 2.
Begg's funnel plot for the publication bias test. Each circle denotes an independent study for the indicated association. Log [OR], natural logarithm of OR. Horizontal line stands for mean effect size. (A) GG versus TT; (B) GG versus GT; (C) GG+GT versus TT; (D) GG versus GT+TT.
Sensitivity analysis
We deleted one single study from the overall pooled analysis each time to check the influence of the removed data set to the overall ORs. The pooled ORs and 95% CIs were not significantly altered when any part of the study was omitted, which indicated that any single study had little impact on the overall ORs.
Discussion
In the present study, an association of the rs2376100 with lung cancer risk was evaluated by the pooled results from six published studies. The results demonstrated that the rs2376100 GG genotype was associated with an increased risk for lung cancer.
Rs2736100 is located in intron 2 of TERT and, on the basis of the evolutionary and sequence pattern extraction through the reduced representation (ESPERR) score (Taylor et al., 2006), is located within a putative regulatory region (Landi et al., 2009). This polymorphism has also been linked to idiopathic pulmonary fibrosis, a disease associated with increased risk of developing lung cancer (Wang et al., 2009). It is also the most studied polymorphism of the TERT gene, as it was described in several previous studies (Chen et al., 2010, 2012; Miki et al., 2010; Wang et al., 2010; Bae et al., 2012; Brenner et al., 2013; Myneni et al., 2103). This polymorphism was reported associated not only with lung cancer (Chen et al., 2010, 2012; Miki et al., 2010; Wang et al., 2010; Bae et al., 2012; Brenner et al., 2013; Myneni et al., 2103), but also with other types of cancer (Wang et al., 2003; Carpentier et al., 2007; Hunter et al., 2007; Sagoo et al., 2009; Savage et al., 2007; Yeager et al., 2007). Notably, for testicular cancer, the G allele of rs 2736100 was associated with a decreased disease risk, whereas for all other tumor types, it was associated with an increased disease risk. As for the association of rs2736100 with lung cancer, several previous studies suggested that the GG genotype was associated with an increased disease risk. Our meta-analysis demonstrated these conclusions.
In this meta-analysis, a total of six case–control studies were analyzed to provide a comprehensive assessment of the association between the rs2736100 polymorphism and lung cancer. All of the studies checked genotypes for quality control. Genotype distribution of controls in all studies was consistent with HWE. In addition, exploring heterogeneity is one of the important goals of meta-analysis. In the present study, no significant heterogeneity was found among the included studies. Sensitivity analysis also showed that omission of any single study did not have a significant impact on the combined ORs. Furthermore, the funnel plot did not reflect obvious asymmetry, and the Egger's test further indicated no considerable publication bias in this meta-analysis. This made the results of this meta-study more reliable to some extent.
There are some limitations in the present meta-analysis. In the studies included, the genotyping methods used were not the same. Besides, other clinical factors such as age, sex, and different chemotherapies in each study might lead to bias. Determining whether or not these factors influence the results of this meta-analysis would need further investigation.
In conclusion, our study suggested that SNP rs2736100 of the TERT gene was associated with a significantly increased risk of lung cancer. Larger well-designed epidemiological studies with ethnically diverse populations and functional evaluations are warranted to confirm our findings.
Author Disclosure Statement
No competing financial interests exist.
References
- Bae EY. Lee SY. Kang BK, et al. Replication of results of genome-wide association studies on lung cancer susceptibility loci in a Korean population. Respirology. 2012;17:699–706. doi: 10.1111/j.1440-1843.2012.02165.x. [DOI] [PubMed] [Google Scholar]
- Baird DM. Variation at the TERT locus and predisposition for cancer. Expert Rev Mol Med. 2010;12:e16. doi: 10.1017/S146239941000147X. [DOI] [PubMed] [Google Scholar]
- Bianchi A. Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Brenner DR. Brennan P. Boffetta P, et al. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Hum Genet. 2013;132:579–589. doi: 10.1007/s00439-013-1270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier C. Lejeune J. Gros F, et al. Association of telomerase gene hTERT polymorphism and malignant gliomas. J Neurooncol. 2007;84:249–253. doi: 10.1007/s11060-007-9378-3. [DOI] [PubMed] [Google Scholar]
- Chen W. Zhang S. Zou X. Evaluation on the incidence, mortality and tendency of lung cancer in China. Thorac Cancer. 2010;1:35–40. doi: 10.1111/j.1759-7714.2010.00011.x. [DOI] [PubMed] [Google Scholar]
- Chen XF. Cai S. Chen QG, et al. Multiple variants of TERT and CLPTM1L constitute risk factors for lung adenocarcinoma. Genet Mol Res. 2012;11:370–378. doi: 10.4238/2012.February.16.2. [DOI] [PubMed] [Google Scholar]
- Hunter DJ. Kraft P. Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara C. Otsu A. Shirakawa T, et al. Genetic polymorphisms and lung cancer susceptibility: a review. Lung Cancer. 2002;37:241–256. doi: 10.1016/s0169-5002(02)00107-1. [DOI] [PubMed] [Google Scholar]
- Landi MT. Chatterjee N. Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D. Kubo M. Takahashi A, et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893–896. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- Myneni AA. Chang SC. Niu R, et al. Genetic polymorphisms of TERT and CLPTM1L and risk of lung cancer—a case-control study in a Chinese population. Lung Cancer. 2013;80:131–137. doi: 10.1016/j.lungcan.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhage JL. Friedman KL. Chromosome end maintenance by telomerase. J Biol Chem. 2009;284:16061–16065. doi: 10.1074/jbc.R900011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagoo GS. Little J. Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6:e28. doi: 10.1371/journal.pmed.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA. Chanock SJ. Lissowska J, et al. Genetic variation in five genes important in telomere biology and risk for breast cancer. Br J Cancer. 2007;97:832–836. doi: 10.1038/sj.bjc.6603934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields PG. Harris CC. Cancer risk and low-penetrance susceptibility genes in gene-environment interactions. J Clin Oncol. 2000;18:2309–2315. doi: 10.1200/JCO.2000.18.11.2309. [DOI] [PubMed] [Google Scholar]
- Taylor J. Tyekucheva S. King DC, et al. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Soria JC. Chang YS, et al. Association of a functional tandem repeats in the downstream of human telomerase gene and lung cancer. Oncogene. 2003;22:7123–7129. doi: 10.1038/sj.onc.1206852. [DOI] [PubMed] [Google Scholar]
- Wang Y. Broderick P. Matakidou A, et al. Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5-CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis. 2010;31:234–238. doi: 10.1093/carcin/bgp287. [DOI] [PubMed] [Google Scholar]
- Wang Y. Kuan PJ. Xing C, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fi brosis and lung cancer. Am J Hum Genet. 2009;84:52–59. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M. Orr N. Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]