Abstract
While the importance of membrane microdomains in receptor-mediated activation of lymphocytes has been established, much less is known about the role of receptor ligand distribution on APC and target cells. Detergent-resistant membrane (DRM) domains, into which glycophosphatidylinositol (GPI)-linked proteins partition, are enriched in cholesterol and glycosphingolipids. ULBP1 is a GPI-linked ligand for natural cytotoxicity receptor NKG2D. To investigate how ULBP1 distribution on target cells affects NKG2D-dependent NK cell activation, we fused the extracellular domain of ULBP1 to the transmembrane domain of CD45. Introduction of this transmembrane domain eliminated the association of ULBP1 with the DRM fraction and caused a significant reduction of cytotoxicity and degranulation by NK cells. Clustering and lateral diffusion of ULBP1 was not affected by changes in the membrane anchor. These results show that the partitioning of receptor ligands in discrete membrane domains of target cells is an important determinant of NK cell activation.
Keywords: NK cell, NKG2D, ULBP1, detergent resistant membrane, cytotoxicity, degranulation
Introduction
NK cells are a subset of cytotoxic lymphocytes that recognize and kill tumor cells and virus-infected cells (1). Lysis of target cells is a multistep process including adhesion of NK cells to target cells, synapse formation, polarization of cytolytic granules toward the target cells, and granule exocytosis (2). Whereas binding of LFA-1 on human NK cells to ICAM on target cells induces adhesion and polarization of lytic granules (3), degranulation is triggered by low affinity Fcg receptor CD16 or by synergistic combinations of co-activation receptors, such as 2B4 and NKG2D (4, 5).
NKG2D is a C-type lectin co-activation receptor expressed as a disulfide-linked homodimer on NK cells, NKT cells, and some T cells (6). In humans, NKG2D binds to stress inducible members of the polymorphic MHC-I related chain A/B (MICA/B) family and the multi-gene family of UL16-binding proteins (ULBPs, RAET1A-E). NKG2D ligands are expressed in multiple types of tumors and play an important role in immunosurveillance of cancer (7). However, by shedding NKG2D ligands from their cell surface, tumor cells may escape the antitumor response mediated by NKG2D (6).
Within the lipid bilayer, proteins and lipids are segregated laterally, leading to functional subcompartmentalization of the plasma membrane (8). Lipid rafts are membrane microdomains enriched in glycosphingolipids, sphingomyelins, and cholesterol. The role of membrane microdomains in promoting receptor-mediated lymphocyte activation has been well established (9, 10). Much less is known about how the distribution of receptor ligands on target cells affects lymphocyte function, although studies have suggested that it may be an important parameter for lymphocyte activation: For example, the cytoplasmic tail of CD80 (B7-1), a CD28 ligand expressed in APC, is required for proper segregation of CD28 at the immunological synapse and for full T cell activation (11). Furthermore, while expression of MICA on resistant target cells could overcome MHC class I-dependent inhibitory signaling in NK cells, a truncated form of MICA lacking a potential acylation site could not (12), suggesting that NKG2D ligand distribution may play a role in overcoming NK cell inhibition.
Here, we tested ULBP1 as a ligand to investigate the role of ligand distribution in NKG2D-dependent human NK cell activation. To do so independently of HLA class I ligands for inhibitory receptors, we expressed ULBP1 in a mouse cell line. A chimera consisting of the extracellular portion of ULBP1 and the transmembrane region of CD45 was generated. Its expression resulted in the localization of the normally GPI-linked ULBP1 from detergent-resistant membrane fractions to detergent-soluble fractions. This redistribution of ULBP1 caused a reduction in cytotoxicity and degranulation by NK cells, implying a role for receptor ligand distribution in the activation of NK cell responses.
Materials and Methods
Cells
Resting human NK cells were isolated from peripheral blood cells by negative selection using an NK cell isolation kit (Stem Cell Technologies). Freshly isolated resting NK cells (95-99% CD3-CD56+) were resuspended in Iscove's modified Dulbecco medium (IMDM, Invitrogen) supplemented with 10% human serum (Valley Biomedical) and used 1-2 days after isolation. Polyclonal IL-2 activated NK cells were cultured as described (3). P815 cells were cultured in IMDM medium supplemented with 10% heat inactivated fetal bovine serum (FBS, Thermo Scientific). For phospholipase treatment, P815-ULBP1 and P815-ULBP1-CD45TM cells were treated with 2 IU/ml Phosphatidylinositol-Specific Phospholipase C (PI-PLC) (Sigma) for 1 hour at 37°C and 5% CO2 and surface protein levels were measured by flow cytometry using a R-Phycoerythrin (RPE)-conjugated ULBP1 mAb (R&D). The ULBP1 mAb was conjugated with a Phycolink® RPE kit (Prozyme).
Transfection of P815 cells
P815 cells were transfected with human ULB1 or ULBP1-CD45TM (Fig. S1) using the BioRad Gene PulserTM (10 μg of each DNA, 260 V, 960 μF). Transfected cells were selected in IMDM supplemented with 10% heat inactivated FBS and 800 μg/ml Geneticin (Invitrogen), and subcloned. Different clones were tested for ULBP1-expression and in functional assays, and representative clones from each cell line were selected for further use.
Detergent resistant membrane (DRM) preparation
DRM preparation was performed as described (13), except that OptiPrep (Axis-Shield) was used instead of sucrose. Fractions 4 to 11 were separated on 12% SDS NuPAGE gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membrane (Invitrogen). The membrane was blocked with Odyssey blocking buffer (LI-COR Biosciences) for 1 h at room temperature and incubated with Biotinylated goat Anti-human ULBP1 Antibody (R&D) overnight at room temperature. After washing, the membrane was stained with IRDye 680 Labeled Streptavidin (LI-COR Biosciences) for 1 h at room temperature and bands were detected using the Odyssey infrared imaging system (LI-COR Biosciences). In the case of Latrunculin A treatment, cells were incubated with either 0.3% DMSO carrier or 3 μM Latrunculin A (Calbiochem) for 40 min at 37°C and 5% CO2.
TIRF microscopy
Cell surface ULBP1 or ULBP1-CD45TM on P815 cells was fluorescently labeled with PE-conjugated ULBP1 mAb (R&D). For Latrunculin A treatment, labeled cells were incubated for 40 min at 37°C and 5% CO2 with either 0.3% DMSO carrier or 3 μM Latrunculin A prior to analysis by total internal reflection (TIRF) microscopy. TIRF imaging and analysis was performed as described (14).
Cellular assays
Cytotoxicity assays were performed as described (15). Degranulation assays were performed as described (4) with minor changes. Briefly, 2 × 105 NK cells were added to 4 × 105 P815 target cells in a total volume of 200 μl IMDM medium supplemented with 10% heat inactivated FBS, 6 μg/ml Monensin (Calbiochem), 20 μl/ml FITC-conjugated CD107a mAb (Becton Dickinson), and 20 μl/ml PE-conjugated CD56 mAb (Becton Dickinson). Cells were mixed and incubated for 1 h at 37°C and 5% CO2. Afterwards cells were spun down and expression of CD107a on CD56 positive cells was determined by flow cytometry. In case of Latrunculin A pre-treatment of the target cells, cells were incubated with either 0.3% DMSO carrier (Sigma) or 3 μM Latrunculin A (Calbiochem) for 40 min at 37°C at 5% CO2 and washed extensively prior to use in the assay.
Results and Discussion
Linking the extracellular portion of ULBP1 to the transmembrane region of CD45 changes its localization within the membrane
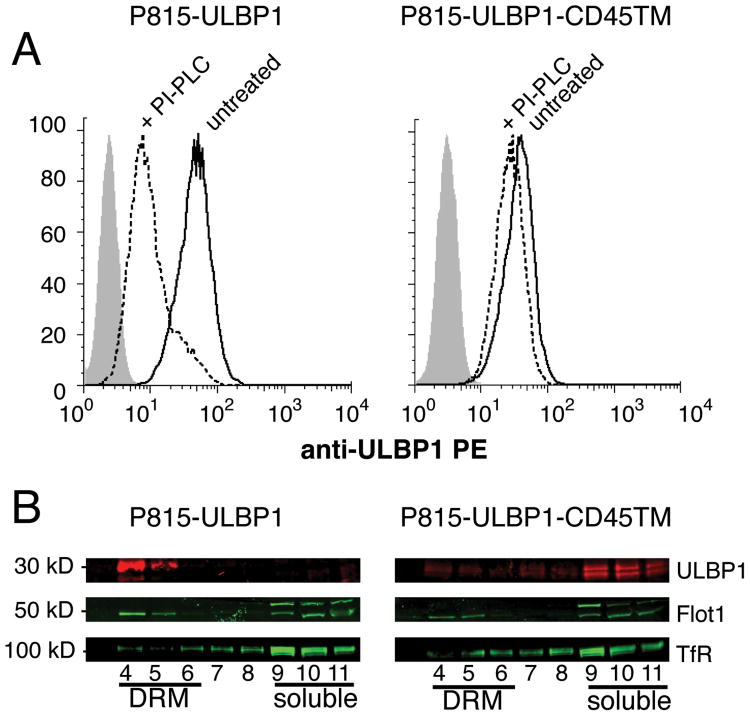
To change the distribution of the NKG2D ligand ULBP1 within the plasma membrane, we generated a chimera consisting of the extracellular portion of ULBP1 and the transmembrane region of CD45 (ULBP1-CD45TM) (Fig. S1). Briefly, the extracellular portion of ULBP1 including the GPI-anchor site (Fig. S1B) was fused to the transmembrane region of CD45 that includes only two extracellular amino acids and four amino acids in the cytosolic portion for anchoring purposes (Fig. S1B). The GPI-linked ULBP1 and the recombinant ULBP1-CD45TM were transfected into the mouse mastocytoma cell line P815, and clones with similar expression levels were selected (Fig. 1A). To verify that ULBP1-CD45TM had lost the GPI anchor, P815 cells expressing ULBP1 and ULBP1-CD45TM were treated with phosphatidylinositol phospholipase C (PI-PLC), which cleaves GPI-anchors. ULBP1 was sensitive to PI-PLC but ULBP1-CD45TM was not, indicating a loss of the GPI anchor in the chimera (Fig. 1A). The incomplete cleavage of ULBP1 (Fig. 1A), which could be due to limited accessibility to phospholipase, is consistent with the low amount of ULBP1 shedding observed in other target cells (16).
Figure 1.
Fusion of the ULBP1 extracellular domain with the transmembrane domain of CD45 alters its distribution in P815 cells. A, P815 cells transfected with GPI-linked ULBP1 (left panel) or ULBP1-CD45TM (right panel), were either untreated (solid line) or PI-PLC treated (dashed line), and analyzed with a PE-conjugated ULBP1 antibody, or an IgG2A isotype control (shaded). Data are representative of 9 individual experiments. B, Fractions 4 to 11 of DRM preparations from P815 cells expressing either GPI-linked ULBP1 (left panel) or ULBP1-CD45TM (right panel) were analyzed by immunoblotting with Abs to ULBP1 (upper lane), raft-associated Flotillin-1 (Flot-1, middle lane), and detergent-soluble membrane transferring receptor (TfR, bottom lane). Data are representative of 2 individual experiments.
To test if linkage to the transmembrane domain of CD45 changed the localization of ULBP1, detergent-resistant membrane (DRM) fractions were prepared from P815-ULBP1 cells and P815-ULBP1-CD45TM cells. Whereas ULBP1 was almost exclusively localized in the DRM fraction, the ULBP1-CD45TM protein was associated with the soluble fraction (Fig. 2B). Association of ULBP1 with the DRM fraction was even stronger than that of the DRM marker flotillin 1 (Fig. 2B). Therefore, linking the extracellular portion of ULBP1 to the transmembrane region of CD45 changed its localization from the DRM fraction to the detergent-soluble membrane fraction.
Figure 2.
Exclusion of ULBP1 from the DRM fraction of target cells reduces the cytotoxicity and degranulation of NK cells. Susceptibility of P815 (open circles), P815-ULBP1 (black circles), and P815-ULBP1-CD45TM (gray circles) cells by freshly isolated resting (A) or IL-2 activated NK cells (B), was measured in a 2 h assay. Bars indicate SD of independent experiments with 3 NK cell donors. Degranulation of freshly isolated resting NK cells (C) or IL-2 activated NK cells (D) in response to P815, P815-ULBP1, and P815-ULBP1-CD45TM cells was measured in a 1 h degranulation assay with CD107a mAb. Each symbol represents an individual donor among 3 (C) or 5 (D), which were tested in independent experiments. Paired t-test was performed. (*) indicates a p value ≤ 0.01.
Targeting of ULBP1 to the detergent-soluble membrane fraction reduces the sensitivity of target cells to lysis by NK cells
It has been shown that redistribution of ICAM-2 on tumor cells via ezrin renders these cells more sensitive to lysis by NK cells (17). To test whether distribution of ULBP1 in either the DRM fraction or the detergent-soluble membrane fraction had any functional consequence for sensitivity to NKG2D-dependent cytotoxicity, P815-ULBP1 and P815-ULBP1-CD45TM cells were used as targets in a 2 h lysis assay (Fig. 2). Expression of ULBP1 on P815 cells rendered them more sensitive to lysis by primary, resting NK cells (Fig. 2A), and IL-2 activated NK cells (Fig. 2B). Expression of ULBP1-CD45TM on P815 cells resulted in a lower sensitivity to lysis by NK cells, as compared to ULBP1 on P815 cells (Fig. 2). These results indicate that distribution of ULBP1 within the membrane may be important for proper NK cell function.
We next tested which step in NK cell cytotoxicity was sensitive to changes in the distribution of ULBP1. Expression of ULBP1 in the detergent-soluble membrane fraction of P815 cells resulted in reduced degranulation of resting NK cells (Fig. 2C) and of IL-2 activated NK cells (Fig. 2D). Expression of ULBP1 and ULBP1-CD45TM on P815 cells had only a minor enhancing effect on the polarization of lytic granules toward the NK–P815 cell contact (not shown). We conclude that a change of the localization of ULBP1 from the DRM domains to the detergent-soluble membrane fraction reduces cytotoxicity of NK cells at the level of degranulation.
Clustering and lateral diffusion of ULBP1 and ULBP1-CD45TM in the plasma membrane are similar
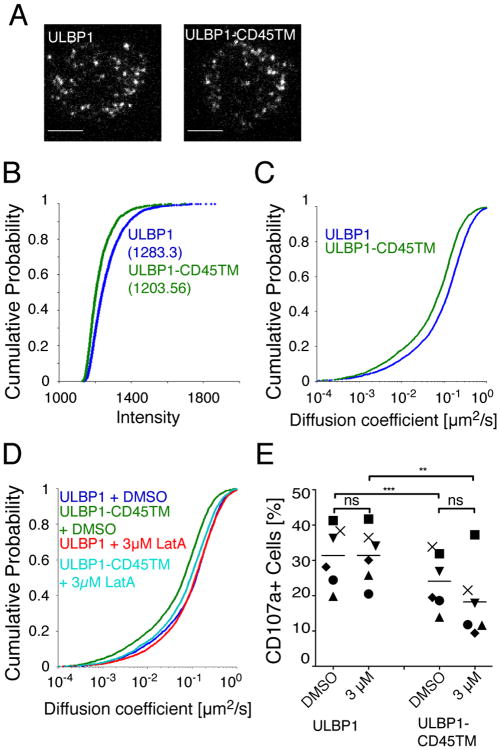
We next tested how the segregation of ULBP1 into DRM and detergent-soluble domains affected the extent of ULBP1 clustering and lateral mobility within the plasma membrane. To investigate these parameters, ULBP1 and ULBP1-CD45TM were labeled with a PE-conjugated antibody to ULBP1 and their distribution and mobility were visualized by total internal reflection fluorescence (TIRF) microscopy. TIRF microscopy is a spatially limited, high contrast technique that eliminates interference from bulk fluorescence, which may be present within cells, and selectively detects fluorophores proximal to and within the plasma membrane of cells on glass coverslips (18). Both ULBP1 and ULBP1-CD45TM were distributed into small clusters at the surface of P815 cells (Fig. 3A). Although individual ULBP1 and ULBP1-CD45TM proteins were labeled with a single PE-fluorophore, photobleaching characteristics (the presence of multiple-step bleaching events over long track length, data not shown) of fluorescent PE-labeled particles suggested that ULBP1 and ULBP1-CD45TM were observed primarily as clusters and not single molecules. Cluster analysis by fluorescence intensity measurements revealed no significant difference in ULBP1 and ULBP1-CD45TM clusters, with a median intensity of 1238 and 1204, respectively (Fig. 3B). Therefore, the distribution of ULBP1 in different membrane domains did not have a detectable impact on the number of ULBP1 molecules per cluster. Furthermore, the cluster intensity for both molecules was homogenous (Fig. 3B). We conclude that the difference in sensitivity to NKG2D-dependent cytotoxicity is not due to a change in the number of ULBP1 molecules per cluster.
Figure 3.
Lateral diffusion of ULBP1 and ULBP1-CD45TM is similar. A, Pictures of P815-ULBP1 (left) and P815-ULBP1-CD45TM (right) cells labeled with PE-conjugated Ab to ULBP1. Scale bars represent 5 µm. B, The intensity of ULBP1 (blue) and ULBP1-CD45 (green) particles was measured in a 5 × 5 pixel grid centered over the peak of the Gaussian distribution calculated for the particle in the first frame in which it appeared in the tracking algorithm. Average particle intensities are shown in cumulative probability plots. Median intensities are indicated in parenthesis. C, The movement of ULBP1 and ULBP1-CD45TM particles labeled with PE-conjugated Ab to ULBP1 was tracked by capturing TIRF images at 35 frames per second for 100 frames. Diffusion coefficients of ULBP1 (blue, n = 3532) and ULBP1-CD45TM (green, n = 2295) particles are shown in cumulative probability plots. Data are from one representative experiment out of 7. D, Movement of DMSO-treated P815-ULBP1 (purple, n = 2826) and P815-ULBP1-CD45TM (green, n = 2921), and Latrunculin A treated P815-ULBP1 (red, n = 4114) and P815-ULBP1-CD45TM (aqua, n = 3240) cells was tracked by TIRF as described in Fig. 3B. Data are from one out of 8 independent experiments. E, Degranulation of IL-2 activated NK cells stimulated with P815-ULBP1 and P815-ULBP1-CD45TM cells pre-treated with DMSO carrier or 3 μM Latrunculin A, as described in Fig. 2B. Each symbol represents an individual donor among 6, which were tested in independent experiments. Paired t-test was performed. (**) indicates a p value ≤ 0.0025 and (***) indicates a p value ≤ 0.0004.
The lateral movement of labeled ULBP1 and ULBP1-CD45TM particles recorded by TIRF microscopy was tracked automatically using an algorithm developed for MatLab software (19), which was further modified to refine particle positioning with a 2D Gaussian fit (20). Short-range mean square displacements (MSDs) were determined from positional coordinates of particles tracked for five frames (over 160 ms; (20)) and were linearly dependent on time under all conditions measured, consistent with a simple diffusion model for this range of movement. Short-range diffusion coefficients were then determined for thousands of particles in multiple cells and graphed either in cumulative probability plots (also known as cumulative distribution function) to represent the frequency of diffusion coefficients for the entire population of tracked particles (Fig. 3C), or median scattered plots (Fig. S2A). Each one of several thousand particles is represented as a separate point in the cumulative probability plot. This type of graph can visually resolve small differences between samples even when extensive overlap occurs. ULBP1 clusters displayed a high lateral mobility at the surface of P815 cells, with a median diffusion coefficient of 0.122 μm2/s. Lateral mobility of ULBP1-CD45TM, with a median diffusion coefficient of 0.075 μm2/s, was reduced compared to ULBP1. Single particle tracking experiments have shown that diffusion rate at the plasma membrane is reduced when proteins associate with lipid rafts (21) or with protein complexes (22). The distribution of ULBP1-CD45TM in the detergent-soluble membrane fraction may have resulted in intermolecular interactions that reduced ULBP1 mobility even further than the association of ULBP1 with DRM domains.
To test if the lateral mobility of ULBP1 and ULBP1-CD45TM was controlled by the actin cytoskeleton, we tracked mobility on P815 cells treated with either DMSO carrier alone or 3 μM Latrunculin A (Fig. 3D, Fig. S2B). Whereas the mobility of ULBP1 did not change after treatment with Latrunculin A, the mobility of ULBP1-CD45TM increased from a median diffusion coefficient of 0.065 μm2/s to a diffusion coefficient of 0.093 μm2/s, which was close to the mobility of ULBP1 (0.118 μm2/s) after treatment (Fig. 3D). A higher dose of Latrunculin A (10 μM) did not increase the mobility of ULBP1-CD45TM any further and had no effect on the mobility of ULBP1 (data not shown).
Previous work from our group has shown that immobilization of ICAM on target cells rather than its clustering, promotes proper LFA-1 dependent conjugate formation and granule polarization in primary NK cells (14). To test whether changes in lateral diffusion of ULBP1 were responsible for the difference in sensitivity to lysis by NK cells (Fig. 2), we took advantage of the similar lateral diffusion of ULBP1 and ULBP1-CD45TM after treatment with Latrunculin A. If lateral diffusion of ULBP1 was the main determinant of the functional difference (i.e. greater ULBP1 mobility leading to increased NKG2D-dependent degranulation), treatment of the target cells with Latrunculin A should equalize the response to P815-ULBP1 and P815-ULBP1-CD45TM cells. As seen earlier in the absence of DMSO (Fig. 2), in the presence of DMSO, ULBP1 induced significantly greater degranulation than ULBP1-CD45TM (Fig. 3E). Treatment of P815 cells with Latrunculin A did not equalize the response induced by ULBP1 and ULBP1-CD45TM: Degranulation induced by ULBP1 did not change, but that induced by ULBP1-CD45TM was reduced even further (Fig. 3E). Therefore, NKG2D-dependent cytotoxicity is controlled by the distribution of NKG2D ligands into separate membrane domains, independently of the number of ligand molecules per cluster, and of the lateral mobility of clusters in the plasma membrane.
The molecular basis for the change in NKG2D-dependent responses when ULBP1 is moved to a different membrane environment is still unknown. This change could be relevant, as the related NKG2D ligand ULBP2 is expressed as both a GPI-linked form and a transmembrane form (23). However, expression of the transmembrane form of ULBP2 on the NK-sensitive CHO cell line had a similar small enhancing effect on sensitivity to lysis by NK cells, as expression of both forms (23). Potential cis interactions of ULBP1 with cell surface proteins are different in the DRM domains than in the rest of the plasma membrane. The slower mobility of ULBP1-CD45TM, as compared to ULBP1, and the recovery to a similar lateral mobility after inhibition of F-actin suggest that ULBP1-CD45TM interacts with molecules tethered to the cytoskeleton. Receptor ligands on target cells may often exist within the context of larger protein complexes, and the role of these complexes in ligand recognition should be given greater consideration. The overall extent of basal ULBP1 clustering, prior to contact of target cells with NK cells, was virtually identical for ULBP1 and ULBP1-CD45TM. Nevertheless, despite a similar number of molecules per cluster, the molecular and biophysical properties of ULBP1 and ULBP1-CD45TM clusters may be different, due to the unique properties of membrane subdomains. Such differences may change the interaction of receptor NKG2D with its ligands at immunological synapses, the organization of which plays an important role in lymphocyte responses (24). Distribution of ULBP1 either within or outside of DRM domains may also affect trogocytosis, a process by which cell surface proteins are transferred between target cells and lymphocytes (25-27). Whether intercellular transfer of a ligand for an activation receptor from a target cell to an effector cell leads to amplification of the response or to desensitization of the receptor is unclear. It would be interesting to investigate if the distribution of ULBP1 in different membrane domains has an impact on its transfer to NK cells.
Our data suggests that ligand distribution into distinct membrane domains in general may play an underappreciated role in the activation of NK cells. Given the potential of tumor cells or virus-infected cells to alter ligand distribution at the plasma membrane and to escape immune responses, it will be important to investigate how the distribution of other ligands impacts the activation of lymphocytes.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIAID/NIH.
We thank M. Vales-Gomez and H. Reyburn for advice and comments, and M. Peterson and M. March for discussion.
Abbreviations used in this paper
- NK cell
- NKG2D
- ULBP1
- detergent resistant membrane
- cytotoxicity
- degranulation
References
- 1.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orange JS. Formation and function of the lytic NK-cell immunological synapse. Nat Rev Immunol. 2008;8:713–725. doi: 10.1038/nri2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber DF, Faure M, Long EO. LFA-1 contributes an early signal for NK cell cytotoxicity. J Immunol. 2004;173:3653–3659. doi: 10.4049/jimmunol.173.6.3653. [DOI] [PubMed] [Google Scholar]
- 4.Bryceson YT, March ME, Barber DF, Ljunggren HG, Long EO. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med. 2005;202:1001–1012. doi: 10.1084/jem.20051143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NM, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 9.Dykstra M, Cherukuri A, Sohn HW, Tzeng SJ, Pierce SK. Location is everything: Lipid rafts and immune cell signaling. Annu Rev Immunol. 2003;21:457–481. doi: 10.1146/annurev.immunol.21.120601.141021. [DOI] [PubMed] [Google Scholar]
- 10.Harder T, Rentero C, Zech T, Gaus K. Plasma membrane segregation during T cell activation: probing the order of domains. Curr Opin Immunol. 2007;19:470–475. doi: 10.1016/j.coi.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Tseng SY, Liu ML, Dustin ML. CD80 cytoplasmic domain controls localization of CD28, CTLA-4, and protein kinase Ctheta in the immunological synapse. J Immunol. 2005;175:7829–7836. doi: 10.4049/jimmunol.175.12.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eleme K, Taner SB, Onfelt B, Collinson LM, McCann FE, Chalupny NJ, Cosman D, Hopkins C, Magee AI, Davis DM. Cell surface organization of stress-inducible proteins ULBP and MICA that stimulate human NK cells and T cells via NKG2D. J Exp Med. 2004;199:1005–1010. doi: 10.1084/jem.20032194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherukuri A, Tzeng SJ, Gidwani A, Sohn HW, Tolar P, Snyder MD, Pierce SK. Isolation of lipid rafts from B lymphocytes. Methods Mol Biol. 2004;271:213–224. doi: 10.1385/1-59259-796-3:213. [DOI] [PubMed] [Google Scholar]
- 14.Gross CC, Brzostowski JA, Liu D, Long EO. Tethering of intercellular adhesion molecule on target cells is required for LFA-1-dependent NK cell adhesion and granule polarization. J Immunol. 2010;185:2918–2926. doi: 10.4049/jimmunol.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson ME, Long EO. Inhibitory receptor signaling via tyrosine phosphorylation of the adaptor Crk. Immunity. 2008;29:578–588. doi: 10.1016/j.immuni.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Messina L, Ashiru O, Boutet P, Aguera-Gonzalez S, Skepper JN, Reyburn HT, Vales-Gomez M. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J Biol Chem. 2010;285:8543–8551. doi: 10.1074/jbc.M109.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helander TS, Carpen O, Turunen O, Kovanen PE, Vaheri A, Timonen T. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 1996;382:265–268. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- 18.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- 19.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolar P, Hanna J, Krueger PD, Pierce SK. The constant region of the membrane immunoglobulin mediates B cell-receptor clustering and signaling in response to membrane antigens. Immunity. 2009;30:44–55. doi: 10.1016/j.immuni.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 22.Vale RD. Microscopes for fluorimeters: the era of single molecule measurements. Cell. 2008;135:779–785. doi: 10.1016/j.cell.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Messina L, Ashiru O, Aguera-Gonzalez S, Reyburn HT, Vales-Gomez M. The human NKG2D ligand ULBP2 can be expressed at the cell surface with or without a GPI anchor and both forms can activate NK cells. J Cell Sci. 2011;124:321–327. doi: 10.1242/jcs.076042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taner SB, Onfelt B, Pirinen NJ, McCann FE, Magee AI, Davis DM. Control of immune responses by trafficking cell surface proteins, vesicles and lipid rafts to and from the immunological synapse. Traffic. 2004;5:651–661. doi: 10.1111/j.1600-0854.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 25.Sprent J. Swapping molecules during cell-cell interactions. Sci STKE. 2005;2005:pe8. doi: 10.1126/stke.2732005pe8. [DOI] [PubMed] [Google Scholar]
- 26.Davis DM. Intercellular transfer of cell-surface proteins is common and can affect many stages of an immune response. Nat Rev Immunol. 2007;7:238–243. doi: 10.1038/nri2020. [DOI] [PubMed] [Google Scholar]
- 27.Daubeuf S, Aucher A, Bordier C, Salles A, Serre L, Gaibelet G, Faye JC, Favre G, Joly E, Hudrisier D. Preferential transfer of certain plasma membrane proteins onto T and B cells by trogocytosis. PLoS One. 2010;5:e8716. doi: 10.1371/journal.pone.0008716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.