Abstract
Recent technological advances provide new tools to manipulate cells and biological agents in micro/nano-liter volumes. With precise control over small volumes, the cell microenvironment and other biological agents can be bioengineered; interactions between cells and external stimuli can be monitored; and the fundamental mechanisms such as cancer metastasis and stem cell differentiation can be elucidated. Technological advances based on the principles of electrical, magnetic, chemical, optical, acoustic, and mechanical forces lead to novel applications in point-of-care diagnostics, regenerative medicine, in vitro drug testing, cryopreservation, and cell isolation/purification. In this review, we first focus on the underlying mechanisms of emerging examples for cell manipulation in small volumes targeting applications such as tissue engineering. Then, we illustrate how these mechanisms impact the aforementioned biomedical applications, discuss the associated challenges, and provide perspectives for further development.
1. Introduction
This review focuses on emerging micro- and nano-scale bioengineering and biomedical microfluidic technology platforms at the convergence of engineering, biology and materials science with an emphasis on broad biotechnology applications in medicine. Strategies have been sought to manipulate cells spatially to develop a better understanding of living systems. A broad range of technologies driven by numerous mechanisms such as fluid mechanics,1–3 chemical affinity,4 magnetic,5–8 electrical,9–11 optical,12–15 and acoustic fields16,17 have been presented to manipulate cells and provide potential solutions to diseases. For instance, achieving high cell density and structural complexity is of great importance for engineering artificial tissues. With broad applications of hydrogels in regenerative medicine and biomedical research,18 manipulating cell-encapsulating hydrogels via non-invasive external fields has emerged as a method to construct tissue models through a bottom-up engineering approach.19 Bioprinting different cell types is another approach to create functional tissue constructs.20,21 The ability to encapsulate and pattern single cells at physiologically relevant cell densities would allow engineering of complex tissue structures in vitro.18,22 Creation of in vitro three-dimensional (3-D) functional tissue constructs mimicking the role, micro-architecture, and functional properties of native biological systems and human tissues could potentially reduce pre-clinical testing on animals.19,23,24 Cost-effectiveness of gathering clinically relevant information is significantly improved through in vitro multivariate testing systems (e.g., cell microarrays for drug screening) due to reduced reagent consumption and increased mass exchange rates between cells and their microenvironment.23 With high-density microwells on a chip, isolation of single cells is achievable, which can be explored to elucidate the underlying mechanisms of cancer pathogenesis and metastasis, as well as to screen drug candidates at high throughput.23
Approaches for manipulating cells in micro/nano-scale volumes have been widely explored to improve point-of-care (POC) testing. Isolation of cells from whole blood (e.g., circulating tumor cells (CTCs) for cancer research,25 cluster of differentiation 4 positive (CD4+) T-lymphocytes for human immunodeficiency virus (HIV) monitoring26–28) has been demonstrated using microfluidic systems via immunocapture by antibodies specific to cell surface proteins.26,28–30 Because of their versatility and affordability, these devices can be used at primary care settings to facilitate clinical decisions. On the other hand, selective capture and on-demand release of stem cells in microfluidic channels provide an attractive alternative to enrich rare cells from complex biological samples, thus creating the possibility for downstream proteomic and genomic analyses.29,30
Here, we present state-of-the-art technologies employed to manipulate cells at the micro-scale level for applications in medicine. This review aims to provide a comprehensive overview with a broad perspective in physics, biology, engineering and medicine by highlighting the most significant approaches to date in cell manipulation for widespread applications. We provide examples focusing on clinical applications that were enabled by these technologies from a broad range of fields including diagnostics, regenerative medicine, reproductive medicine, and biopreservation. Although these fields seem to be apart from each other, we underline how they have been impacted by these emerging micro/nano-scale technologies sharing the same core competencies enabled by our evolving technological ability to command cells and their microenvironment in micro/nano-scale volumes.
2. Theories for modelling cell manipulation
There are several methods based on magnetic, optical, electrical, and mechanical principles to manipulate cells31 (Table 1). For instance, magnetic particles can be selectively attached to cells for cell separation or purification in microfluidic devices. There is a growing interest in optical tweezers for parallel, non-contact, and contamination-free manipulation of cells.32,33 Manipulation of target cells can also be achieved by microfabricated structures such as microfilters,34 microwells,35 microgrippers,36 dam structures,37 and sandbag structures.38 Electrical forces can be employed through electrophoresis and dielectrophoresis to manipulate cells. Dielectrophoretic forces arise from polarizability of cells, while electrophoresis arises from the interaction of cell charges and an electric field.39,40 Here, we describe underlying mechanisms for a set of emerging techniques41 where cell-encapsulating femto-to-nano-liter hydrogels are assembled and/or patterned into complex geometries for applications including tissue engineering.8,19,42
Table 1.
Principles, advantages, and disadvantages of main technologies for cell manipulation in micro-scale volumes for applications in medicine
| Applications | Principles | Advantages | Limitations |
|---|---|---|---|
| Diagnostic applications and applications in isolation, purification, and enrichment of cells | Magnetic:230–232 Magnetic field gradients can be employed to capture the super paramagnetic beads (10–100 nm in diameter), and cells that are selectively attached to beads. | High specificity, high efficiency, non-contact, clean, versatile, and non-invasive. | Difficulty in separating captured cells from magnetic beads when needed. |
| Electrical:39 Dielectrophoresis (DEP) is the motion imparted on uncharged particles due to polarization when subjected to a non-uniform electric field. DEP is the electronic analog of optical tweezers. | High efficiency, high selectivity, parallel manipulation. | Low survival rate of cells, complex instrumentation. | |
| Chemical:26,28–30,130 Antibody immobilization onto microfluidic channels. | High selectivity and high capture efficiency. | Chemical waste disposal, specific antibody development, lengthy procedure for antibody immobilization. | |
| Mechanical:34–38 Separation of target cells can be achieved by microfabricated structures. Advantages and disadvantages are separately written in the next columns for: (1) size-dependent filter-based microfluidic devices; (2) microgrippers; (3) dam and sandbag structures. | (1) High labeling efficiency, short detection time, high reproducibility based on simple and robust experimental procedures, high detection sensitivity at cell level; (2) manipulation of single cells with minimal damage; (3) relatively easier fabrication, and can efficiently immobilize cells with minimal stress. | (1) Poor selectivity; (2) Complex fabrication procedures, poor selectivity; (3) Poor selectivity. | |
| Optical:32,33 Biological molecules can be tethered to dielectric spheres, which can be captured at the focal point of an electric field gradient. | High-resolution, non-contact, contamination-free. | Limited manipulation area, complex optical setup, complex operation, and expensive instrumentation. | |
| Applications in tissue engineering and regenerative medicine | Magnetic assembly:7,8 Magnetic field gradients can be employed to actuate magnetic nano-particle (MNP) and/or free/stable radical-encapsulating hydrogels. | Non-contact, contamination-free, high throughput, low adverse effects on cells, high spatial resolution. | Although there are FDA approved magnetic particles and their release from micro-scale hydrogels are proven,233 release from large assembled tissue constructs has to be proven. |
| Acoustic assembly:16 Acoustic fields can be used to agitate the liquid surface where cell-encapsulating hydrogels assemble. | Non-contact, contamination-free, high throughput, low adverse effects on cells, low complexity. | Low spatial resolution. | |
| Surface tension driven assembly:4 Tendency of multiphase liquid–liquid systems to minimize the surface area and to reach a lower energy configuration is used. | Low complexity. | Low spatial resolution, low throughput. | |
| Microfluidic assembly:234–236 Microfabricated fluidic channels can be used as rails to transport hydrogel blocks. | High spatial resolution, low adverse effects on cells. | Complex fabrication, low throughput. | |
| Ratchet assembly:237 Unidirectional surfaces offer to assemble cell-encapsulating microgels via sequential assembly processes. | Low adverse effects on cells. | Complex fabrication procedures. | |
| Bioprinting: Several bioprinting methods based on acoustic,17,79 inkjet,80,81 valvebased,47,78,82,83 and laser printing13,22,84 technologies have been used to manipulate cell-encapsulating hydrogels. | High spatial resolution, high throughput. | High complexity, medium potential adverse effects on cells, medium cell density. |
2.1. Magnetic manipulation of cell-encapsulating hydrogels
Micro/nano-scale technologies have a significant impact on modern medicine.43–47 To manipulate cells in micro/nano-scale volumes, magnetic fields have been exploited in a variety of ways including direct cellular manipulation, cell sorting, 3-D cell culture, local hyperthermia therapy, and clinical imaging applications.43,48–56 For instance, magnetic fields have been utilized to manipulate cells to create 3-D tissue culture models leveraging magnetic levitation.57 Magnetic nanoparticles (MNPs) have been utilized to create two-dimensional (2-D) surface patterns,43,58–60 as well as 3-D cell culture arrays61 and characterize cell-membrane mechanical properties.62 In these magnetic methods, cells were first mixed directly with ferro-fluids or functionalized MNPs, and then exposed to external magnetic fields, allowing controlled manipulation. Besides, methods to encapsulate MNPs in hydrogels have been developed based on microfluidics51,63–65 and applied to multiplexed bioassays, e.g. rapid sensing of nucleic acids.66
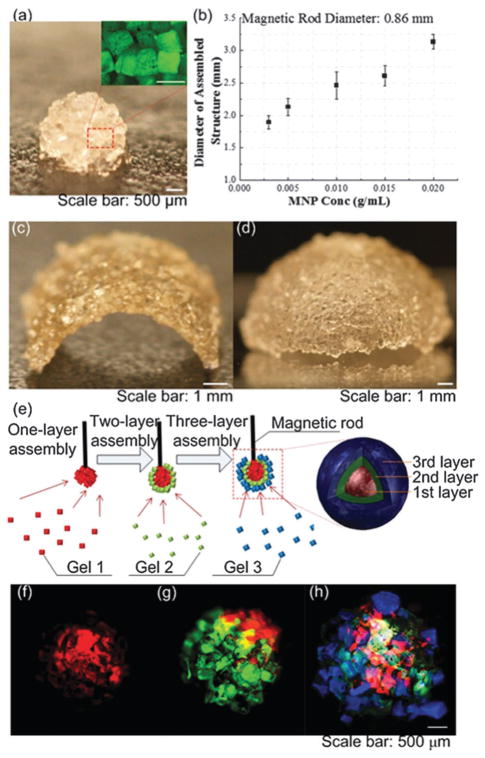
Assembly of cell encapsulating micro-scale hydrogels has been performed by utilizing MNPs (Fig. 1).7 Gels were also shown to present paramagnetic properties without MNPs due to formation of free radicals during photocrosslinking.8 These hydrogels were directed on a fluid reservoir by using a permanent magnet.8 Magnetic transport of hydrogels can be affected by several factors including: (i) viscous drag, (ii) hydrogel–fluid interactions (agitations to the flow field), (iii) gravitational field, (iv) buoyancy force due to density difference, and (v) thermal effects. Here, it is assumed that the active forces are the magnetically-induced forces and fluidic drag forces. The motion of cell-encapsulating hydrogels can be modeled using Newton’s law,
Fig. 1.
Magnetic manipulation and assembly of cell-encapsulating hydrogels. (a) Magnified image of the assembled single-layer spheroid construct. (b) Maximum spheroid assembly size versus MNP concentration. (c) and (d) Images of engineered arc and dome constructs by employing a flexible surface. (e) 3-D assembly of fluoresencently tagged gels. Layers were stained with rhodamine-B (f), FITC-dextran (g), and TPB (1,1,4,4-tetraphenyl-1,3-butadiene) (h), respectively from inner to outer shell. Reproduced with permission.7
| (1) |
where m and vm are the mass and velocity of the hydrogel, and Fm and Ff are the magnetic and fluidic drag forces, respectively. The magnetic force on a magnetized hydrogel by utilizing MNPs can be written as,
| (2) |
where V and M are the volume and magnetization of the hydrogel, respectively and H is the applied magnetic field. The fluidic drag force is approximated using Stokes’ law for the drag on a sphere,
| (3) |
where η and vm are the viscosity and the velocity of the hydrogel, respectively. Using these active forces, kinematics of floating hydrogels can be predicted,8 and assembly performance can be evaluated and improved.
2.2. Capillary driven manipulation of cell-encapsulating hydrogels
Lateral capillary forces were utilized for assembly of cell encapsulating micro-scale hydrogels,4 and evaluated mathematically to assess the limitations that the size of objects imposes on the assembly process.67 The assembly of two objects at the interface of two fluids (e.g., air and mineral oil,4 or two liquids67) can be evaluated by calculating the change in interfacial free energy. As two surfaces move from infinite separation to some finite separation, d (Fig. 2), the height h of the interface between the two objects can be calculated using the linearized Laplace equation:67
Fig. 2.
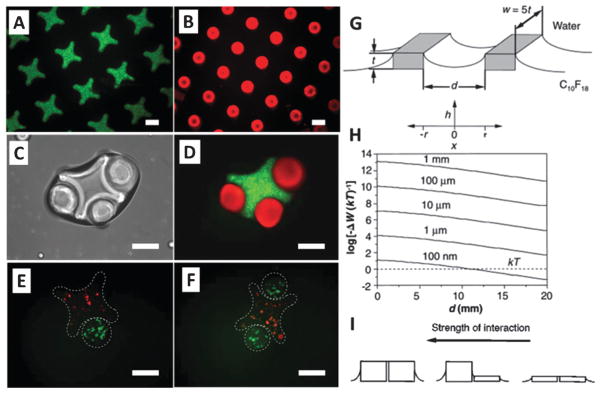
Capillary driven manipulation and assembly of cell-encapsulating hydrogels. (A and B) Fluorescence images of cross-shaped and rod-shaped hydrogels stained with FITC-dextran and Nile red, respectively. (C) Phase-contrast image of lock-and-key assemblies with three rods per cross. (D) Fluorescence image of lock-and-key assemblies. (E and F) Fluorescence images of single rod and two rod assemblies (scale bars are 200 μm). (G) The schematic of two objects and the coordinate system for eqn (4) and (5). The objects have a height of t and a width of w = 5t, and their proximate surfaces are separated by d. (H) For a range of object height, t = 1 mm to 100 nm, the change in interfacial free energy (non-dimensionalized with thermal energy, kT) in bringing two surfaces from infinite separation to a finite separation, d, is plotted. (I) The strength of interaction is correlated with the height of capillaries. Reproduced with permission.4,67
| (4) |
where γ is the interfacial free energy, Δρ is the density difference between the two fluids, g is the gravitational acceleration, and ΔP0 is the change in pressure across the interface at x = 0. The solution of eqn (4) is:67
| (5) |
where xc can be set as the capillary length (γ/Δρg)1/2. The interfacial energy can be calculated as a function of distance between objects. The alteration in interfacial free energy, ΔW, can be set to 5Δlγt (Fig. 2).67 Results showed that interfacial free energy for capillary self-assembly is favorable for flat objects with thickness as small as 100 nm (Fig. 2).67 For 2-D assembly of spherical objects, the radius of object at which interfacial free energy is close to thermal energy (kT) has been calculated to be on the order of 1 to 10 μm.68,69
2.3. Bioprinting cell encapsulating droplets
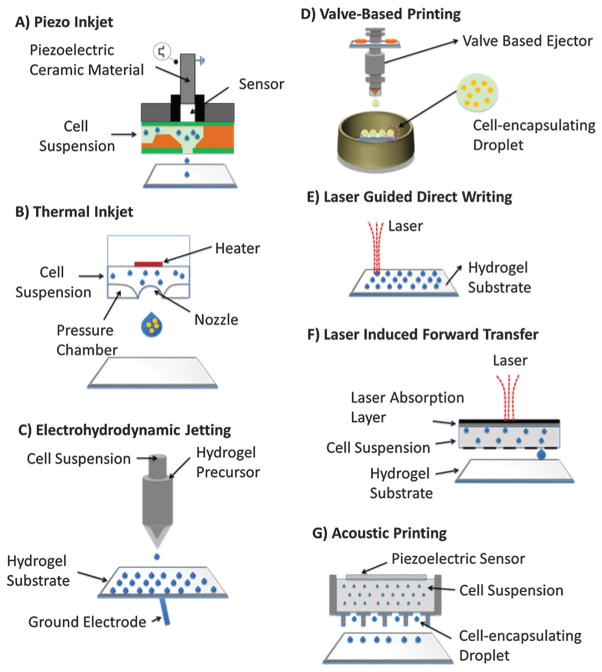
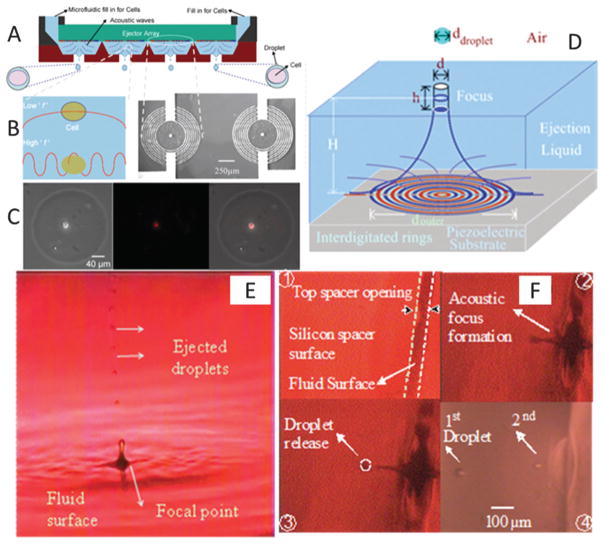
Droplet generation techniques based on microelectromechanical systems (MEMS) have been originally developed for applications in semiconductor industry.70–77 To address the needs in complex tissue engineering applications, these techniques have also been used for cell encapsulation17,78 and patterning (Fig. 3).17,21,78 These technologies offer significant advantages over existing top-down scaffolding methods. Among the different techniques of bioprinting are acoustic,17,79 inkjet,80,81 valve-based,47,78,82,83 and laser printing13,22,84 technologies. For instance, cell-encapsulating picoliter droplets were acoustically generated from an open-pool without a nozzle, which eliminates challenges associated with nozzle shear or clogging (Fig. 4). These devices can eject a broad range of materials including viscous polymers, fluids and cells simultaneously from the same ejector array. Further, they offer micro-scale precision over the cell position with droplet directionality. Pico-liter to nano-liter droplets (as small as 3–200 μm in diameter) can be generated with acoustic patterning technologies.76,85
Fig. 3.
Bioprinting technologies. (A) Piezo inkjet printing. (B) Thermal printing. (C) Electrohydrodynamic jetting. (D) Valve-based printing. (E) Laser guided direct writing. (F) Laser induced forward transfer. (G) Acoustic printing.
Fig. 4.
Schematic for acoustic nozzles droplet generation technology. (A) Multiple ejectors and ejected cell encapsulating droplets from different reservoirs. (B) Comparison of acoustic wavelength with the cell diameter. (B) The interdigitated circular micromachined device. Scale bar is 250 μm. (C) Computerized printing of single cell encapsulating droplets via xyz stage control. (D) Cylindrical acoustic focus at the fluid surface. (E and F) 28 μm droplets ejected upward from an open pool. Droplets ejected downward from a 100 μm wide microfluidic channel spacer opening. The ejector generated single droplets drop-on-demand without satellites. Reproduced with permission.17,71
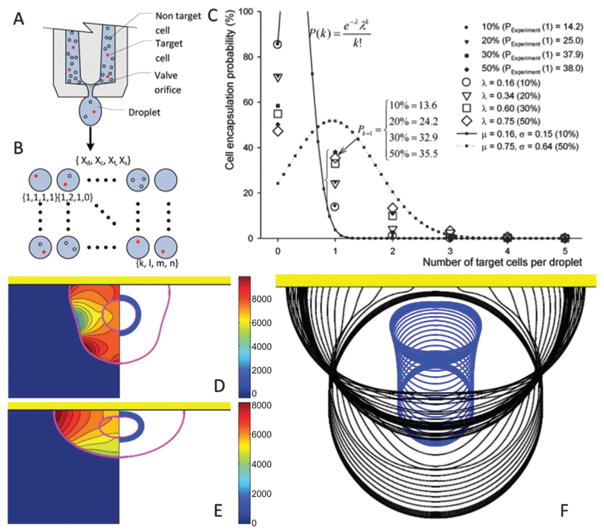
Bioprinting can be divided into two steps: (i) formation of cell-encapsulating droplets from cell suspension, and (ii) impact and subsequent relaxation of droplets on the receiving surface. Cell encapsulation is a highly probabilistic phenomenon as there are several governing parameters such as cell density and cell distribution in suspension. Statistical methodologies can provide a better understanding of the cell encapsulation process. A reliable and repeatable control can be gained over the parameters that characterize the cell encapsulation process. For several target cell concentrations and types of cell loading, the encapsulation process was statistically analyzed.86 As shown in Fig. 5A–C, while the percentage of target cells and homogeneity decreased in cell suspension solutions, the encapsulation probability of a target cell P(Xt) decreased. More recently, a statistical model based on negative binomial regression has been also presented to demonstrate how (i) cell concentration in the ejection fluid, (ii) droplet size, and (iii) cell size affect the number of cells encapsulated in an ejected droplet.87
Fig. 5.
Statistical and computational modeling of cell encapsulation and the printing process. (A–C) P(Xt) (eqn (3.6) in ref. 86) are cell encapsulation probability functions for the heterogeneous cell mixture including several cell loading concentrations. (A) Heterogeneous mixture of target and non-target cells inside an ejector reservoir. (B) Four parameters were distributed onto a matrix: (Xd) the number of droplets that contain cells, (Xc) number of cells per droplet, (Xt) number of target cells, and (Xs) droplets encapsulating a single target cell. (C) Cell encapsulation probability, P(Xt), as a function of number of target cells per droplet for cell concentration = 1.5 × 105 cells per mL. (D–F) Inner droplet representing the cell was assumed to be a highly viscous fluid and non-wetting (not sticking to the surface) while encapsulating droplets partially wetted the substrate. A moving contact line model90,238 was utilized to predict the dynamic contact angle. (D and E) Pressure contours and pressure distribution on the cell were plotted at the left half and the right half, respectively. Shear stresses peaked in the vicinity of the triple point during the initial phase of droplet–surface interaction. Triple point is the point where outer droplets, receiving substrate and ambient air, coincide. Maximum pressure was located near the contact line just before recoiling, and migrated to the distal end from the receiving surface where it stayed there until the recoil phase. Governing non-dimensional numbers are: We = 0.5, Re = 30, do/di = 2.85, σo/σi = 2541, μc/μd = 10. (F) Sequential impact images of cell encapsulating droplets. (A–C) are reproduced with permission86 and (D–F) are reproduced with permission.89
Similarly, for cell deposition, computational models enable researchers to develop an understanding of how parameters such as ejection speed and viscosity of cell suspension affect cell viability.88–90 Mechanical factors, e.g., shear stresses, hydrodynamic pressures, capillary forces, may amplify and cause deformation of cells. These factors can be controlled experimentally by adjusting ejection speed or by replacing cell suspension fluid with those having material properties (e.g., viscosity, density, and surface tension) that lead to less shear stress on cells. Cell viability also depends on receiving surface characteristics such as hydrophobicity, roughness, and elasticity of the surface. Prediction of cell deformation and viability via computational methods can be complementary to fulfil several design goals for building complex viable tissue constructs. A finite difference front tracking model was developed to model the deposition of viscous compound droplets (i.e., a smaller droplet encapsulated in a larger droplet) onto a receiving surface as a model for a cell printing process89,91 (Fig. 5D–F). In this work, the settings that result in least cell deformation and the smallest rate of deformation were identified. The analyses were performed for a set of nondimensional numbers, i.e., Reynolds number (Re), Weber number (We), viscosity ratio (μc/μd), surface tension ratio (σo/σi), diameter ratio (do/di), and equilibrium contact angle (θe). “Re” and “We” are non-dimensional numbers in fluid mechanics to evaluate the ratio of inertial forces compared to viscous forces and surface tension, respectively.
The computational results demonstrated that cell deformation gradually increased as: (i) Re increased; (ii) do/di decreased; (iii) σo/σi increased; (iv) μc/μd decreased; or (v) θe decreased. On the other hand, a local minimum, at least, of maximum geometrical deformation was obtained at We = 2. Cell viabilities were linked to cell deformation by employing an experimental correlation of compression of cells between parallel plates92 (Fig. 5F). Results showed that θe and μc/μd were highly correlated with cell viability.
3. Applications of micro-scale manipulation of cells
3.1. Diagnostic applications
The capability of manipulating cells (e.g., capture/isolation) at the micro-scale level, in combination with rapid detection technologies, creates the potential for inexpensive, portable and disposable devices for POC testing. Particularly, there is an urgent need for such diagnostic tools in resource-constrained settings where well-trained technicians, basic laboratory infrastructure and sustained financial support are not available.28,93,94 Additionally, patients would benefit from rapid healthcare technologies similar to rapid glucose tests. Such tests can enable close monitoring of patients suffering from a broad range of diseases ranging from renal diseases to infectious diseases such as HIV. Microfluidic technologies for cell manipulation in micro-scale volumes have enabled new approaches to monitor and detect various diseases. Here, we focus on (i) CTC capture and detection, (ii) CD4+ T lymphocyte count in blood, (iii) pathogen detection and cancer biomarker detection, and (iv) sperm monitoring and sorting.
3.1.1. Capture/detection of CTCs in microfluidic devices
Cancer related deaths account for approximately 13% of all deaths each year,95 primarily due to metastasis of CTCs in distant organs and subsequent metastatic diseases.96 As such, isolation of rare CTCs (at a ratio of one cell per 109 blood cells) from whole blood can potentially achieve cancer detection and reduce cancer-related mortality, as well as provide target cells for molecular characterization, drug screening and treatment monitoring. Earlier, antibody coated magnetic beads have been used to develop a system to isolate and quantify CTCs from blood using the CellSearch System (Veridex LLC, Raritan, NJ).97 Microfluidic devices have adapted this antibody-based isolation approach to isolate and enrich CTCs from peripheral blood.25,98,99 Generally, microfluidic based approaches rely on the interactions between proteins on the surface of CTCs and corresponding ligands immobilized in microfluidic devices. Depending on the downstream applications, captured cells on-chip are subsequently subject to immunostaining and molecular analyses to provide biological insights into metastasis and to identify new biomarkers for anti-cancer treatment monitoring.
For example, isolation of CTCs from whole blood using a microchip was reported.25 This microchip consists of micro-posts and the microchannel surface is coated with an anti-Epithelial cell adhesion molecule (EpCAM). On the surface of CTCs, the adhesion protein EpCAM is up-regulated compared to healthy blood cells. Under the optimal flow conditions, CTCs from metastatic lung, prostate, pancreatic, breast and colon cancer patients were captured on-chip due to the difference in the expression level of EpCAM.25 The captured CTCs were subsequently immunostained for confirmation. Under controlled laminar flow conditions, the authors achieved capture effciency of approximately 60% at a flow rate of 1–2 mL per hour and a purity of around 50%. With the advantage of continuous flow, this technology detected CTCs from 115 patients out of 116 in a study, indicative of the potential for management of cancer at clinical settings. This technology has been further developed by integrating an automated quantitative imaging system, which improved the analytical throughput.99
In addition to immunocapture, physical characteristics have been utilized for isolation of CTCs. CTCs are generally softer and more deformable than healthy cells,100 which allows the detection of carcinoma cells using impedimetric transducers to measure the increase in volume of cancer cells under hypsometric pressure.101 Alternatively, the size differences between cancer and benign cells can also be used to detect CTCs, based on conductance measurements.102 These methods allow detection of cancer cells from whole blood without pre-labeling, potentially realizing serial monitoring of anti-cancer therapy at the primary care settings in the future.
Captured CTCs can also be coupled with molecular techniques to monitor the regulation of protein expression that is associated with cancer metastasis. For example, the over-expression of epidermal growth factor receptor gene (EFGR) has a high correlation with cancer metastasis.103 In a small cohort study, genomic analysis of those isolated CTCs from non-small-cell lung cancer patients was performed.98 It was observed that the number of isolated CTCs was highly correlated with therapeutic responses and/or tumor progression. Particularly, the emergence of EGFR mutations was identified in some cases with progressive tendency. Constant molecular monitoring of cancer patients receiving treatment has potential to provide valuable information on prognostics and treatment efficacy.
3.1.2. CD4+ T lymphocyte count
CD4+ T lymphocyte cell count has been widely used as a gateway method to enroll acquired immune deficiency syndrome (AIDS) patients to initiate antiretroviral therapy (ART) in developing countries. Although there are significant advances in developing preventative microbicide vehicles,104–110 HIV/AIDS has caused 25 million deaths worldwide since the first case was reported in 1981.111 HIV primarily infects host immune cells such as CD4+ T lymphocytes and leads to immunodeficiency syndrome. Over time, CD4+ cell count gradually decreases and this reflects the host immune status against HIV replication. The World Health Organization (WHO) guidelines recommend a clinical cutoff of 350 cells per μL to initiate ART and/or switch effective treatment regimens if immunological failure occurs. Although commercial flow cytometers can be used to measure CD4+ cell count, prohibitive cost of instruments, consumables, and maintenance challenges significantly limit the wide use of CD4+ cell count to initiate ART in developing countries.112 In addition, CD4+ cell count needs to be monitored regularly to evaluate treatment response and patient adherence. Without regular monitoring (e.g., average every 4–6 months), HIV may evade effective drug suppression and lead to emergence of drug-resistant strains in infected individuals, which thus facilitate the spread of drug-resistant strains in the extended population. Clearly, there is an urgent need to develop rapid, simple and inexpensive POC CD4 assays to control the AIDS pandemic in resource-constrained settings.
So far, a variety of CD4+ cell counting methods have been implemented using microfluidic devices,26–28,44,113 which require precise manipulation of blood on-chip. The common approach employed by these devices is to capture, isolate and detect CD4+ T lymphocytes with a combination of mechanical, electrical, optical and cell-adhesion mechanisms. CD4+ T lymphocytes can be captured with efficiency up to 70.2 ± 6.5% in a straight microchannel via the aid of anti-CD4+ antibody immobilized on the device surface. For rapid detection, the captured CD4+ T lymphocytes can be stained using two specific biomarkers, CD4+ and CD3+ molecules, on the cell membrane. By dual-labeling, CD4+ T lymphocytes can be detected under a fluorescence microscope, and the number of CD4+ T lymphocytes can be counted manually under an optical microscope. To speed up the detection process, automated counting methods that use image recognition algorithms were developed.26,28,114 These algorithms enabled 100 times faster outcomes than manual counting for the fluorescent stained cells, especially when a large number of cells are present in the microchannels on the order of thousands. These approaches significantly increase the efficiency and accuracy to obtain CD4+ cell count. However, it should be noted that the need for a fluorescence microscope is not practical for POC testing in resource-constrained settings. To address this barrier, new technologies were developed including shadow-based lensless imaging,26–28,112,115 electrical sensing of lysed cells after isolation116 integrated with microfluidic chips where the cells were selectively captured. These advances led to point-of-care microfluidic detection methods that provide a CD4+ count using fingerprick volume of whole blood without any sample preprocessing steps.26 Specifically, to suit the need in resource-limited settings, label-free detection and counting strategies have been developed using light microscopy, impedance measurements and lensless imaging. In an initial effort, a light microscope was used to count captured CD4+ cells in a microfluidic device which involves a tedious manual counting step.117 To overcome this drawback, a rapid CD4+ cell counting method was developed using impedance measurement.118 This method eliminated the need for a fluorescence microscope and significantly shortened the quantification time as required in manual counting, which can be potentially implemented at the POC settings. Recently, CD4+ cell counts can be obtained using a lensless, ultra wide-field cell array based on shadow imaging (LUCAS) (Fig. 6),112 which enables rapid CD4+ cell counting within 10 minutes.27 Further, the LUCAS system has been validated in Tanzania with clinical samples and the results show positive correlation with the gold standard, i.e., flow cytometry, showing the potential of pairing with POC testing to facilitate clinical on-site decision-making.26
Fig. 6.
Microfluidic CD4 cell count performed by minimally trained personnel at MUHAS. (a) Microfluidic chips. (b) Preparation of surface chemistry with antibody injection. (c) Injection of blood sample. (d) Lensless imaging detection of CD4 cell count on-chip. Reproduced with permission.26
3.1.3. Microfluidics based POC applications
Similarly, microfluidic devices can also be used to detect multiple pathogens such as bacteria and viruses as well as smaller molecules such as proteins, demonstrating the versatility for developing POC assays in a miniaturized format. For example, microchips have been developed to selectively capture E. coli from various samples such as whole blood, phosphate buffered saline (PBS), milk, and spinach.119 This method can be used to monitor potential food poisoning and open wound infections without using conventional bacterial detection methods such as agar plate culture or polymerase chain reaction, as these conventional methods are time-consuming and instrument-dependent. In addition, agar plating, the gold standard for bacterial detection, requires 48 to 72 hours to complete,120 and it is often associated with false negative results ranging from 7.2 to 21.2%.121,122 The immuno-based microchip captured E. coli via lipopolysaccharide binding protein with a detection limit of 50, 50, 50, and 500 CFUs per mL in PBS, blood, milk, and spinach samples, respectively. The presented technology can be broadly applied to detect other pathogens, creating new avenues for POC diagnosis and monitoring of infectious diseases.
Enabled by the reliable immunocapture of pathogens in microfluidic devices, this technology was extended to address the urgent need for developing rapid HIV viral load assays, which is essential to diagnose early HIV infection and monitor AIDS patients on antiretroviral therapy (ART).94 Also, HIV viral load assays can be used to prevent mother to child transmission (MTCT), since traditional immunoassays cannot reliably detect HIV infection in infants until the age of 18 months due to passively transferred maternal antibodies.123 Previously, an on-chip detection method was developed to selectively capture HIV from 10 μL unprocessed HIV-infected patient whole blood.124 The method leveraged the photo-stability of quantum dots (Qdots) and tagged the captured HIV particles with dual-color labeling. Two types of Qdots (Qdot525 and Qdot655) were used to label envelope glycoprotein gp120 and high-mannose glycans for dual-staining. Recently, the on-chip capture mechanism was used to capture various HIV subtypes125 that are dominant in developing countries and constitute a significant challenge for commercial Ribonucleic acid (RNA) viral load assays.126,127 The results showed that anti-gp120 antibodies immobilized via Protein G surface chemistry captured subtypes A, B and C spiked in whole blood with efficiencies of 73.2 ± 13.6, 74.4 ± 14.6 and 78.3 ± 13.3% at a viral load of 1000 copies per mL, and with efficiencies of 74.6 ± 12.9, 75.5 ± 6.7 and 69.7 ± 9.5% at a viral load of 10 000 copies per mL. In another recent study, an on-chip sample preparation method that can remove blood cells from whole blood based on size-exclusion was developed.128 This lab-on-a-chip filter (2 μm pore size) device isolated HIV at high recovery efficiencies of 89.9 ± 5.0%, 80.5 ± 4.3% and 78.2 ± 3.8%, for the clinically relevant viral load levels of 1000, 10 000 and 100 000 copies per mL, respectively, while retaining 81.7 ± 6.7% of red blood cells (RBCs) and 89.5 ± 2.4% of white blood cells (WBCs) on the filter membrane. The chip can be operated by a single manual pipetting step without requiring any sample pre-processing steps. These devices can be potentially integrated with other detection mechanisms such as RT-PCR or enzyme-linked immunosorbent assay (ELISA) to achieve sample-in answer-out for HIV viral load monitoring and early diagnosis in resource-constrained settings. Recently, an electrical sensing method was developed to measure the impedance change resulted from the HIV viral lysate in a disposable microchip, which showed promising results for the detection of multiple HIV-subtypes including A, B, C, D, E, G, Panel at an early stage.40
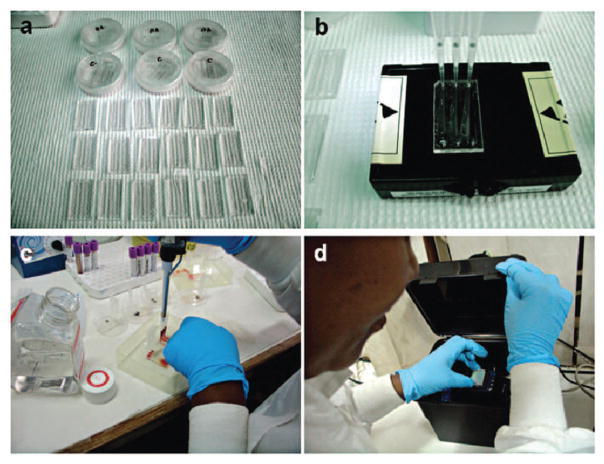
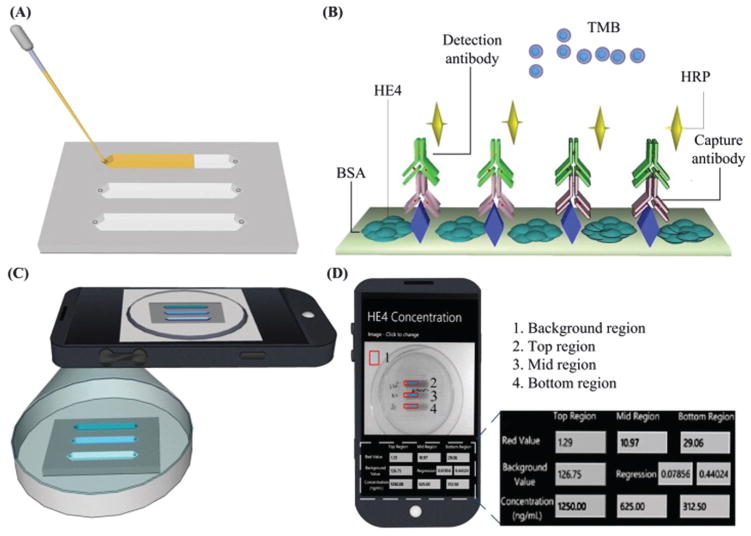
Additionally, microfluidic devices can be used to miniaturize conventional laboratory-based detection methods such as ELISA to detect protein biomarkers.129,130 For instance, ovarian cancer is asymptomatic at the early stages and it often leads to a poor 5 year survival rate of 33% at stages III and IV, when patients present with symptoms.131 To address the urgent need for ovarian cancer detection, an ELISA microchip was developed to detect human epididymis protein 4 (HE4) from urine.130 The microchip ELISA results were detected using a cell phone via integration of a mobile application that reports the concentration of HE4 on a phone screen (Fig. 7). As shown, the level of HE4 in urine samples from cancer patients (n = 19) detected was significantly elevated than the healthy controls (n = 20) (p < 0.001). Receiver operating characteristic (ROC) analyses showed that the microchip ELISA had a sensitivity of 89.5% at a specificity of 90%. This approach offers a quantitative ELISA test using a simple cell phone based colorimetric detection instead of an expensive, bulky spectrophotometer, thus potentially offering an attractive POC platform. In addition, the use of multi-channel microfluidic design allows detection of multiple biomarkers simultaneously, as well as the control.
Fig. 7.
Microchip ELISA integrated with cell phone application for detection of ovarian cancer from urine. (A) Loading of a small volume (100 μL) of urine sample into the microchannel. (B) Principle of direct ELISA for detection of HE4 on-chip. (C) The color development in microchannels was imaged using a cell phone built-in camera. (D) The concentration of HE4 in urine reported on the cell phone screen via a mobile application. Reproduced with permission.130
3.1.4. Microfluidic based sperm monitoring and sorting for reproductive medicine
6.1 million American couples (approximately 10% of American couples of childbearing age) are affected by infertility.132 Male originated issues are reported in almost half of the infertility cases, which require evaluation of parameters defining sperm quality, such as motility and sperm count. To address male based infertility issues, in vitro fertilization (IVF) with or without intra cytoplasmic sperm injection (ICSI) has been the most preferred assisted reproductive technology (ART) in clinics. One of the challenges of IVF and ICSI is identification and isolation of the most motile and the healthiest sperm from semen samples that have poor sperm counts (i.e., oligozoospermia) and/or poor motility (i.e., oligospermaesthenia).
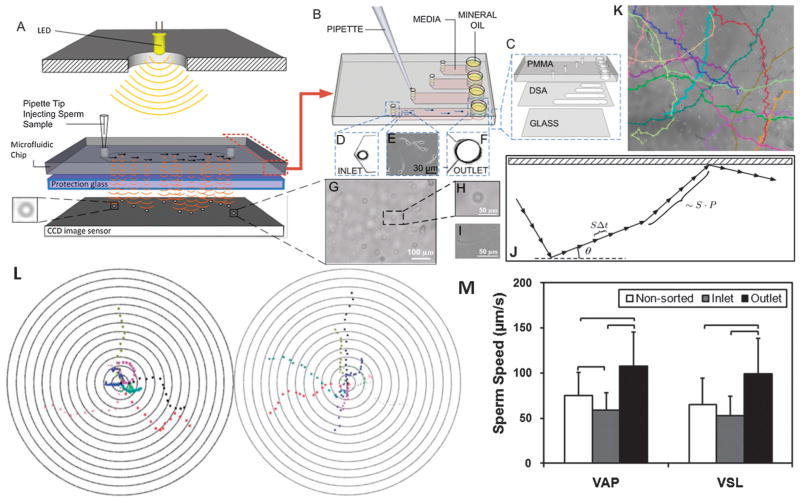
The applications of micro/nano-scale technologies to reproductive medicine have been explored.115,133–137 Microfluidic technologies have been employed to separate motile sperm with a higher efficiency compared to conventional sperm sorting techniques.115,134–139 A microfluidic device with an outlet junction was developed to separate motile sperm from directionally moving non-motile sperm.138 More recently, a lensless imaging system was integrated with a microfluidic chip to achieve automatic and wide field-of-view (FOV) imaging of a small population of sperm inside a microfluidic channel (Fig. 8).115 Using a lensless charge-coupled device (CCD) system, sperm swimming inside a channel were directly imaged as shadow patterns and sperm movements were tracked (Fig. 8). Since the field of view of the lensless sensor (4 mm × 5.3 mm) is approximately 20 times that of a conventional 10× objective lens (1 mm2), sperm stay within the field of view. Further, larger CCD systems (37.25 mm × 25.70 mm) have been earlier shown to reliably detect immobilized cells in microchannels.27,112 These systems could be used to record sperm over an entire channel and even in multiple parallel or serial channels. As the sperm is known to be responsive to gravitational stimuli,140 sperm motion in both horizontal and vertical microchip configurations was recorded, and results were displayed in bull’s eye plots presenting the sperm movement on chip (Fig. 8L). In both configurations, sperm displayed great diversity in their patterns of motion and direction. The swimming paths were tracked and the kinematic parameters that define sperm motility, including average path velocity (VAP), straight-line velocity (VSL) and straightness (VSL/VAP) were quantified (Fig. 8M). VAP refers to the average distance that a sperm covers in the direction of movement per unit time. VSL is the ratio of the straight-line distance between the start and end points of a sperm trajectory to the elapsed time until the sperm reaches the end point. Sperm were tracked in both horizontal and vertical configurations and the motilities were measured. The microfluidic channels create an environment that mimics the natural swim paths of sperm. Under real physiological conditions, sperm move through the vaginal mucus, cervical mucus and the cervical canal similar to a microfluidic channel. Once the mucus on the vaginal surface transforms into a less viscous watery phase, microchannels are formed. Such a space constrained microenvironment (i.e., the length scale in the flow direction is significantly larger than that in other directions) directs sperm towards the oocyte along with other contributing factors such as chemotaxis.115 To demonstrate microchip-based sperm sorting, sperm motilities at the outlet and inlet were compared to that of non-sorted sperm controls. As shown in Fig. 8M, sperm motility at the outlet was significantly higher than those of non-sorted sperm (p < 0.01), indicating that the microchip can be used to sort the most motile sperm, which can then be collected from the outlet. In addition, given the wide range of sperm velocities even after sorting, single cell based processing and monitoring enables separation of highest quality motile sperm from the rest utilizing either vertical or horizontal configurations. Recently, the exhaustion of mouse sperm (30 min) and human sperm (>1 hour) were quantitatively demonstrated and experimentally validated using microfluidics as an enabling technology.141
Fig. 8.
Sperm sorting. (A) Lensless imaging platform (LUCAS) integrated with a microchip for sperm tracking as highlighted by Nature Photonics.239 Shadows of the sperm generated by diffraction can be imaged using CCD in one second. (B) Loading sperm samples into microchannels of the space-constrained microfluidic sorting (SCMS) system from the inlets. The SCMS system with different channel lengths is assessed for effective sperm sorting. (C) The chip has three layers: PMMA, double-sided adhesive film (DSA), and glass coverslip. (D) Image of the channel inlet with a diameter of 0.65 mm under a 2×. (E) Image of sperm swimming inside a microchannel under a 10× objective. (F) The channel outlet with 2 mm diameter viewed using a 2× objective. (G–I) Sperm shadows on LUCAS. (J) A schematic of the trajectory of a sperm performing a Persistent Random Walk (PRW), where S is the velocity, P is the persistence time, Δt is the time step, and θ is the angle the trajectory makes with the x-axis. (K) Sperm tracks from image analysis. (L) Bull’s eye plot showing sperm motility vectors in the horizontal (left) and vertical (right) configurations. The distance between the adjacent concentric circles is 100 μm. (M) Comparison of Average Path Velocity (VAP) and Straight Line Velocity (VSL) of sperm for non-sorted conditions, and at the inlet and outlet of the 7 mm long microfluidic channel. The VAP and VSL were observed to be significantly greater for the sperm cells imaged at the outlet of the microfluidic channel compared to non-sorted sperm and the sperm at the inlet. Therefore the microfluidic sperm tracking system presented here shows potential to be also used as a sorting platform (n = 33–66, brackets indicate statistical significance with p < 0.01 between the groups). Reproduced with permission.115
3.2. Applications in isolation, purification, and enrichment of cells
Isolation, purification and enrichment of rare and specific target cells from heterogeneous populations and manipulation of cells in micro-scale volumes have enabled advancement of fields such as: cell based diagnostics,27,29,117 genomic and proteomic analyses,25,142,143 clonal and population studies,144,145 stem cell isolation for tissue engineering,29,30,146,147 and circulating tumor cell isolation for cancer research.25,148 Fluorescence-activated cell sorting and magnetic-activated sorting are commonly used cell manipulation technologies for isolation, which require preliminary processing and labeling of cells with fluorophore or magnetic particle conjugated antibodies. While these methods are powerful and successfully separate cells from heterogeneous mixtures, the cost, complexity and requirements for infrastructure (e.g., facility and reagents) limit their use.149 Miniaturization of these systems has been attempted by reducing them to simpler microfluidic versions.150,151 However, the required peripheral equipment still remains large in size and costly. Furthermore, genomic studies on captured cells in microfluidic systems are mainly hampered by the loss of genomic material, which can adhere to the channel walls due to the large surface to volume ratio. These limitations can be addressed by the release and downstream processing of captured cells in microfluidic channels through manipulation in micro-scale volumes.29,30
Release of the selectively captured live cells (e.g., CTC, CD4+, CD34+, endothelial cells) in micro-scale volumes would enable post-culturing, clonal and molecular characterization studies. However, challenges remain in effectively releasing the captured cells in microfluidic channels without compromising the viability of the captured cells.152–154 To minimize adverse effects on cells, manipulation and recovery of the captured cells need to be performed without using chemical or physical factors, which can affect cellular characteristics. For example, using enzymatic or fluid shear based detachment is known to adversely affect cell viability and function.155,156 Alginate based hydrogels have recently been used in combination with poly(ethylene glycol) and conjugated antibodies in microfluidic channels to capture endothelial progenitor cells from blood.157 The captured cells were then released by dissolving the hydrogel in the channels with a chelator, e.g., ethylenedi-aminetetraacetic acid (EDTA). Alginate hydrogels, however, allow a high level of non-specific binding154,158 and offer limited numbers of sites for the conjugating antibodies, which significantly reduce the capture efficiency.157 Furthermore, coating and chelation of hydrogels in microfluidic channels add extra steps and cost, lengthy processing time, operational complexities and additional reagents, which render this approach challenging to apply, especially at the bedside or resource-constrained settings.
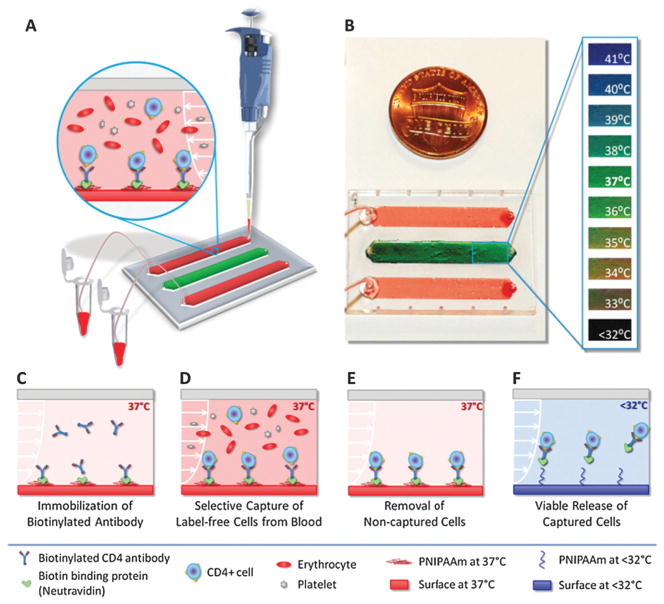
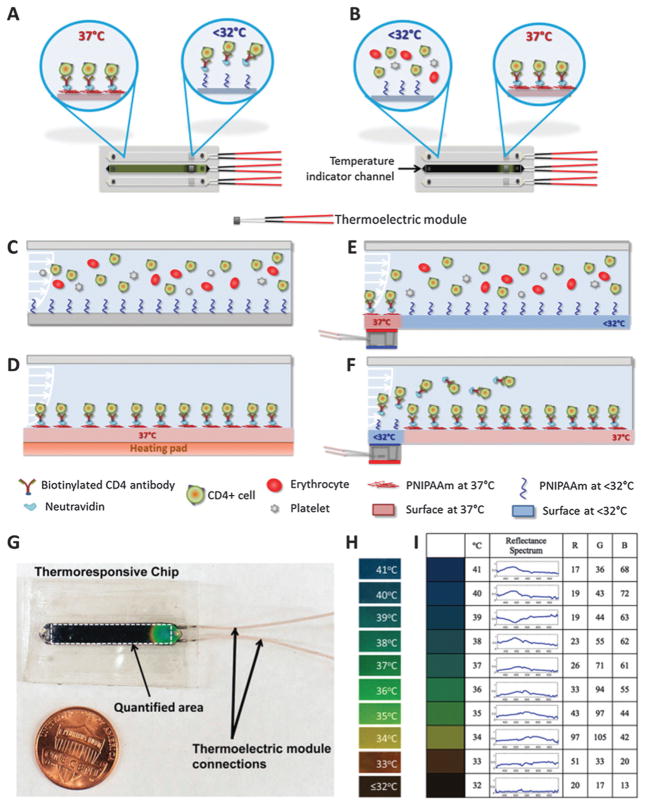
Most of the downstream applications of cell capture require the captured cells to be lysed on-chip for genomic and proteomic analysis. However, cellular materials are lost to the microchannel surfaces due to the large surface to volume ratio. Additionally, in most cases the captured cells are so rare in concentration that they need to be expanded over culture for following biological analysis steps. Hence, manipulation and release of captured cells in microfluidic channels is a significant enabling biotechnology with broad applications. It was previously shown that poly(N-isopropylacrylamide) (PNIPAAm) coated surfaces interact strongly with peptides and proteins (e.g., insulin chain A, serum albumin) above lower critical solution temperature (LCST).159,160 Whereas, when the temperature of the surface is reduced below the LCST, a complete desorption of the adsorbed proteins is possible.159,160 It was shown that the protein adsorption–desorption mechanism on a temperature-responsive polymer (PNIPAAm)160 can be modified to develop a biotin-binding protein and biotinylated antibody based surface chemistry to rapidly and selectively capture and controllably release cells on-demand in microfluidic channels.29,30 Thermoresponsive microfluidic channels were developed by functionalizing surfaces with biotin-binding protein and biotinylated antibody that can selectively capture specific cells from blood and controllably release with high viability and specificity, which provide a new platform to manipulate cells in micro-scale volumes (Fig. 9). More recently, a microfluidic system with sufficient micro-scale control to locally release a selected set of captured cells on-chip would significantly increase specificity (Fig. 10).30 As microfluidic cell capture methods suffer from non-specific binding events, being able to manipulate and selectively release the undesired or desired cells in channels30 would improve the purity of captured cells especially for subsequent genomic/proteomic analysis161 and biomarker discovery.162
Fig. 9.
Isolation, purification, and enrichment of cells using thermoresponsive microfluidic channels for on-demand releasing the selectively captured cells from heterogeneous mixtures.29 (A) The thermoresponsive microfluidic chip is composed of three channels. The middle channel was used as an indicator of the temperature. (B) The indicator channel was covered with liquid crystal dye. (C) Schematic representing the mechanism of label-free selective capture of cells and on-demand release of cells in thermoresponsive microchannels. (D) Sample at 37 °C (e.g., blood) is injected into the channel, and the target cells (e.g., CD4+ or CD34+ cells) are captured. (E) The non-target cells in channels, which are not captured, are then washed off. (F) The channels are then cooled down below 32 °C. Upon cooling, the released cells are rinsed off the channels and collected at the outlet. Reproduced with permission.29
Fig. 10.
Manipulation of cells in microchannels through local capture and on-demand release. (A) Local release of captured cells. (B) Local capture of specific cells in microchannels. (C–F) Mechanism of local manipulation of cells in microchannels in the absence and presence of thermoelectric heating elements. (G) Local control of temperature thermoresponsive microchannels and a photograph of a microchip with a local temperature control. (H) Temperature responsive dye works between 32 °C to 41 °C, and displays green color at 37 °C, the temperature at which cells were captured. The dye appears black below 32 °C, at which on-demand local cell release is achieved. (I) Baseline RGB values represent the colors displayed by the temperature indicator channel. Reproduced with permission.30
3.3. Applications in tissue engineering and regenerative medicine
Cells in tissues and organs are intricately organized in a 3-D architecture at high densities. The capability to precisely position individual cells in a 3-D architecture at high densities can contribute to recapitulation of more physiologically similar tissues through manipulation of cells in micro-scale volumes. Over the years, there have been significant efforts to generate tissue-engineered organs.163–172 The following characteristics of native tissues can be mimicked by employing technologies to manipulate cells in micro/nano-scale volumes: complex 3-D cellular and extracellular architecture, high cell density and complex vascular network. A crucial aspect of tissue engineering is to reproduce the body’s architectural intricacies that would facilitate the vital cell–cell and cell–extracellular matrix (ECM) interactions in creating an in vivo-like environment.173–178 Current methods for generating 3-D tissue constructs offer a limited control over cell density, which is well below the physiologically relevant densities.22 For instance, cardiac tissue is comprised of a high density of cardiomyocytes, cardiofibroblasts and endothelial cells (~108 cells per cm3).179 There is a need for a nutrient and oxygen rich environment for the cells residing deep in tissues. Therefore, a crucial challenge in tissue engineering of organs is to supply sufficient oxygen and nutrients to sustain cells located deep within the 3-D tissue construct with a porous or vascular network.180–182 Current engineered 3-D tissues are developed to culture cells within biodegradable natural or synthetic scaffolds.164,183–186 These engineered scaffolds function as 3-D structures within which cells grow, interact with the ECM and communicate with other cells in the proximity.164,187 Scaffolding techniques have traditionally been used to form degradable porous, polymer scaffolds (e.g., polyethylene glycol, collagen hydrogels and agarose) that are seeded with cells forming a 3-D construct.163,188–190 Even though these methods are commonly used, scaffold-based technologies face challenges in precisely patterning cells in specific architectures to form highly organized and high cell density tissue constructs. Stepwise brick-by-brick building block techniques constitute an alternative approach to build 3-D tissue constructs.190 In this approach, 3-D tissue structures are built by using micro-scale hydrogels (200–500 μm in size).
Various bioprinting methods have been developed to control cell seeding in 2-D and 3-D settings, such as inkjet and laser printing21,80,81,191 to gain control over the spatial cellular and ECM composition, cell viability and functionality, and spatial density.192 Cell manipulation technologies in micro-scale volumes can have a clinical impact on medicine, such as in cardiac and neural tissue regeneration. Cardiovascular diseases affect 70 million Americans, resulting in an economic burden of $300 billion and accounting for nearly 40% of all deaths in the US.18 Conventional treatments of cardiac injury cannot achieve myocardial regeneration. Engineered cardiac tissue grafts can be surgically placed into the defect site to facilitate cardiac regeneration.18 Additionally, neurodegenerative diseases,193–196 such as Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD), Amyotrophic lateral sclerosis (ALS), and Traumatic brain injury (TBI), lead to impairment of the neural system, loss of cognitive abilities, and permanent paralysis. Currently, there is no effective treatment for neurodegenerative diseases, or brain and spinal cord injuries.197,198 Neural tissue is extremely complex, contains many different cell types with connections forming an intricate network. The neural circuits of the brain are 3-D networks that are diverse in cellular and molecular composition, as well as morphology. For example, for brain tissue engineering, which has thousands of different kinds of cells arranged in complex 3-D organization, new kinds of engineering techniques and micro-scale cell manipulation methods are required.42,199–203
3.4. High-throughput in vitro drug testing applications
Cell microarrays have been widely used to screen for drug candidates in a high-throughput manner.23 Compared to traditional screening methods in a 384-well plate, cell microarrays have advantages such as reduction of reagent consumption and shortened assay time. In a typical cell microarray, high-density cell spots are deposited via micro-scale cell manipulation methods (e.g., bioprinting, soft lithography) with precise temporal and spatial control. To test the drug efficacy and toxicity on cell microarrays, millions of drug candidates can be screened simultaneously. As reported, a variety of methods have been developed to deposit cells in micro-scale volumes onto microarrays, including patterning,204 stamping,205 microfluidic-based drug loading206 and aerosol sprays.207,208 These studies clearly demonstrated the widespread applications of 3-D cell microarrays for drug screening with significantly increased cost-effectiveness.209–212
3.5. Applications in biopreservation and cryoprinting
Biopreservation aims to preserve cells and tissues by cooling them to sub-zero temperatures at which biological activity is slowed down or completely ceased.213,240 Following the cryopreservation time of interest, frozen tissues and cells are thawed, and ideally samples resume to their biological activity. During this process, cryoprotectant agents (CPAs) are employed to avoid cryoinjury of cells by cooling to temperatures at which intracellular ice formation takes place. The current practice for cryopreservation are slow freezing and vitrification using various CPAs such as dimethylsulphoxide (DMSO), 1,2-propane-diol (PROH) and ethylene glycol (EG), sucrose, trehalose and mannitol. The slow freezing approach214 is an established method, in which samples are cryopreserved at controlled freezing rates. Low levels of CPAs are used (e.g., 1.5 M) in the slow freezing method to avoid intracellular ice formation and minimize cell membrane215 and cytoskeleton damage.216 However, this technique is not ideal due to the low post-thaw cell survivability and functionality. On the other hand, vitrification transforms cells into a glass-like solidification state,217 and has emerged as a potentially ideal alternative method because of the higher survival rate by eliminating ice crystal formation. However, high levels of CPA (e.g., 6–8 M) have to be used for rapid freezing necessary for vitrification (e.g., −1500 °C min−1), which may also cause uncontrolled differentiation and smaller viability of cells.218–220
Integration of nano-liter droplet cell encapsulation technologies with vitrification is promising.221–223 Single to few cells encapsulated in droplets can be directly ejected into a liquid nitrogen reservoir utilizing bioprinting technologies for vitrification. These droplets can also be placed on a thin film and then directly dipped into liquid nitrogen.224 This process enables minimized CPA volumes for vitrification, and, hence, benefits from enhanced warming and cooling rates due to the larger surface to volume ratio. Moreover, biopreservation technologies may need to achieve processing of large sample volumes at high-throughput to enable clinical relevance and practical use. For instance, the cryopreservation of blood is of great importance to public health during natural disasters, global issues, warfare, and in clinical settings due to fluctuations in supply and demand.225 Also, the existing approaches employing high CPA concentrations may require tedious manual steps during handling of cells between CPA loading and unloading solutions. Therefore, integration of nano-liter cell encapsulation technologies with vitrification can reduce manual steps, handling errors and operator variability.
Recently, a droplet ejection technology was developed to vitrify oocyte encapsulating nano-liter droplets.226 This platform was utilized to vitrify and cryopreserve oocytes, which were encapsulated in nano-liter droplets at high-throughput.226 During this vitrification process, morphologies of the mouse oocytes, their parthenogenetic development, and survival rates were compared to fresh oocytes cultured in the potassium simplex optimized medium (KSOM) right after retrieval. Similar survival rates were reported between the results of the droplet encapsulation method and the control group (i.e., fresh oocytes after 24 h in culture). Morphologies of the oocytes and the rate of parthenogenetic activation were also comparable between the oocytes collected after vitrification/thawing and the fresh oocytes. Another important application of cryopreservation is blood banking.224 Recently, RBC encapsulating nano-liter droplets (e.g., radius < 100 μm) were vitrified at high throughput (Fig. 11).224 A co-flow droplet ejection system (i.e., nitrogen and RBC flow) was used to generate RBC encapsulating droplets. The effects of the experimental parameters: (1) the RBC flow rate (RBCs were loaded with CPA) (160 to 220 mL min−1), (2) the nitrogen gas flow rate (3.2 to 4.8 L min−1), and (3) the distance between the ejector tip and droplet receiving film (60 to 90 mm), were evaluated (Fig. 11).224 To enable vitrification of RBCs with low CPA levels, nano-liter droplets were created in a continuous manner by adjusting the nitrogen gas flow rate. This approach minimized the possibility of negative osmotic and toxic effects by increasing cooling rates and using a low level of CPAs.227,228 On the other hand, stem cells have been clinically essential due to their pluripotent potential and their role in regeneration of damaged tissues.185 For stem cells, high CPA levels can cause uncontrolled differentiation and smaller viability.219,220 Vitrification results in higher viabilities of stem cells compared to other methods including slow freezing.229,240 Low CPA-level vitrification in nanoliter volumes has immense potential for stem cells compared to other methods in preserving their functionality.219,220,229 Long-term biopreservation of cells in micro-scale volumes has a broad impact on multiple fields including tissue engineering, regenerative medicine, stem cells, blood banking, animal strain preservation (biodiversity protection), clinical sample storage, transplantation medicine and in vitro drug testing.
Fig. 11.

A schematic of the blood cryopreservation platform. (a) Ejection and deposition of RBC encapsulating droplets on receiving paper (top) and droplet pictures (bottom). Scale bar is 500 μm. (b) Average size of ejected droplets is plotted for a range of sheath gas flow rates (3.2, 4.0, 4.8 L m−1), blood flow rates (160, 180, 200, and 220 mL min−1), and droplet collecting distances (60, 75, and 90 mm). Reproduced with permission.224
4. Conclusions and prospects
Manipulating cells in small volumes while maintaining viability and biological functionality has been a significant challenge for various applications in medicine. Advanced micro-scale manipulation technologies emerge at the convergence of multiple fields including basic sciences, engineering, and medicine. To address these challenges, several approaches have been developed based on magnetic, electrical, mechanical, acoustic, and fluidic principles. Among these approaches, bioprinting technologies are promising to achieve manipulation of cells and biological agents for applications in drug discovery, drug toxicity and efficacy testing, biopreservation and cancer research. On the other hand, development of microfluidic technologies has enabled miniaturization of traditional methods of manipulating, isolating, detecting and quantifying rare cells and molecules from bodily fluids. Due to simplicity, low cost and portability, microfluidic devices can be utilized widely across many disciplines to manipulate cells and biological agents in micro/nano-scale volumes. However, when a vast number of cells, different cell types and multiple scales are involved, challenges remain to be addressed in high-throughput applications of these technologies. Current challenges in micro-manipulation of biological samples are the need for improved speed, flexibility, higher levels of automation, and contact-free operation. These challenges need to be addressed to make them widely available biological tools reducing manual operations without compromising accuracy and cost. To develop widely applicable methods that allow integration of various miniaturized devices on a single system, and parallel processing, a better understanding of microscale manipulation technologies is needed. Further, technologies are needed that have advantages including lower reagent and power consumption, portability, shorter reaction time, and lower cost to manipulate cells in micro-scale volumes. Future studies should also address the need for versatility, ease-of-use, scalability, and high-throughput utilization. These various challenges could be potentially addressed at the convergence of multiple fields.
Acknowledgments
We would like to acknowledge U54 EB015408, R21 HL112114, R01 AI093282, R01 AI081534, R21 AI087107, R21 HL095960, and R01 EB015776.
Biographies
 Savas Tasoglu
Savas Tasoglu
Dr Tasoglu received his PhD from UC Berkeley. His current research interests at Harvard are magnetics, neurotechnologies, microfluidics, cell and tissue mechanics, regenerative medicine, microbicides, cryopreservation, and cell-based diagnostics for point-of-care. Dr Tasoglu’s achievements in research and teaching have been recognized with numerous fellowships and awards including Chang-Lin Tien Fellowship in Mechanical Engineering, Allen D. Wilson Memorial Scholarship, and UC Berkeley Institute Fellowship for Preparing Future Faculty. Dr Tasoglu’s three articles with first authorship featured the cover of prestigious journals such as Advanced Materials, Trends in Biotechnology, and Physics of Fluids.
 Umut Atakan Gurkan
Umut Atakan Gurkan
Dr Gurkan is leading the CASE Biomanufacturing and Microfabrication Laboratory in Mechanical and Aerospace Engineering at Case Western Reserve University. Dr Gurkan’s research interests include micro/nano-engineered biological systems with applications in medicine. Dr Gurkan’s collaborative clinical research and teaching have been recognized with various prestigious awards, including the IEEE-Engineering in Medicine and Biology Society Wyss Award for Translational Research and the Partners in Excellence Award for Outstanding Community Contributions. Dr Gurkan’s research has been highlighted by international and national news agencies, newspapers, and scientific publishers, including MIT News, Science Daily, Wired News, Reuters, Hurriyet News, Lab Chip, and Nature Photonics.
 ShuQi Wang
ShuQi Wang
Dr ShuQi Wang obtained his medical degree from Bengbu Medical College, China, in 2000. He further pursued his master’s degree in Molecular Biology and Immunology at the Institute of Dermatology, Peking Union Medical College, Chinese Academy of Medical Sciences. In 2009, he received his PhD from the University of Cambridge, UK, focusing on rapid nucleic acid amplification technologies for HIV viral load monitoring in resource-limited settings. Presently, he is working on the development of rapid immunoassays for point-of-care diagnostics using microfluidic approaches.
 Utkan Demirci
Utkan Demirci
Utkan Demirci, PhD, is an Assistant Professor of Medicine and Health Sciences and Technology at Harvard University Medical School and Brigham and Women’s Hospital. Dr Demirci creates nano- and micro-scale technologies providing solutions for real world problems in medicine. Dr Demirci is an internationally renowned scientist and his work has been recognized with numerous national and international awards, and highlighted in Wired Magazine, Science Daily, Reuters Healthnews, Nature Photonics, MIT Technology Review, AIP News, BioTechniques, and Biophotonics.
Notes and references
- 1.Kessler JO. Nature. 1985;313:218–220. [Google Scholar]
- 2.Sundararajan N, Pio MS, Lee LP, Berlin AA. J Microelectromech Syst. 2004;13:559–567. [Google Scholar]
- 3.Lee GB, Hwei BH, Huang GR. J Micromech Microeng. 2001;11:654–661. [Google Scholar]
- 4.Du Y, Lo E, Ali S, Khademhosseini A. Proc Natl Acad Sci U S A. 2008;105:9522–9527. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H, Purdon AM, Westervelt RM. Appl Phys Lett. 2004;85:1063–1065. [Google Scholar]
- 6.Yan J, Skoko D, Marko JF. Phys Rev E: Stat Nonlinear Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 7.Xu F, Wu CA, Rengarajan V, Finley TD, Keles HO, Sung Y, Li B, Gurkan UA, Demirci U. Adv Mater. 2011;23:4254–4260. doi: 10.1002/adma.201101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tasoglu S, Kavaz D, Gurkan UA, Guven S, Chen P, Zheng R, Demirci U. Adv Mater. 2013;25:1137–1143. doi: 10.1002/adma.201200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srigunapalan S, Eydelnant IA, Simmons CA, Wheeler AR. Lab Chip. 2012;12:369–375. doi: 10.1039/c1lc20844f. [DOI] [PubMed] [Google Scholar]
- 10.Bogojevic D, Chamberlain MD, Barbulovic-Nad I, Wheeler AR. Lab Chip. 2012;12:627–634. doi: 10.1039/c2lc20893h. [DOI] [PubMed] [Google Scholar]
- 11.Hughes MP. Electrophoresis. 2002;23:2569–2582. doi: 10.1002/1522-2683(200208)23:16<2569::AID-ELPS2569>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Nahmias YK, Gao BZ, Odde DJ. Appl Opt. 2004;43:3999–4006. doi: 10.1364/ao.43.003999. [DOI] [PubMed] [Google Scholar]
- 13.Nahmias Y, Schwartz RE, Verfaillie CM, Odde DJ. Biotechnol Bioeng. 2005;92:129–136. doi: 10.1002/bit.20585. [DOI] [PubMed] [Google Scholar]
- 14.Grier DG. Nature. 2003;424:810–816. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 15.MacDonald MP, Spalding GC, Dholakia K. Nature. 2003;426:421–424. doi: 10.1038/nature02144. [DOI] [PubMed] [Google Scholar]
- 16.Xu F, Finley TD, Turkaydin M, Sung Y, Gurkan UA, Yavuz AS, Guldiken RO, Demirci U. Biomaterials. 2011;32:7847–7855. doi: 10.1016/j.biomaterials.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demirci U, Montesano G. Lab Chip. 2007;7:1139–1145. doi: 10.1039/b704965j. [DOI] [PubMed] [Google Scholar]
- 18.Lanza RP, Langer RS, Vacanti J. Principles of tissue engineering. 3. Elsevier Academic Press; Amsterdam, Boston: 2007. [Google Scholar]
- 19.Gurkan UA, Tasoglu S, Kavaz D, Demirel MC, Demirci U. Adv Healthcare Mater. 2012;1:149–158. doi: 10.1002/adhm.201200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mironov V, Visconti RP, Kasyanov V, Forgacs G, Drake CJ, Markwald RR. Biomaterials. 2009;30:2164–2174. doi: 10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasoglu S, Demirci U. Trends Biotechnol. 2013;31:10–19. doi: 10.1016/j.tibtech.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillotin B, Souquet A, Catros S, Duocastella M, Pippenger B, Bellance S, Bareille R, Remy M, Bordenave L, Amedee J, Guillemot F. Biomaterials. 2010;31:7250–7256. doi: 10.1016/j.biomaterials.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 23.Xu F, Wu JH, Wang SQ, Durmus NG, Gurkan UA, Demirci U. Biofabrication. 2011;3:034101. doi: 10.1088/1758-5082/3/3/034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malafaya PB, Silva GA, Reis RL. Adv Drug Delivery Rev. 2007;59:207–233. doi: 10.1016/j.addr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon S, Gurkan UA, Blander J, Fawzi WW, Aboud S, Mugusi F, Kuritzkes DR, Demirci U. PLoS One. 2011;6:e21409. doi: 10.1371/journal.pone.0021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon S, Keles HO, Ozcan A, Khademhosseini A, Haeggstrom E, Kuritzkes D, Demirci U. Biosens Bioelectron. 2009;24:3208–3214. doi: 10.1016/j.bios.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurkan UA, Moon S, Geckil H, Xu F, Wang S, Lu TJ, Demirci U. Biotechnol J. 2011;6:138–149. doi: 10.1002/biot.201000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurkan UA, Anand T, Tas H, Elkan D, Akay A, Keles HO, Demirci U. Lab Chip. 2011;11:3979–3989. doi: 10.1039/c1lc20487d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurkan UA, Tasoglu S, Akkaynak D, Avci O, Unluisler S, Canikyan S, MacCallum N, Demirci U. Adv Healthcare Mater. 2012;1:661–668. doi: 10.1002/adhm.201200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi C, Li CW, Ji S, Yang M. Anal Chim Acta. 2006;560:1–23. [Google Scholar]
- 32.Chiou PY, Ohta AT, Wu MC. Nature. 2005;436:370–372. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 33.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Annu Rev Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 34.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 35.Khademhosseini A, Yeh J, Jon S, Eng G, Suh KY, Burdick JA, Langer R. Lab Chip. 2004;4:425–430. doi: 10.1039/b404842c. [DOI] [PubMed] [Google Scholar]
- 36.Chronis N, Lee LP. J Microelectromech Syst. 2005;14:857–863. [Google Scholar]
- 37.Yang J, Li CW, Yang MS. Lab Chip. 2004;4:53–59. doi: 10.1039/b309940g. [DOI] [PubMed] [Google Scholar]
- 38.Li CW, Cheung CN, Yang J, Tzang CH, Yang MS. Analyst. 2003;128:1137–1142. doi: 10.1039/b304354a. [DOI] [PubMed] [Google Scholar]
- 39.Voldman J. Annu Rev Biomed Eng. 2006;8:425–454. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 40.Shafiee H, Jahangir M, Inci F, Wang S, Willenbrecht RBM, Giguel FF, Tsibris AMN, Kuritzkes DR, Demirci U. Small. 2013 doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldberg M, Langer R, Jia XQ. J Biomater Sci, Polym Ed. 2007;18:241–268. doi: 10.1163/156856207779996931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, Parlakgul G, Bernstein J, Xing W, Boyden ES, Demirci U. Adv Mater. 2013;25:1192–1198. doi: 10.1002/adma.201203261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krebs MD, Erb RM, Yellen BB, Samanta B, Bajaj A, Rotello VM, Alsberg E. Nano Lett. 2009;9:1812–1817. doi: 10.1021/nl803757u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, Hsiang M, Peter T, Zavahir S, Thior I, Romanovicz D, Bernard B, Goodey AP, Walker BD, McDevitt JT. PLoS Med. 2005;2:e182. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voldman J, Gray ML, Schmidt MA. Annu Rev Biomed Eng. 1999;1:401–425. doi: 10.1146/annurev.bioeng.1.1.401. [DOI] [PubMed] [Google Scholar]
- 46.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 47.Song YS, Adler D, Xu F, Kayaalp E, Nureddin A, Anchan RM, Maas RL, Demirci U. Proc Natl Acad Sci U S A. 2010;107:4596–4600. doi: 10.1073/pnas.0914059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kose AR, Fischer B, Mao L, Koser H. Proc Natl Acad Sci U S A. 2009;106:21478–21483. doi: 10.1073/pnas.0912138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frasca G, Gazeau F, Wilhelm C. Langmuir. 2009;25:2348–2354. doi: 10.1021/la8030792. [DOI] [PubMed] [Google Scholar]
- 50.Yellen BB, Hovorka O, Friedman G. Proc Natl Acad Sci U S A. 2005;102:8860–8864. doi: 10.1073/pnas.0500409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CH, Abate AR, Lee D, Terentjev EM, Weitz DA. Adv Mater. 2009;21:3201–3204. [Google Scholar]
- 52.Pankhurst Q, Connolly J, Jones SK, Dobson J. J Phys D: Appl Phys. 2003;36:R167–R181. [Google Scholar]
- 53.Alsberg E, Feinstein E, Joy MP, Prentiss M, Ingber DE. Tissue Eng. 2006;12:3247–3256. doi: 10.1089/ten.2006.12.3247. [DOI] [PubMed] [Google Scholar]
- 54.Souza GR, Molina JR, Raphael RM, Ozawa MG, Stark DJ, Levin CS, Bronk LF, Ananta JS, Mandelin J, Georgescu MM, Bankson JA, Gelovani JG, Killian TC, Arap W, Pasqualini R. Nat Nanotechnol. 2010;5:291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Renard PE, Jordan O, Faes A, Petri-Fink A, Hofmann H, Rufenacht D, Bosman F, Buchegger F, Doelker E. Biomaterials. 2010;31:691–705. doi: 10.1016/j.biomaterials.2009.09.091. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Huang G, Zhang X, Li B, Chen Y, Lu T, Lu TJ, Xu F. Adv Funct Mater. 2013;23:660–672. [Google Scholar]
- 57.Coleman CB, Gonzalez-Villalobos RA, Allen PL, Johanson K, Guevorkian K, Valles JM, Hammond TG. Biotechnol Bioeng. 2007;98:854–863. doi: 10.1002/bit.21526. [DOI] [PubMed] [Google Scholar]
- 58.Dobson J. Nat Nanotechnol. 2008;3:139–143. doi: 10.1038/nnano.2008.39. [DOI] [PubMed] [Google Scholar]
- 59.Ito A, Ino K, Kobayashi T, Honda H. Biomaterials. 2005;26:6185–6193. doi: 10.1016/j.biomaterials.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 60.Akiyama H, Ito A, Kawabe Y, Kamihira M. Biomed Microdevices. 2009;11:713–721. doi: 10.1007/s10544-009-9284-x. [DOI] [PubMed] [Google Scholar]
- 61.Okochi M, Takano S, Isaji Y, Senga T, Hamaguchi M, Honda H. Lab Chip. 2009;9:3378–3384. doi: 10.1039/b909304d. [DOI] [PubMed] [Google Scholar]
- 62.Meyer CJ, Alenghat FJ, Rim P, Fong JHJ, Fabry B, Ingber DE. Nat Cell Biol. 2000;2:666–668. doi: 10.1038/35023621. [DOI] [PubMed] [Google Scholar]
- 63.Hwang DK, Dendukuri D, Doyle PS. Lab Chip. 2008;8:1640–1647. doi: 10.1039/b805176c. [DOI] [PubMed] [Google Scholar]
- 64.Pregibon DC, Toner M, Doyle PS. Langmuir. 2006;22:5122–5128. doi: 10.1021/la0534625. [DOI] [PubMed] [Google Scholar]
- 65.Yuet KP, Hwang DK, Haghgooie R, Doyle PS. Langmuir. 2010;26:4281–4287. doi: 10.1021/la903348s. [DOI] [PubMed] [Google Scholar]
- 66.Bong KW, Chapin SC, Doyle PS. Langmuir. 2010;26:8008–8014. doi: 10.1021/la904903g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bowden N, Terfort A, Carbeck J, Whitesides GM. Science. 1997;276:233–235. doi: 10.1126/science.276.5310.233. [DOI] [PubMed] [Google Scholar]
- 68.Kralchevsky PA, Paunov VN, Denkov ND, Ivanov IB, Nagayama K. J Colloid Interface Sci. 1993;155:420–437. [Google Scholar]
- 69.Paunov VN, Kralchevsky PA, Denkov ND, Nagayama K. J Colloid Interface Sci. 1993;157:100–112. [Google Scholar]
- 70.Demirci U. Rev Sci Instrum. 2005;76:065103. [Google Scholar]
- 71.Demirci U. J Microelectromech Syst. 2006;15:957–966. [Google Scholar]
- 72.Demirci U. Appl Phys Lett. 2006;88:144104. [Google Scholar]
- 73.Demirci U, Ergun AS, Oralkan O, Karaman M, Khuri-Yakub BT. IEEE Trans Ultrason Ferroelectr Freq Control. 2004;51:887–895. doi: 10.1109/tuffc.2004.1320749. [DOI] [PubMed] [Google Scholar]
- 74.Demirci U, Ozcan A. Electron Lett. 2005;41:1219–1220. [Google Scholar]
- 75.Demirci U, Toner M. Appl Phys Lett. 2006;88:053117. [Google Scholar]
- 76.Demirci U, Yaralioglu GG, Haeggstrom E, Khuri-Yakub BT. IEEE Trans Semicond Manuf. 2005;18:709–715. [Google Scholar]
- 77.Demirci U, Yaralioglu GG, Haeggstrom E, Percin G, Ergun S, Khuri-Yakub BT. IEEE Trans Semicond Manuf. 2004;17:517–524. [Google Scholar]
- 78.Demirci U, Montesano G. Lab Chip. 2007;7:1428–1433. doi: 10.1039/b705809h. [DOI] [PubMed] [Google Scholar]
- 79.Fang Y, Frampton JP, Raghavan S, Sabahi-Kaviani R, Luker G, Deng CX, Takayama S. Tissue Eng, Part C. 2012;18:647–657. doi: 10.1089/ten.tec.2011.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boland T, Xu T, Damon B, Cui X. Biotechnol J. 2006;1:910–917. doi: 10.1002/biot.200600081. [DOI] [PubMed] [Google Scholar]
- 81.Nakamura M, Kobayashi A, Takagi F, Watanabe A, Hiruma Y, Ohuchi K, Iwasaki Y, Horie M, Morita I, Takatani S. Tissue Eng. 2005;11:1658–1666. doi: 10.1089/ten.2005.11.1658. [DOI] [PubMed] [Google Scholar]
- 82.Moon S, Hasan SK, Song YS, Xu F, Keles HO, Manzur F, Mikkilineni S, Hong JW, Nagatomi J, Haeggstrom E, Khademhosseini A, Demirci U. Tissue Eng, Part C. 2010;16:157–166. doi: 10.1089/ten.TEC.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moon S, Kim YG, Dong L, Lombardi M, Haeggstrom E, Jensen RV, Hsiao LL, Demirci U. PLoS One. 2011;6:e17455. doi: 10.1371/journal.pone.0017455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Odde DJ, Renn MJ. Trends Biotechnol. 1999;17:385–389. doi: 10.1016/s0167-7799(99)01355-4. [DOI] [PubMed] [Google Scholar]
- 85.Elrod SA, Hadimioglu B, Khuriyakub BT, Rawson EG, Richley E, Quate CF, Mansour NN, Lundgren TS. J Appl Phys. 1989;65:3441–3447. [Google Scholar]
- 86.Moon S, Ceyhan E, Gurkan UA, Demirci U. PLoS One. 2011;6:e21580. doi: 10.1371/journal.pone.0021580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ceyhan E, Xu F, Gurkan UA, Emre AE, Turali ES, El Assal R, Acikgenc A, Wu CM, Demirci U. Lab Chip. 2012;12:4884–4893. doi: 10.1039/c2lc40523g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang W, Huang Y, Grujicic M, Chrisey DB. J Manuf Sci Eng. 2008;130:021012. [Google Scholar]
- 89.Tasoglu S, Kaynak G, Szeri AJ, Demirci U, Muradoglu M. Phys Fluids. 2010;22:082103. doi: 10.1063/1.3475527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muradoglu M, Tasoglu S. Comput Fluids. 2010;39:615–625. [Google Scholar]
- 91.Muradoglu M, Tasoglu S. ASME. 2009;2009:1095–1106. [Google Scholar]
- 92.Takamatsu H, Rubinsky B. Cryobiology. 1999;39:243–251. doi: 10.1006/cryo.1999.2207. [DOI] [PubMed] [Google Scholar]
- 93.Wang S, Inci F, De Libero G, Singhal A, Demirci U. Biotechnol Adv. 2013 doi: 10.1016/j.biotechadv.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang S, Xu F, Demirci U. Biotechnol Adv. 2010;28:770–781. doi: 10.1016/j.biotechadv.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.World Health Organization. [Accessed on February 4, 2013];2009 [Google Scholar]
- 96.Zetter BR. Annu Rev Med. 1998;49:407–424. doi: 10.1146/annurev.med.49.1.407. [DOI] [PubMed] [Google Scholar]
- 97.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. New Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 98.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. New Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, Moore AL, Tsukrov DI, Kempner ME, Dahl DM, Wu CL, Iafrate AJ, Smith MR, Tompkins RG, Sequist LV, Toner M, Haber DA, Maheswaran S. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cross SE, Jin YS, Rao J, Gimzewski JK. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 101.de la Rica R, Thompson S, Baldi A, Fernandez-Sanchez C, Drain CM, Matsui H. Anal Chem. 2009;81:10167–10171. doi: 10.1021/ac9021049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J Am Chem Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang H, Berezov A, Wang Q, Zhang G, Drebin J, Murali R, Greene MI. J Clin Invest. 2007;117:2051–2058. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, Kharsany ABM, Sibeko S, Mlisana KP, Omar Z, Gengiah TN, Maarschalk S, Arulappan N, Mlotshwa M, Morris L, Taylor D On behalf of the CAPRISA 004 Trial Group, . Science. 2010;329:1168–1174. [Google Scholar]
- 105.Tasoglu S, Park SC, Peters JJ, Katz DF, Szeri AJ. J Non-Newtonian Fluid Mech. 2011;166:1116–1122. doi: 10.1016/j.jnnfm.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tasoglu S, Peters JJ, Park SC, Verguet S, Katz DF, Szeri AJ. Phys Fluids. 2011;23:093101. doi: 10.1063/1.3633337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tasoglu S, Szeri AJ, Katz DF. Biophys J. 2011;100:489. [Google Scholar]
- 108.Tasoglu S, Katz DF, Szeri AJ. J Non-Newtonian Fluid Mech. 2012;187:36–42. doi: 10.1016/j.jnnfm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Szeri AJ, Park SC, Tasoglu S, Verguet S, Gorham A, Gao Y, Katz DF. Biophys J. 2010;98:604. [Google Scholar]
- 110.Tasoglu S, Rohan LC, Katz DF, Szeri AJ. Phys Fluids. 2013;25:031901. doi: 10.1063/1.4793598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.UNAIDS. [Accessed: January 2027th 2013.];Report on the global AIDS epidemic. Available at http://data.unaids.org/pub/GlobalReport/2008/JC1510_2008GlobalReport_en.zip.
- 112.Ozcan A, Demirci U. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 113.Jokerst JV, Floriano PN, Christodoulides N, Simmons GW, McDevitt JT. Lab Chip. 2008;8:2079–2090. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alyassin MA, Moon S, Keles HO, Manzur F, Lin RL, Haeggstrom E, Kuritzkes DR, Demirci U. Lab Chip. 2009;9:3364–3369. doi: 10.1039/b911882a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang XH, Khimji I, Gurkan UA, Safaee H, Catalano PN, Keles HO, Kayaalp E, Demirci U. Lab Chip. 2011;11:2535–2540. doi: 10.1039/c1lc20236g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheng X, Liu Y-s, Irimia D, Demirci U, Yang L, Zamir L, Rodriguez WR, Toner M, Bashir R. Lab Chip. 2007;7:746–755. doi: 10.1039/b705082h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheng X, Irimia D, Dixon M, Ziperstein JC, Demirci U, Zamir L, Tompkins RG, Toner M, Rodriguez WR. J Acquired Immune Defic Syndr. 2007;45:257–261. doi: 10.1097/QAI.0b013e3180500303. [DOI] [PubMed] [Google Scholar]
- 118.Cheng XH, Gupta A, Chen CC, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2009;9:1357–1364. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang S, Inci F, Chaunzwa TL, Ramanujam A, Vasudevan A, Subramanian S, Chi Fai Ip A, Sridharan B, Gurkan UA, Demirci U. Int J Nanomed. 2012;7:2591–2600. doi: 10.2147/IJN.S29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sanderson MW, Sreerama S, Nagaraja TG. Curr Microbiol. 2007;55:158–161. doi: 10.1007/s00284-007-0083-4. [DOI] [PubMed] [Google Scholar]
- 121.Fan ST, Teoh-Chan CH, Lau KF. Eur J Clin Microbiol Infect Dis. 1989;8:142–144. doi: 10.1007/BF01963898. [DOI] [PubMed] [Google Scholar]
- 122.Quilici N, Audibert G, Conroy MC, Bollaert PE, Guillemin F, Welfringer P, Garric J, Weber M, Laxenaire MC. Clin Infect Dis. 1997;25:1066–1070. doi: 10.1086/516090. [DOI] [PubMed] [Google Scholar]
- 123.World Health Organization. [Accessed: January 27th 2013.];2010 http://www.who.int/hiv/pub/paediatric/diagnosis/en/index.html.
- 124.Kim YG, Moon S, Kuritzkes DR, Demirci U. Biosens Bioelectron. 2009;25:253–258. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang S, Esfahani M, Gurkan UA, Inci F, Kuritzkes DR, Demirci U. Lab Chip. 2012;12:1508–1515. doi: 10.1039/c2lc20706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tang N, Huang S, Salituro J, Mak WB, Cloherty G, Johanson J, Li YH, Schneider G, Robinson J, Hackett J, Jr, Swanson P, Abravaya K. J Virol Methods. 2007;146:236–245. doi: 10.1016/j.jviromet.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 127.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. New Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang S, Sarenac D, Chen M, Huang S, Giguel F, Kuritzkes D, Demirci U. Int J Nanomed. 2012;7:5019–5028. doi: 10.2147/IJN.S32579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang M, Sun S, Kostov Y, Rasooly A. Lab Chip. 2010;10:1011–1017. doi: 10.1039/b923996k. [DOI] [PubMed] [Google Scholar]
- 130.Wang S, Zhao X, Khimji I, Akbas R, Qiu W, Edwards D, Cramer DW, Ye B, Demirci U. Lab Chip. 2011;11:3411–3418. doi: 10.1039/c1lc20479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clarke-Pearson DL. New Engl J Med. 2009;361:170–177. doi: 10.1056/NEJMcp0901926. [DOI] [PubMed] [Google Scholar]
- 132.Centers for disease control and prevention. [Accessed: January 27th 2013.]; available at http://www.cdc.gov/reproductivehealth/infertility/
- 133.Saragusty J, Arav A. Reproduction. 2011;141:1–19. doi: 10.1530/REP-10-0236. [DOI] [PubMed] [Google Scholar]
- 134.Cho BS, Schuster TG, Zhu XY, Chang D, Smith GD, Takayama S. Anal Chem. 2003;75:1671–1675. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- 135.Hyakutake T, Hashimoto Y, Yanase S, Matsuura K, Naruse K. Biomed Microdevices. 2009;11:25–33. doi: 10.1007/s10544-008-9207-2. [DOI] [PubMed] [Google Scholar]
- 136.Ma R, Xie L, Han C, Su K, Qiu T, Wang L, Huang G, Xing W, Qiao J, Wang J, Cheng J. Anal Chem. 2011;83:2964–2970. doi: 10.1021/ac103063g. [DOI] [PubMed] [Google Scholar]
- 137.Lopez-Garcia MD, Monson RL, Haubert K, Wheeler MB, Beebe DJ. Biomed Microdevices. 2008;10:709–718. doi: 10.1007/s10544-008-9182-7. [DOI] [PubMed] [Google Scholar]
- 138.Schuster TG, Cho B, Keller LM, Takayama S, Smith GD. Reprod BioMed Online. 2003;7:75–81. doi: 10.1016/s1472-6483(10)61732-4. [DOI] [PubMed] [Google Scholar]
- 139.Xie L, Ma R, Han C, Su K, Zhang Q, Qiu T, Wang L, Huang G, Qiao J, Wang J, Cheng J. Clin Chem. 2010;56:1270–1278. doi: 10.1373/clinchem.2010.146902. [DOI] [PubMed] [Google Scholar]
- 140.Winet H, Bernstein GS, Head J. J Reprod Fertil. 1984;70:511–523. doi: 10.1530/jrf.0.0700511. [DOI] [PubMed] [Google Scholar]
- 141.Tasoglu S, Safaee H, Zhang X, Kingsley JL, Catalano PN, Gurkan UA, Nureddin A, Kayaalp E, Anchan RM, Maas L, Tüzel E, Demirci U. Small. 2013 doi: 10.1002/smll.201300020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, Russom A, Warner EA, Moldawer LL, De A, Bankey PE, Petritis BO, Camp DG, Rosenbach AE, Goverman J, Fagan SP, Brownstein BH, Irimia D, Xu W, Wilhelmy J, Mindrinos MN, Smith RD, Davis RW, Tompkins RG, Toner M. Nat Med. 2010;16:1042–1047. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Salehi-Reyhani A, Kaplinsky J, Burgin E, Novakova M, Demello AJ, Templer RH, Parker P, Neil MAA, Ces O, French P, Willison KR, Klug D. Lab Chip. 2011;11:1256–1261. doi: 10.1039/c0lc00613k. [DOI] [PubMed] [Google Scholar]
- 144.McKenna BK, Selim AA, Bringhurst FR, Ehrlich DJ. Lab Chip. 2009;9:305–310. doi: 10.1039/b811889b. [DOI] [PubMed] [Google Scholar]
- 145.Ramadan Q, Samper V, Poenar D, Liang Z, Yu C, Lim TM. Sens Actuators, B. 2006;113:944–955. [Google Scholar]
- 146.Hosokawa M, Arakaki A, Takahashi M, Mori T, Takeyama H, Matsunaga T. Anal Chem. 2009;81:5308–5313. doi: 10.1021/ac900535h. [DOI] [PubMed] [Google Scholar]
- 147.Wu HW, Lin CC, Lee GB. Biomicrofluidics. 2011;5:26. doi: 10.1063/1.3528299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu Y, Phillips JA, Yan JL, Li QG, Fan ZH, Tan WH. Anal Chem. 2009;81:7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang K, Cometti B, Pappas D. Anal Chim Acta. 2007;601:1–9. doi: 10.1016/j.aca.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 150.Sung-Yi Y, Suz-Kai H, Yung-Ching H, Chen-Min C, Teh-Lu L, Gwo-Bin L. Meas Sci Technol. 2006;17:2001. [Google Scholar]
- 151.Fu AY, Chou HP, Spence C, Arnold FH, Quake SR. Anal Chem. 2002;74:2451–2457. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- 152.Zhang X, Jones P, Haswell SJ. Chem Eng J. 2008;135:S82–S88. [Google Scholar]
- 153.Wankhede SP, Du Z, Berg JM, Vaughn MW, Dallas T, Cheng KH, Gollahon L. Biotechnol Prog. 2006;22:1426–1433. doi: 10.1021/bp060127d. [DOI] [PubMed] [Google Scholar]
- 154.Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Lab Chip. 2009;9:1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- 155.Jung K, Hampel G, Scholz M, Henke W. Cell Physiol Biochem. 1995;5:353–360. [Google Scholar]
- 156.Fujioka N, Morimoto Y, Takeuchi K, Yoshioka M, Kikuchi M. Appl Spectrosc. 2003;57:241–243. doi: 10.1366/000370203321535187. [DOI] [PubMed] [Google Scholar]
- 157.Hatch A, Hansmann G, Murthy SK. Langmuir. 2011;27:4257–4264. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Suárez-González D, Barnhart K, Saito E, Vanderby R, Hollister SJ, Murphy WL. J Biomed Mater Res, Part A. 2010;95:222–234. doi: 10.1002/jbm.a.32833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kanazawa H, Yamamoto K, Kashiwase Y, Matsushima Y, Takai N, Kikuchi A, Sakurai Y, Okano T. J Pharm Biomed Anal. 1997;15:1545–1550. doi: 10.1016/s0731-7085(96)02004-3. [DOI] [PubMed] [Google Scholar]
- 160.Huber DL, Manginell RP, Samara MA, Kim BI, Bunker BC. Science. 2003;301:352–354. doi: 10.1126/science.1080759. [DOI] [PubMed] [Google Scholar]
- 161.Lion N, Rohner TC, Dayon L, Arnaud IL, Damoc E, Youhnovski N, Wu ZY, Roussel C, Josserand J, Jensen H, Rossier JS, Przybylski M, Girault HH. Electrophoresis. 2003;24:3533–3562. doi: 10.1002/elps.200305629. [DOI] [PubMed] [Google Scholar]
- 162.Tarasow TM, Penny L, Patwardhan A, Hamren S, McKenna MP, Urdea MS. Bioanalysis. 2011;3:2233–2251. doi: 10.4155/bio.11.224. [DOI] [PubMed] [Google Scholar]
- 163.Langer R, Vacanti JP. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 164.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci U S A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kose GT, Kenar H, Hasirci N, Hasirci V. Biomaterials. 2003;24:1949–1958. doi: 10.1016/s0142-9612(02)00613-0. [DOI] [PubMed] [Google Scholar]
- 166.Unger RE, Sartoris A, Peters K, Motta A, Migliaresi C, Kunkel M, Bulnheim U, Rychly J, Kirkpatrick CJ. Biomaterials. 2007;28:3965–3976. doi: 10.1016/j.biomaterials.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 167.Salgado AJ, Coutinho OP, Reis RL. Macromol Biosci. 2004;4:743–765. doi: 10.1002/mabi.200400026. [DOI] [PubMed] [Google Scholar]
- 168.Gurkan UA, Cheng XG, Kishore V, Uquillas JA, Akkus O. J Biomed Mater Res, Part A. 2010;94:1070–1079. doi: 10.1002/jbm.a.32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gurkan UA, Gargac J, Akkus O. Tissue Eng, Part A. 2010;16:2295–2306. doi: 10.1089/ten.TEA.2009.0565. [DOI] [PubMed] [Google Scholar]
- 170.Gurkan UA, Kishore V, Condon KW, Bellido TM, Akkus O. Calcif Tissue Int. 2011;88:388–401. doi: 10.1007/s00223-011-9467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gurkan UA, Krueger A, Akkus O. Tissue Eng, Part A. 2011;17:417–428. doi: 10.1089/ten.TEA.2010.0193. [DOI] [PubMed] [Google Scholar]
- 172.Christenson EM, Anseth KS, van den Beucken L, Chan CK, Ercan B, Jansen JA, Laurencin CT, Li WJ, Murugan R, Nair LS, Ramakrishna S, Tuan RS, Webster TJ, Mikos AG. J Orthop Res. 2007;25:11–22. doi: 10.1002/jor.20305. [DOI] [PubMed] [Google Scholar]
- 173.Cho TJ, Gerstenfeld LC, Einhorn TA. J Bone Miner Res. 2002;17:513–520. doi: 10.1359/jbmr.2002.17.3.513. [DOI] [PubMed] [Google Scholar]
- 174.Le AX, Miclau T, Hu D, Helms JA. J Orthop Res. 2001;19:78–84. doi: 10.1016/S0736-0266(00)00006-1. [DOI] [PubMed] [Google Scholar]
- 175.Tatsuyama K, Maezawa Y, Baba H, Imamura Y, Fukuda M. Eur J Histochem. 2000;44:269–278. [PubMed] [Google Scholar]
- 176.Muraglia A, Martin I, Cancedda R, Quarto R. Bone. 1998;22:131s–134s. doi: 10.1016/s8756-3282(98)00009-x. [DOI] [PubMed] [Google Scholar]
- 177.Histing T, Garcia P, Matthys R, Leidinger M, Holstein JH, Kristen A, Pohlemann T, Menger MD. J Orthop Res. 2010;28:397–402. doi: 10.1002/jor.21008. [DOI] [PubMed] [Google Scholar]
- 178.Thompson Z, Miclau T, Hu D, Helms JA. J Orthop Res. 2002;20:1091–1098. doi: 10.1016/S0736-0266(02)00017-7. [DOI] [PubMed] [Google Scholar]
- 179.Radisic M, Vunjak-Novakovic G. J Serb Chem Soc. 2005;70:541–556. [Google Scholar]
- 180.Borenstein JT, Terai H, King KR, Weinberg EJ, Kaazempur-Mofrad MR, Vacanti JP. Biomed Microdevices. 2002;4:167–175. [Google Scholar]
- 181.Bursac N, Parker KK, Iravanian S, Tung L. Circ Res. 2002;91:e45–e54. doi: 10.1161/01.res.0000047530.88338.eb. [DOI] [PubMed] [Google Scholar]
- 182.Fidkowski C, Kaazempur-Mofrad MR, Borenstein J, Vacanti JP, Langer R, Wang Y. Tissue Eng. 2005;11:302–309. doi: 10.1089/ten.2005.11.302. [DOI] [PubMed] [Google Scholar]
- 183.Li RK, Yau TM, Weisel RD, Mickle DA, Sakai T, Choi A, Jia ZQ. J Thorac Cardiovasc Surg. 2000;119:368–375. doi: 10.1016/S0022-5223(00)70193-0. [DOI] [PubMed] [Google Scholar]
- 184.Masters KS, Shah DN, Leinwand LA, Anseth KS. Biomaterials. 2005;26:2517–2525. doi: 10.1016/j.biomaterials.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 185.Geckil H, Xu F, Zhang XH, Moon S, Demirci U. Nanomedicine. 2010;5:469–484. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee KY, Money DJ. Chem Rev. 2001;101:1869. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 187.Griffith LG, Swartz MA. Nat Rev: Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 188.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 189.Tibbitt MW, Anseth KS. Biotechnol Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yang S, Leong KF, Du Z, Chua CK. Tissue Eng. 2002;8:1–11. doi: 10.1089/107632702753503009. [DOI] [PubMed] [Google Scholar]
- 191.Barron JA, Wu P, Ladouceur HD, Ringeisen BR. Biomed Microdevices. 2004;6:139–147. doi: 10.1023/b:bmmd.0000031751.67267.9f. [DOI] [PubMed] [Google Scholar]
- 192.Xu T, Gregory CA, Molnar P, Cui X, Jalota S, Bhaduri SB, Boland T. Biomaterials. 2006;27:3580–3588. doi: 10.1016/j.biomaterials.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 193.Sinden RR. Nature. 2001;411:757–758. doi: 10.1038/35081234. [DOI] [PubMed] [Google Scholar]
- 194.Ross CA, Poirier MA. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 195.Barnham KJ, Masters CL, Bush AI. Nat Rev Drug Discovery. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 196.Wong E, Cuervo AM. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bradbury EJ, McMahon SB. Nat Rev Neurosci. 2006;7:644–653. doi: 10.1038/nrn1964. [DOI] [PubMed] [Google Scholar]
- 198.Woerly S, Awosika O, Zhao P, Agbo C, Gomez-Pinilla F, de Vellis J, Espinosa-Jeffrey A. Neurochem Res. 2005;30:721–735. doi: 10.1007/s11064-005-6866-8. [DOI] [PubMed] [Google Scholar]
- 199.Lavik E. Neural Tissue Engineering. Springer; New York: 2011. [Google Scholar]
- 200.Musick K, Khatami D, Wheeler BC. Lab Chip. 2009;9:2036–2042. doi: 10.1039/b820596e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Silver J, Miller JH. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 202.Kim DH, Viventi J, Amsden JJ, Xiao JL, Vigeland L, Kim YS, Blanco JA, Panilaitis B, Frechette ES, Contreras D, Kaplan DL, Omenetto FG, Huang YG, Hwang KC, Zakin MR, Litt B, Rogers JA. Nat Mater. 2010;9:511–517. doi: 10.1038/nmat2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS. Nat Mater. 2011;10:799–806. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- 204.Bailey SN, Sabatini DM, Stockwell BR. Proc Natl Acad Sci U S A. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lee MY, Park CB, Dordick JS, Clark DS. Proc Natl Acad Sci U S A. 2005;102:983–987. doi: 10.1073/pnas.0406755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Upadhyaya S, Selvaganapathy PR. Lab Chip. 2010;10:341–348. doi: 10.1039/b918291h. [DOI] [PubMed] [Google Scholar]
- 207.Gosalia DN, Diamond SL. Proc Natl Acad Sci U S A. 2003;100:8721–8726. doi: 10.1073/pnas.1530261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ma H, Horiuchi KY, Wang Y, Kucharewicz SA, Diamond SL. Assay Drug Dev Technol. 2005;3:177–187. doi: 10.1089/adt.2005.3.177. [DOI] [PubMed] [Google Scholar]
- 209.Starkuviene V, Pepperkok R, Erfle H. Expert Rev Proteomics. 2007;4:479–489. doi: 10.1586/14789450.4.4.479. [DOI] [PubMed] [Google Scholar]
- 210.Chen DS, Davis MM. Curr Opin Chem Biol. 2006;10:28–34. doi: 10.1016/j.cbpa.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 211.Yarmush ML, King KR. Annu Rev Biomed Eng. 2009;11:235–257. doi: 10.1146/annurev.bioeng.10.061807.160502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gidrol X, Fouque B, Ghenim L, Haguet V, Picollet-D’hahan N, Schaack B. Curr Opin Pharmacol. 2009;9:664–668. doi: 10.1016/j.coph.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 213.Guven S, Demirci U. Nanomedicine. 2012;7:1787–1789. doi: 10.2217/nnm.12.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Windrum P, Morris TC, Drake MB, Niederwieser D, Ruutu T. Bone Marrow Transplant. 2005;36:601–603. doi: 10.1038/sj.bmt.1705100. [DOI] [PubMed] [Google Scholar]
- 215.Desrosiers P, Légaré C, Leclerc P, Sullivan R. Fertil Steril. 2006;85:1744–1752. doi: 10.1016/j.fertnstert.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 216.Fabbri R, Porcu E, Marsella T, Rocchetta G, Venturoli S, Flamigni C. Hum Reprod. 2001;16:411–416. doi: 10.1093/humrep/16.3.411. [DOI] [PubMed] [Google Scholar]
- 217.Xu F, Moon SJ, Emre AE, Turali ES, Song YS, Hacking SA, Nagatomi J, Demirci U. Biofabrication. 2010;2:014105. doi: 10.1088/1758-5082/2/1/014105. [DOI] [PubMed] [Google Scholar]
- 218.Song YS, Moon S, Hulli L, Hasan SK, Kayaalp E, Demirci U. Lab Chip. 2009;9:1874–1881. doi: 10.1039/b823062e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ji L, de Pablo JJ, Palecek SP. Biotechnol Bioeng. 2004;88:299–312. doi: 10.1002/bit.20243. [DOI] [PubMed] [Google Scholar]
- 220.Hegner B, Weber M, Dragun D, Schulze-Lohoff E. J Hypertens. 2005;23:1191–1202. doi: 10.1097/01.hjh.0000170382.31085.5d. [DOI] [PubMed] [Google Scholar]
- 221.Stachecki JJ, Cohen J. Reprod BioMed Online. 2004;9:152–163. doi: 10.1016/s1472-6483(10)62124-4. [DOI] [PubMed] [Google Scholar]
- 222.Ludwig M, Al-Hasani S, Felberbaum R, Diedrich K. Hum Reprod. 1999;14:162–185. doi: 10.1093/humrep/14.suppl_1.162. [DOI] [PubMed] [Google Scholar]
- 223.Demirci U, Khademhosseini A, Langer R, Blander J. Microfluidic Technologies for Human Health. World Scientific Publishing Company; 2012. [Google Scholar]
- 224.Samot J, Moon S, Shao L, Zhang X, Xu F, Song Y, Keles HO, Matloff L, Markel J, Demirci U. PLoS One. 2011;6:e17530. doi: 10.1371/journal.pone.0017530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Fuller BJ. CryoLetters. 2004;25:375–388. [Google Scholar]
- 226.Zhang X, Khimji I, Shao L, Safaee H, Desai K, Keles HO, Gurkan UA, Kayaalp E, Nureddin A, Anchan RM, Maas RL, Demirci U. Nanomedicine. 2012;7:553–564. doi: 10.2217/nnm.11.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Meryman HT, Hornblower M. Transfusion. 1972;12:145–156. doi: 10.1111/j.1537-2995.1972.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 228.Tullis JL, Ketchel MM, Pyle HM, Pennell RB, Gibson JG, 2nd, Tinch RJ, Driscoll SG. JAMA, J Am Med Assoc. 1958;168:399–404. doi: 10.1001/jama.1958.03000040035008. [DOI] [PubMed] [Google Scholar]
- 229.Tan FCK, Lee KH, Gouk SS, Magalhaes R, Poonepalli A, Hande MP, Dawe GS, Kuleshova LL. CryoLetters. 2007;28:445–460. [PubMed] [Google Scholar]
- 230.Berger M, Castelino J, Huang R, Shah M, Austin RH. Electrophoresis. 2001;22:3883–3892. doi: 10.1002/1522-2683(200110)22:18<3883::AID-ELPS3883>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 231.Furdui VI, Harrison DJ. Lab Chip. 2004;4:614–618. doi: 10.1039/b409366f. [DOI] [PubMed] [Google Scholar]
- 232.Inglis DW, Riehn R, Austin RH, Sturm JC. Appl Phys Lett. 2004;85:5093–5095. [Google Scholar]
- 233.Xu F, Inci F, Mullick O, Gurkan UA, Sung Y, Kavaz D, Li B, Denkbas EB, Demirci U. ACS Nano. 2012;6:6640–6649. doi: 10.1021/nn300902w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chung SE, Park W, Shin S, Lee SA, Kwon S. Nat Mater. 2008;7:581–587. doi: 10.1038/nmat2208. [DOI] [PubMed] [Google Scholar]
- 235.Park W, Lee H, Park H, Kwon S. Lab Chip. 2009;9:2169–2175. doi: 10.1039/b904153b. [DOI] [PubMed] [Google Scholar]
- 236.Chung SE, Jung Y, Kwon S. Small. 2011;7:796–803. doi: 10.1002/smll.201001806. [DOI] [PubMed] [Google Scholar]
- 237.Sekeroglu K, Gurkan UA, Demirci U, Demirel MC. Appl Phys Lett. 2011;99:63703–637033. doi: 10.1063/1.3625430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kistler SF, editor. Wettability. Dekker; New York: 1993. [Google Scholar]
- 239.Won R. Nat Photonics. 2011;5:512. [Google Scholar]
- 240.Zhang X, Catalono PN, Gurkan UA, Khimji PI, Demirci U. Nanomedicine. 2011;6:1115–1129. doi: 10.2217/nnm.11.71. [DOI] [PMC free article] [PubMed] [Google Scholar]