Abstract
The formation of patterns that are proportional to the size of the embryo is an intriguing but poorly understood feature of development. Molecular mechanisms controlling such proportionality, or scaling, can be probed through quantitative interrogations of the properties of morphogen gradients that instruct patterning. Recent studies of the Drosophila morphogen gradient Bicoid (Bcd), which is required for anterior-posterior (AP) patterning in the early embryo, have uncovered two distinct ways of scaling. Whereas between-species scaling is achieved by adjusting the exponential shape characteristic of the Bcd gradient profile, namely, its length scale or length constant (λ), within-species scaling is achieved through adjusting the profile’s amplitude, namely, the Bcd concentration at the anterior (B0). Here, we report a case in which Drosophila melanogaster embryos exhibit Bcd gradient properties uncharacteristic of their size. The embryos under investigation were from a pair of inbred lines that had been artificially selected for egg size extremes. We show that B0 in the large embryos is uncharacteristically low but λ is abnormally extended. Although the large embryos have more total bcd mRNA than their smaller counterparts, as expected, its distribution is unusually broad. We show that the large and small embryos develop gene expression patterns exhibiting boundaries that are proportional to their respective lengths. Our results suggest that the large-egg inbred line has acquired compensating properties that counteract the extreme length of the embryos to maintain Bcd gradient properties necessary for robust patterning. Our study documents, for the first time to our knowledge, a case of within-species Bcd scaling achieved through adjusting the gradient profile’s exponential shape characteristic, illustrating at a molecular level how a developmental system can follow distinct operational paths towards the goal of robust and scaled patterning.
Keywords: Bicoid, Canalization, Length constant, Morphogen gradient, Robust patterning, Size scaling
INTRODUCTION
A striking feature of animal development is the formation of body parts that are proportional to an individual’s overall body size (Waddington, 1942). This is reflective of the robust nature of the developmental process in the face of inevitable variations at all levels, ranging from molecular to organismal and environmental levels (Patel and Lall, 2002; Flatt, 2005; Martinez Arias and Hayward, 2006; Hendrikse et al., 2007; Lander, 2007; Lott et al., 2007). A full, mechanistic understanding of scaling requires knowledge about two distinct aspects of development: specification of scaled patterns and coordinated tissue growth (Su and O’Farrell, 1998; Day and Lawrence, 2000; Crickmore and Mann, 2008; Ben-Zvi et al., 2011; Wartlick et al., 2011; Yang and Xu, 2011; Baena-Lopez et al., 2012). Recent quantitative studies have uncovered insights into scaled patterning specification in Drosophila (Houchmandzadeh et al., 2002; Gregor et al., 2005; He et al., 2008; Manu et al., 2009; Cheung et al., 2011). A well-documented feature of patterning is the involvement of regulatory networks (Jaeger et al., 2004; Levine and Davidson, 2005; Bergmann et al., 2007; Lander, 2011). By nature, these networks can confer robustness to a patterning system, reflective of the regulatory power of, for example, feedback loops and cross-regulatory circuits (Manu et al., 2009; Lander, 2011). Recent studies of the Drosophila morphogen gradient of Bicoid (Bcd) show that the initial, maternal input into an embryonic patterning system also exhibits properties of size scaling (Gregor et al., 2007a; He et al., 2008; de Lachapelle and Bergmann, 2010; Cheung et al., 2011), suggesting that quantitative evaluations of morphogen gradients can advance our understanding of developmental robustness and scaling.
The morphogen gradient of Bcd is required for patterning along the anterior-posterior (AP) axis in the early Drosophila embryo (Driever and Nüsslein-Volhard, 1988; Struhl et al., 1989; Ephrussi and St. Johnston, 2004). The protein gradient is derived from the anteriorly localized and maternally deposited bcd mRNA (Berleth et al., 1988). Although mechanistic details of Bcd gradient formation remain a topic of active studies (Gregor et al., 2007b; Spirov et al., 2009; Porcher et al., 2010; Little et al., 2011; Liu and Ma, 2011), it is generally thought that this process can be described, in part, by a simple diffusion model, in which Bcd protein synthesized at the anterior diffuses and decays throughout the embryo (Grimm et al., 2010; Liu et al., 2011). This diffusion model gives rise to a steady-state exponential profile of the morphogen concentration B, such that, B=Ae-x/λ, where A is the amplitude, λ is the length constant (also referred to as the length scale), and x is distance from the source (Rice, 1985; Bergmann et al., 2007; Wartlick et al., 2009). Since bcd mRNA, the source for Bcd protein, is not localized to a single point in the actual embryo (Spirov et al., 2009; Cheung et al., 2011; Little et al., 2011), as it is in the idealized diffusion model, the experimentally measured Bcd profile deviates from the exponential function at the anterior region (Houchmandzadeh et al., 2002; Deng et al., 2010; Dalessi et al., 2011). For an experimentally observed Bcd profile, its concentration at the anterior, B0, can be used as a substitute for amplitude (He et al., 2008). In Drosophila melanogaster, the length constant λ of the observed Bcd gradient profile is ∼100 μm (Gregor et al., 2007a; He et al., 2008; He et al., 2010b; Cheung et al., 2011; Liu et al., 2011). This generally characteristic λ is constrained by, according to the simple diffusion model, the effective diffusivity and stability of Bcd in the early embryo of this species (Gregor et al., 2005; Gregor et al., 2008; Porcher et al., 2010; Drocco et al., 2011; Liu et al., 2011; Liu and Ma, 2011).
Studies of the Bcd gradient properties have uncovered two distinct ways to scale an exponential profile with embryo length (Gregor et al., 2005; He et al., 2008; Cheung et al., 2011). In both cases, the Bcd gradient profiles from large and small embryos become similar to each other (i.e. they converge) as a function of relative AP position or fractional embryo length x/L (He et al., 2008; de Lachapelle and Bergmann, 2010; Cheung et al., 2011). Thus, mechanisms of scaling the Bcd gradient profile can be viewed effectively as ways for the embryos to achieve comparable Bcd concentrations at a given relative AP position (particularly approaching the middle sections of the embryos) regardless of their size. The two distinct ways of Bcd gradient scaling were revealed by comparative studies of differently sized embryos either from distinct species or from a single species. While between-species Bcd gradient scaling is achieved by scaling λ with embryo length L (Gregor et al., 2005), within-species scaling is achieved by scaling B0 with embryo volume V (He et al., 2008; Cheung et al., 2011). In both cases, the Bcd gradient profile in large embryos can ‘reach’ further into the embryo as an absolute distance from the anterior.
In this study, we analyze quantitatively the Bcd gradient profiles in embryos from a pair of Drosophila melanogaster inbred lines that had been artificially selected for egg size extremes (Miles et al., 2011). We show that, although the large embryos have a low B0, which is uncharacteristic for their size, the Bcd profile’s λ is atypically large for Drosophila melanogaster. We show that this unusually large λ, when expressed as a value relative to L, is similar to that observed in the small embryos. These results document the first case of within-species Bcd gradient scaling achieved by adjusting the profile’s exponential shape characteristic λ. We further show that the distribution of bcd mRNA in the large embryos under investigation is abnormally broad, a feature that is not inherent to embryos that are large. Our analysis of hunchback (hb) and even-skipped (eve) reveals robust expression patterns with boundary positions that are proportional to the lengths of the large and small embryos. Together, our results suggest that the large-egg inbred line has acquired compensating properties to counteract the extreme embryo size such that the scaling properties of the Bcd gradient profile are maintained to instruct robust AP patterning. Our current study, in conjunction with a previous report (Cheung et al., 2011), provides an illustration, at a molecular level, of how a developmental system can achieve robust and scaled patterning by following distinct operational paths within a species. It advances our understanding of how a robust ‘end result’ of patterning, or canalization (Waddington, 1942), can be achieved in molecular terms.
RESULTS
Embryos from a pair of selected Drosophila melanogaster inbred lines exhibit Bcd gradient properties uncharacteristic of their size
In a recent study (Cheung et al., 2011), we quantified both Bcd protein and bcd mRNA properties in embryos from a pair of inbred Drosophila melanogaster lines that had been artificially selected for egg size extremes (Miles et al., 2011). We also analyzed embryos from multiple populations (each being genetically heterogeneous) that had been selected for egg size. These experiments led to a general observation that large embryos contain more maternally deposited bcd mRNA than their smaller counterparts. We proposed that a volume-dependent Bcd production rate leads to a higher B0 in larger embryos and, consequently, scaling of the Bcd gradient profile within a species (Cheung et al., 2011). During our analysis of another pair of inbred lines, 2.49.3 and 9.31.2 (henceforth referred to as the large-egg line and small-egg line, respectively), we found that, as detailed below, the embryos exhibited Bcd properties uncharacteristic of their size. In the study reported here, we used quantitative immunofluorescence staining to detect nuclear Bcd in whole-mount embryos. Our staining procedures avoided any nonlinear amplification steps and our imaging was performed under conditions documented to be within a linear range (He et al., 2008; Cheung et al., 2011). We thus use the terms fluorescence intensity and Bcd concentration (both in arbitrary unit, a.u.) interchangeably.
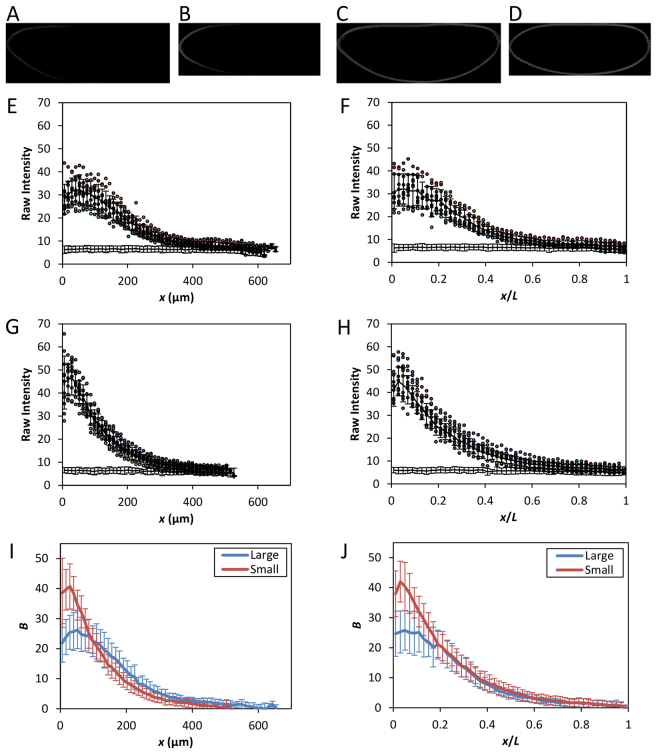
Fig. 1A,B shows raw fluorescence images of the large and small embryos under investigation. The image of the large embryo (Fig. 1A) shows visually lower fluorescence intensity in the anterior than the small embryo (Fig. 1B; see 1C,D for corresponding images of nuclear staining). Quantification of the intensity data (see below) revealed that the Bcd fluorescence intensity at the anterior is B0=24.7±7.5 (mean ± s.d.) in the large embryos (n=10), which is significantly lower than that of the small embryos (40.0±7.6, n=11; P=7.5×10-4). These B0 properties contrast with the size measurements of these embryos: L=626.6±24.5 μm and 506.9±19.3 μm for the large and small embryos, respectively (P=1.3×10-10; all P values in this article were from Student’s t-tests), and V=22±3.0 × 106 μm3 and 11±1.3 × 106 μm3 for the large and small embryos, respectively (P=1.3×10-13), demonstrating that the Bcd gradient properties in these embryos are uncharacteristic of their size. Despite the unusual B0 properties, the large and small embryos develop robust patterns of gene expression along the AP axis (see below), suggesting that quantitatively characterizing the Bcd gradient properties in these embryos may reveal useful new insights relevant to our understanding of how scaled patterning specification is achieved.
Fig. 1.
Bcd gradient properties uncharacteristic of embryo size. (A-D) Shown are midsagittal images of embryos from the large-egg line 2.49.3 (A,C) and the small-egg line 9.31.2 (B,D), detecting either Bcd (A,B) or nuclei (C,D). The intensities in the images shown were adjusted for presentational purposes only. (E-H) Extracted fluorescence intensities are plotted as a function of either x (E,G) or x/L (F,H). n=10 and 11 for lines 2.49.3 (E,F) and 9.31.2 (G,H), respectively. Mean intensity is shown as black line; error bars are s.d. Also shown are the relevant portions of the mean intensity (and s.d.) from bcdE1 embryos. (I-J) Mean Bcd profiles from the large (blue) and small (red) embryos as a function of x (I) or x/L (J). Error bars are s.d.
Bcd gradient profiles in the large and small embryos exhibit scaling properties
To evaluate quantitatively the Bcd gradient profiles from the large and small embryos, we extracted the Bcd fluorescence intensity, B, from individual embryos and expressed it as a function of either absolute distance from the anterior x or fractional embryo length x/L. To ensure proper comparisons of the captured fluorescence intensity data, all of our experimental and imaging steps for these embryos were performed on a side-by-side basis (with different samples mounted on separate slides). In addition, we incorporated embryos lacking Bcd (i.e. embryos from bcdE1 females, referred to as bcdE1 embryos) with the experimental embryos (i.e. embryos were mixed during staining) so that background fluorescence intensities could be measured under identical conditions as reported previously (He et al., 2008; Cheung et al., 2011). Fig. 1E-H shows the raw Bcd intensities captured from individual embryos, with background intensities from bcdE1 embryos also shown at the bottom of these panels. Fig. 1I,J shows the mean B profiles of the large and small embryos as a function of either x or x/L, respectively. Here and throughout this article (except Fig. 1E-H), all B values have been background-subtracted without any further adjustments, unless otherwise indicated. The quantitative data shown in Fig. 1E-J confirmed our visual observation that the large embryos have lower concentrations of Bcd in the anterior than their small counterparts.
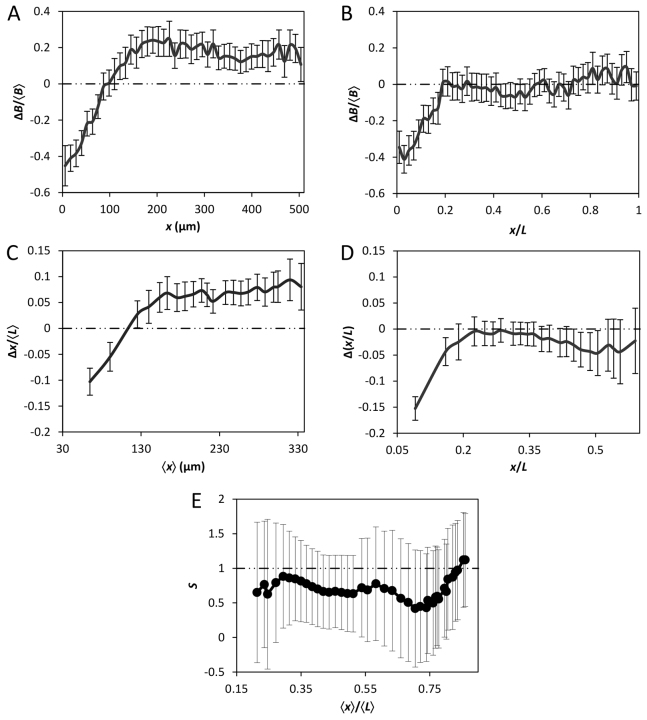
The mean B profiles from the large and small embryos converge upon each other as a function of relative AP position x/L (Fig. 1J). This convergence takes place in broad regions of the embryos along the AP axis (except the anterior) and is a hallmark of Bcd gradient scaling (He et al., 2008; de Lachapelle and Bergmann, 2010; Deng et al., 2010; Cheung et al., 2011). To evaluate this convergence or scaling better, we plotted the differences between the mean B values in these embryos, ΔB, as a function of either x or x/L (Fig. 2A and 2B, respectively). These results show that, despite the lower mean B in the anterior region of the large embryos yielding a negative ΔB, the two mean profiles cross each other at x=∼100 μm to yield a positive ΔB (Fig. 2A; see also Fig. 1I). This positive ΔB is propagated relatively stably through the remainder of the AP length (Fig. 2A). Fig. 2B shows the ΔB profile as a function of relative AP position x/L. It demonstrates that, upon the neutralization of the negative ΔB at x/L=′∼0.2, ΔB becomes greatly reduced (relative to that in Fig. 2A) and stays close to zero for the remainder of the AP length. These results demonstrate quantitatively that the mean Bcd gradient profiles from the large and small embryos become similar to each other as a function of relative AP position (except the anterior), a feature indicative of scaling (see supplementary material Fig. S1 for a further illustration of profile similarity, where the B values from these embryos at their relative positions are plotted against each other).
Fig. 2.
Quantitative evaluation of Bcd gradient scaling. (A,B) Shown are Bcd intensity differences, ΔB, between the large and small embryos as a function of x (A) or x/L (B). ΔB is calculated by subtracting the mean B of line 9.31.2 from the mean B of line 2.49.3, and normalizing to averaged B, ⟨B⟩. (C,D) Shown are differences in positions, Δx, at which mean Bcd gradient profiles cross given thresholds, plotted as a function of x (C) or x/L (D). For panel C, Δx is normalized to the average length of both embryos ⟨L⟩, and ⟨x⟩ denotes averaged position at which the mean profiles cross a threshold. Interpolated mean profiles were used to obtain x at arbitrary B thresholds chosen. Error bars are s.d. of the difference between the sample means (Cheung et al., 2011). (E) Scaling coefficient S as a function of AP position in the large and small embryos. This analysis was performed as described previously (de Lachapelle and Bergmann, 2010; Cheung et al., 2011). Error bars shown represent 95% confidence intervals from linear regression (Cheung et al., 2011).
To evaluate further the scaling properties of the Bcd gradient profiles, we plotted the difference (between the large and small embryos) in positions at which the mean Bcd profiles cross given thresholds. This positional difference, defined as Δx, was plotted as a function of either x or x/L, shown in Fig. 2C and 2D, respectively. For effective visual comparisons, the display windows of these two panels are similar, such that x in Fig. 2C, when normalized to the average length of all embryos ⟨L⟩, is equivalent to x/L shown in Fig. 2D. These results show that Bcd-encoded positional information in the large and small embryos also becomes reduced when the AP position is measured relative to embryo length in comparison with when measured as absolute distance from the anterior. Furthermore, we analyzed our Bcd intensity data pooled from the large and small embryos to estimate the scaling coefficient S at different positions (de Lachapelle and Bergmann, 2010). In this analysis, a perfect scaling is indicated by S=1, with S values greater or smaller than 1 indicating hyper- or hypo-scaling, respectively. Fig. 2E shows that the estimated S values range between 0.5 and 1, suggesting a modest hypo-scaling of the Bcd gradient profiles in these embryos as a group. Together, our results document that, despite the ‘reversed’ B0 in the large and small embryos, their Bcd gradient profiles exhibit scaling properties in a broad region along the AP axis.
The Bcd gradient in the large embryos has an unusually extended length constant
The unusual B0 properties in conjunction with the scaled Bcd gradient profiles suggest that either the large or small embryos (or both) have properties that are uncharacteristic of their size. We suspected, based on our experience with examining Bcd gradient profiles, that the large embryos were somewhat unusual. As noted above, despite the lower B0 in the large embryos, the mean Bcd gradient profile of these embryos crosses above that of the small embryos at x=′∼100 μm (Fig. 1I). These results suggested that these two gradient profiles must have distinct shape characteristics. For an exponential gradient, the parameter depicting its shape characteristic is the length constant λ (also referred to as the length scale), which basically quantifies how quickly a gradient drops off as a function of distance x. We reasoned that, as λ in normal Drosophila melanogaster embryos has a generally characteristic value of ∼100 μm, measurement of this parameter may reveal quantitative information about which of the embryos may have unusual Bcd gradient properties. Towards this end, we first estimated λ values for the Bcd gradient profiles in individual embryos. We found that the large and small embryos have a mean λ of 141.8±19.1 μm and 102.6±16.3 μm, respectively (P=6.7×10-5). These results document quantitatively that the Bcd gradient profiles from these embryos have distinct shape characteristics. They also reveal that λ in the large embryos is atypically large for Drosophila melanogaster.
In the large embryos, the observed λ is extended in proportion to their extreme embryo lengths. In particular, when the length constant was calculated for the Bcd gradient profiles in individual embryos as a relative distance λ/L, we found that the mean values for the large and small embryos become nearly identical to each other (0.22±0.03 and 0.22±0.05, respectively; P=0.98). To generate a visual illustration of the Bcd gradient scaling mechanism under investigation, we plotted the mean B profiles on a logarithmic scale (Fig. 3). In such plots, the length constant λ is the negative reciprocal of the slope. Thus, the slopes of the linear fits in such plots provide a direct visualization of the scaling mechanism for the Bcd gradient profiles in the embryos under investigation. Specifically, we plotted ln(B/Bmax) as a function of x or x/L, as shown in Fig. 3A and 3B, respectively. Although the linear fits for data from the large and small embryos have distinct slopes in the x plot (Fig. 3A), they become parallel to each other in the x/L plot (Fig. 3B; see legend for details). Together, these results show that the large embryos have unusual properties that lead to an expansion of λ in proportion to L, a strategy previously only documented for scaling in embryos from different species (Gregor et al., 2005).
Fig. 3.
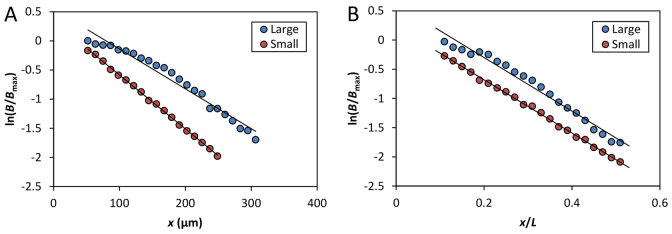
Evaluation of the relationship between Bcd profile’s length constant and embryo length. (A,B) Shown are semi-log scatter plots of mean B from the indicated embryos as a function of x (A) or x/L (B). B is normalized to their respective maximal mean intensity, Bmax. The x/L range of 0.1-0.5 was used to reduce effects of experimental errors in posterior and non-exponential shape in anterior. Linear fits (shown as solid black lines) in A are: y=-0.0071x + 0.60 (R2=0.97) and y=-0.0092x + 0.34 (R2=0.99) for the large and small embryos, respectively. In B, they are: y=-4.58x + 0.62 (R2=0.97) and y=-4.55x + 0.23 (R2=0.99), respectively.
The large embryos have more bcd mRNA than the small embryos
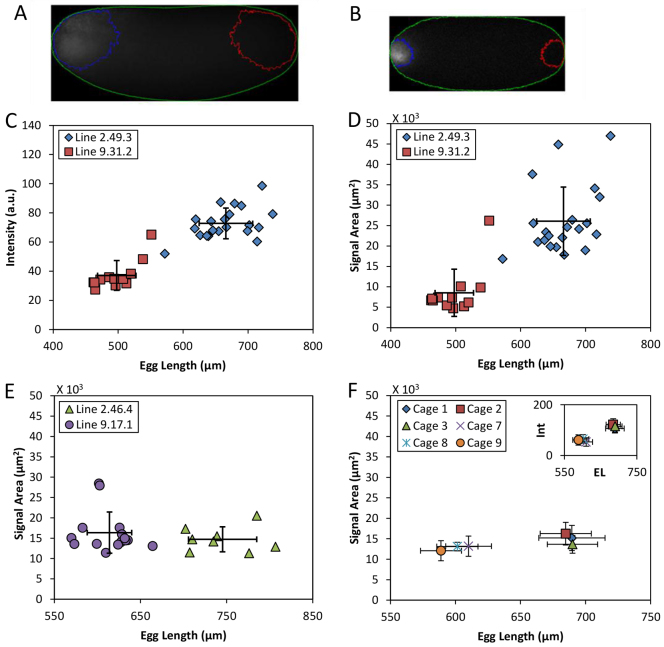
The unusually low B0 in the large embryos raised an important question with regard to the amount of maternally deposited bcd mRNA (Cheung et al., 2011). Specifically, is the large-egg inbred line ‘defective’ in oogenesis such that the amount of bcd mRNA is no longer proportional to egg volume? To address this question, we performed quantitative fluorescence in situ hybridization (FISH) in whole-mount embryos to detect bcd mRNA (see Fig. 4A,B for fluorescence images). Under our reported experimental and analytical framework (Cheung et al., 2011), the captured epifluorescence intensities are known to have a linear relationship with the total amount of bcd mRNA in the embryos. Fig. 4C shows a scatter plot of the detected FISH intensities (in a.u.) against measured embryo length L. These results show that, as in other well-documented cases (Cheung et al., 2011), the large embryos also have more bcd mRNA than the small embryos. The detected epifluorescence intensities in these embryos are 7.27±1.05×105 and 3.70±1.02×105 (a.u.), respectively (P=1.1×10-10), representing a relative difference of 96.5% (see supplementary material Table S1 for relevant values). The measured lengths of the large and small embryos are: L=666±41 and 498±30 μm, respectively (n=21 and 12, respectively; P=1.5×10-13), representing a relative difference of 33.7%. Their calculated volumes are: V=22±3.0×106 μm3 and 11±1.3×106 μm3, respectively, representing approximately a twofold difference. These results are consistent with the proposed general property of egg volume-dependent deposition of bcd mRNA, suggesting that, with regard to this aspect, oogenesis is normal in the large-egg inbred line.
Fig. 4.
Quantification of bcd mRNA in early embryos. (A,B) Immunofluorescence images of embryos detecting bcd mRNA, showing embryo masking (green) and contour outlines for specific signals (blue) or background subtraction (red). Intensities in images shown were adjusted for presentational purposes only. (C) Scatter plot of fluorescence intensities (see Materials and methods) against L for the large (blue) and small (red) embryos under current investigation. (D-F) Scatter plots of the areas containing specific signals against L. Data are from the large (blue) and small (red) embryos under current investigation (D), or embryos from our previously published lines 2.46.4 (green) and 9.17.1 (purple) shown in panel E, and population cages shown in panel F. Inset in F shows mean intensities against mean L. Error bars are s.d.
A broader distribution of bcd mRNA in the large embryos and its impact on Bcd gradient formation evaluated in a theoretical model
Both the depressed B0 and expanded λ in the large embryos suggest an existence of unusual properties relevant to Bcd gradient formation in these embryos. According to the simple diffusion model, λ of a steady-state exponential gradient is a function of the diffusion constant D and degradation rate ω such that 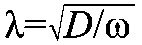 (Wartlick et al., 2009; Grimm et al., 2010; Liu et al., 2011; Liu and Ma, 2011). The unusually large λ suggested that, within the framework of this simple model in its idealized form, either Bcd diffusivity or stability (or both) is enhanced in the large embryos. In addition, modeling studies based on more realistic sources had documented that the distribution characteristics of bcd mRNA can affect the Bcd gradient behavior in the anterior part of the embryo (Deng et al., 2010; Dalessi et al., 2011). Thus, a more flattened Bcd profile in the anterior portion of the large embryos suggested to us that bcd mRNA might be abnormally distributed in these embryos. This suggestion was reinforced by the initial impression that we had on our bcd mRNA FISH data where the large embryos indeed appeared to have an extended distribution of bcd mRNA (see below for quantitative results).
(Wartlick et al., 2009; Grimm et al., 2010; Liu et al., 2011; Liu and Ma, 2011). The unusually large λ suggested that, within the framework of this simple model in its idealized form, either Bcd diffusivity or stability (or both) is enhanced in the large embryos. In addition, modeling studies based on more realistic sources had documented that the distribution characteristics of bcd mRNA can affect the Bcd gradient behavior in the anterior part of the embryo (Deng et al., 2010; Dalessi et al., 2011). Thus, a more flattened Bcd profile in the anterior portion of the large embryos suggested to us that bcd mRNA might be abnormally distributed in these embryos. This suggestion was reinforced by the initial impression that we had on our bcd mRNA FISH data where the large embryos indeed appeared to have an extended distribution of bcd mRNA (see below for quantitative results).
Fig. 4A and 4B show raw epifluorescence images of, respectively, the large and small embryos detecting bcd mRNA. In these figures, we also show the contour outline representing the threshold marking the areas containing the specific signals (see Materials and methods). A quantitative comparison between the large and small embryos (Fig. 4D; supplementary material Fig. S2C and Table S1) revealed a significantly larger signal area in the large embryos [2.6±0.8×104 μm2 and 8.5±0.6×103 μm2, respectively; P=3.9×10-7]. To determine whether a broad distribution of bcd mRNA may be a property that is inherent to large embryos, we evaluated the signal areas in embryos from our previously published pair of inbred lines (Fig. 4E; supplementary material Fig. S2D) as well as those from population cages (Fig. 4F). Our results show that in all these cases the signal areas are mostly insensitive to embryo size (see supplementary material Table S2 for a listing of all relevant measurements and comparisons). In particular, the relative differences in signal areas between these tested embryos do not exceed 35% (Fig. 4E,F; supplementary material Table S2), contrasting the greater than threefold difference in signal areas between the large and small embryos under current investigation (Fig. 4D; supplementary material Table S1). Together, these results document that the large embryos under current investigation have an unusually broad distribution of bcd mRNA, a feature that is not inherent to Drosophila melanogaster embryos that are large.
To investigate potential genetic ‘defects’ of the large-egg line, we sequenced both bcd and the coding regions of candidate genes known to have a role in bcd mRNA localization, staufen (Ferrandon et al., 1994), Vps36 (Irion and St Johnston, 2007), swallow (Nüsslein-Volhard et al., 1987; Hegdé and Stephenson, 1993) and exuperantia (Macdonald et al., 1991; Marcey et al., 1991). Supplementary material Table S4 summarizes deviations between our sequencing data and that of FlyBase (see Materials and methods). We found no changes in the 3′UTR of bcd (Berleth et al., 1988; Macdonald and Struhl, 1988). There were two changes in bcd coding region, one of which is predicted to cause an amino acid (aa) change. The coding regions of swa and exu did not have any ‘mutations’ that are predicted to cause aa changes though silent ‘mutations’ were found in exu. However, we detected a significant number of ‘mutations’ in the coding regions of Vps36 and stau each with multiple predicted aa changes. These results show that the large-egg line is indeed genetically different from the ‘standard wild type’. It remains to be determined whether any, or a combination, of these ‘mutations’ is functionally responsible for the broader distribution of bcd mRNA in the large embryos.
A recent theoretical study by Dalessi et al. has obtained the steady-state solutions for morphogen concentrations produced from an extended source (Dalessi et al., 2012). In addition, Sascha Dalessi and Sven Bergmann (personal communication) suggested that a broader distribution of bcd mRNA could explain our observed Bcd gradient properties in the large embryos under their theoretical framework. We obtained from Sven Bergmann the computer code that they had generated to produce the Bcd gradient profiles shown in Fig. 5 (see legend for details). Like experimentally observed profiles (Fig. 1I,J), the theoretical profiles are also expressed as a function of either x or x/L, shown in Fig. 5A and 5B, respectively. A resemblance of the behaviors of the Bcd gradient profiles in Fig. 1I,J and Fig. 5A,B shows that, under the theoretical framework of Dalessi et al. (Dalessi et al., 2012), a broadly distributed bcd mRNA source, coupled with an elevated D and ω, can recapitulate our experimentally observed Bcd properties in the large embryos (see Fig. 5 legend).
Fig. 5.
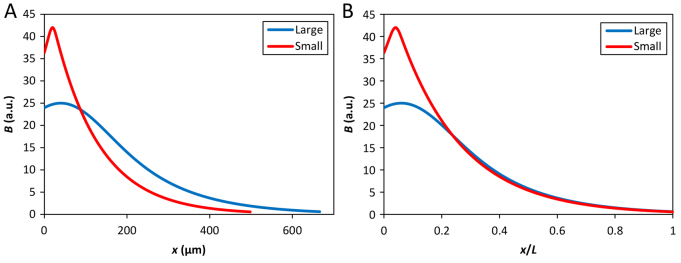
Evaluating the impact of bcd RNA distribution on Bcd gradient formation in a theoretical model. (A,B) Bcd profiles of the large (blue) and small (red) embryos as steady-state concentrations (a.u.) based on equation 14 in Dalessi et al. (Dalessi et al., 2012), plotted as a function of x (A) or x/L (B). In the Dalessi et al. model, the ‘spread’ of a normally distributed source is characterized by σ (in units of L). σ=0.12 and 0.01 was used for the large and small embryos, respectively. The respective ‘center’ locations of the source (in units of L) were assigned as 0.06 and 0.04, and experimental estimates of L=666 μm and 498 μm, and λ/L=0.22 (for both embryos) were used. The production rates for the large and small embryos (in units of their respective decay rates) were estimated according to the observed Bmax ratio (′25:42). The ratio of the detected bcd mRNA intensities in the large and small embryos (7.27:3.70) allowed the calculation of the ratios of both the diffusion constants (3.2) and decay rates (1.8). The results shown were based on an analysis by Sascha Dalessi and Sven Bergmann and the computer code that they had generated (personal communication).
Robust expression patterns of hunchback and even-skipped in the large embryos
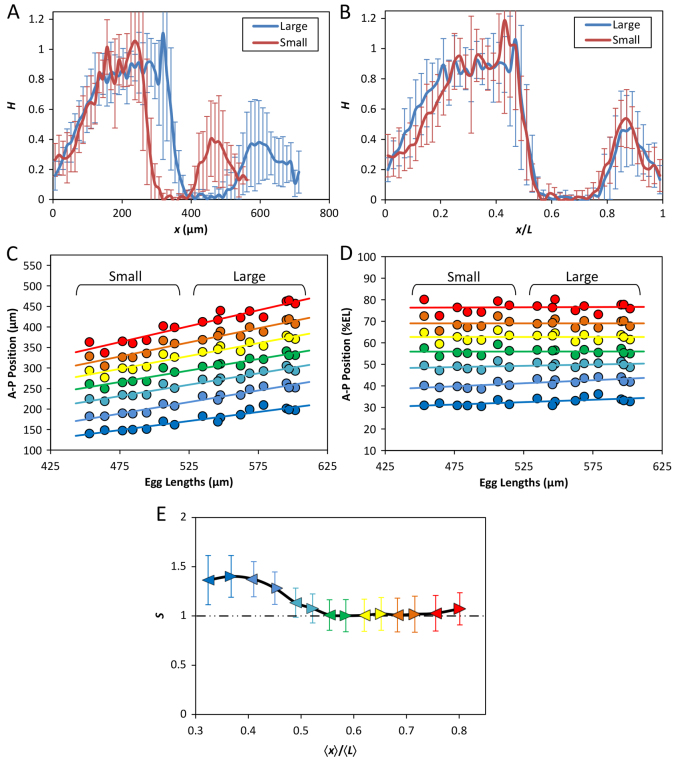
To investigate the functional impact of the observed Bcd gradient properties on AP patterning, we first analyzed the expression pattern of the Bcd target gene hb. We performed quantitative FISH to detect mature hb mRNA in whole-mount embryos. Fig. 6A and 6B show the mean hb FISH intensity profiles in the large and small embryos as a function of x or x/L, respectively (see supplementary material Fig. S3 for corresponding intensity data extracted from individual embryos). The mean hb boundary position, xhb, which is defined as the position at which the hb mRNA FISH intensity level is at its half maximal, is 340.5±20.5 μm and 271.0±15.9 μm for the large and small embryos, respectively (n=10 and 7; P=1.9×10-6). However, when AP position was expressed as a relative value x/L, the mean hb boundary positions in these embryos become basically identical to each other (0.50±0.03 and 0.50±0.02 for the large and small embryos, respectively; P=0.78). These results show that a scaled gene expression boundary is formed near mid embryo.
Fig. 6.
Robust expression patterns of hunchback and even-skipped. (A,B) Mean profiles (with s.d.) of FISH intensities detecting hb mRNA as a function of x (A) or x/L (B). n=10 and 7 for the large and small embryos, respectively. See supplementary material Fig. S3 for data from individual embryos. (C,D) Scatter plots of detected eve expression boundary positions against L. Either absolute (C) or relative (D) AP positions of the anterior boundaries of each of the seven stripes are shown. Linear regression is shown for each boundary. (E) Scaling coefficient profiles of eve expression boundaries in the large and small embryos. For this analysis, both the anterior and posterior boundaries (shown as arrowheads) of each eve stripe were used, with color code being the same as in C and D. Error bars represent 95% confidence intervals from linear regression (Cheung et al., 2011).
To investigate the patterning properties in a broader portion along the AP axis, we determined the expression profiles of the pair-rule gene eve (see Materials and methods). Fig. 6C and 6D show scatter plots of the anterior boundary positions (in x or x/L, respectively) of eve expression stripes against L, documenting a correlation between these boundaries and L. To improve quantification of eve stripe boundaries in relation to L, we calculated the scaling coefficient S using data pooled from the large and small embryos. Here we analyzed both the anterior and posterior boundaries for each of the seven eve stripes. Fig. 6E shows that, although eve boundaries in the anterior are modestly hyper-scaled (S>1), those near mid embryo and beyond are nearly perfectly scaled (S∼1). The modest hyper-scaling in the anterior is also visually exhibited in Fig. 6D by the slight incline of the relative boundary positions of stripes 1 and 2 when plotted against L. A comparison of the S profiles shown in Fig. 2E and Fig. 6E reveals that, despite differences in their absolute values and profile smoothness, they exhibit a broadly similar trend within the x/L range of ∼0.3 to ∼0.6. Considering the inherent limitations to accurate measurement of Bcd concentrations, this similarity in trend suggests that Bcd has a role in regulating the scaling properties within this portion of the embryos. Meanwhile, the differences between the scaling properties of the Bcd gradient profiles and eve expression boundaries are supportive of the important roles of other maternal inputs and cross-regulatory mechanisms in the evolution and refinement of AP patterns (Manu et al., 2009; Ochoa-Espinosa et al., 2009; Löhr et al., 2010; Porcher and Dostatni, 2010; Chen et al., 2012; Jaeger et al., 2012).
DISCUSSION
The current work builds on our previous investigation of mechanisms for Bcd gradient scaling within a species (Cheung et al., 2011). Both studies take advantage of Drosophila melanogaster inbred lines that had been selected for egg size extremes (Miles et al., 2011). Although stochastic variations in egg size within an established laboratory line tend to be small (Gregor et al., 2007a; Lott et al., 2007; He et al., 2008), the egg size differences between the selected lines are greatly enhanced. This enhances our ability to identify specific ways by which within-species Bcd gradient scaling can be achieved (Cheung et al., 2011). The embryos from the pair of inbred lines under current investigation exhibit Bcd gradient properties uncharacteristic of their size. In particular, the large embryos have a lower B0 than the small embryos (Fig. 1E-J). However, the Bcd gradient profiles in broad regions of these embryos converge when expressed as a function of relative AP position x/L (Fig. 1I,J). Such convergence is indicative of scaling as it reduces the differences (between the large and small embryos) in both Bcd concentrations and Bcd-encoded positional information (Fig. 2A-D). Unlike the previously documented within-species scaling, where B0 is scaled with embryo volume (He et al., 2008; Cheung et al., 2011), the Bcd gradient scaling reported here is achieved by extending λ in the large embryos such that its relative length scale is similar to that of the small embryos (Fig. 3). This article thus provides a first documentation of a case of within-species Bcd gradient scaling through a mechanism previously suggested to be deployed only by embryos from different species (Gregor et al., 2005).
Our results suggest that the large-egg line described in this report, not the small-egg line, has acquired special Bcd gradient properties to counterbalance the more extreme length of the embryos. In Drosophila melanogaster, the Bcd gradient profiles have a generally characteristic λ of ∼100 μm, a value that is constrained by the species-dependent properties relevant to Bcd gradient formation; namely, the effective diffusion constant D and effective degradation rate ω of Bcd in the early embryo. As expected, the large embryos that we study here have more bcd mRNA than the small counterparts (Fig. 4C; supplementary material Fig. S2A), suggesting a normal maternal deposition of bcd mRNA during oogenesis with regard to its total amount (Cheung et al., 2011). But these embryos have a broader distribution of bcd mRNA (Fig. 4D; supplementary material Fig. S2C), a property not inherent to embryos that are large (Fig. 4E,F; supplementary material Fig. S2D and Table S2). Under the theoretical framework of Dalessi et al. (Dalessi et al., 2012), a broader distribution of bcd mRNA, in conjunction with a larger D and ω, can readily explain the observed Bcd gradient properties in the large embryos (Fig. 5). The precise mechanism of Bcd gradient formation remains a controversial topic. One model suggests that a bcd mRNA gradient dictates the formation and shape of the protein gradient (Lipshitz, 2009; Spirov et al., 2009), but more recent studies argue strongly against this model and document a role of both Bcd protein diffusion and decay in gradient formation (Deng et al., 2010; Little et al., 2011; Liu and Ma, 2011). It has been suggested that bcd mRNA is subject to translational inhibition by Nanos (Wharton and Struhl, 1991; Gavis and Lehmann, 1992; Simpson-Brose et al., 1994; Johnstone and Lasko, 2001; Gamberi et al., 2002). Such an action of Nanos would be accentuated in the large embryos owing to the more extended distribution of bcd mRNA. It remains to be seen how the introduction of a role of Nanos will influence future experimental and theoretical investigations of Bcd gradient formation from an extended source. For example, it will be interesting to determine, with a consideration of a role of Nanos, whether the large-egg line indeed has abnormal Bcd diffusivity and stability as predicted by the Dalessi et al. model (Dalessi et al., 2012) (Fig. 5, legend) or whether the broad distribution of bcd mRNA represents the only underlying ‘defect’.
An important finding of this study is the robustness of hb expression pattern, which has a scaled boundary position in the large and small embryos (Fig. 6A,B). This is remarkable considering the significant size difference between these embryos and the considerable shape difference between their Bcd gradient profiles. Gap genes such as hb are the earliest zygotic genes to respond to the maternal Bcd gradient input and they sit at the top of the regulatory hierarchy in the AP patterning network. The hb expression boundary is located near mid embryo. Thus, the positioning of the hb boundary can influence the further ‘territorial divisions’ that take place on both sides of this boundary, permitting this boundary to exert a more extended impact on AP patterning (Howard and ten Wolde, 2005; de Lachapelle and Bergmann, 2010; Liu et al., 2011). The hb boundary is a montage (Liu and Ma, 2013a) of two overlapping boundaries that have distinct temporal dynamics and respond to distinct regulatory inputs. But our recent studies show that perturbations of Bcd gradient properties can directly lead to corresponding changes in hb expression (He et al., 2008; He et al., 2010a; He et al., 2010b; He et al., 2011; Liu et al., 2011; Liu and Ma, 2011; He et al., 2012), suggesting that the positioning of the hb boundary is determined primarily by the Bcd gradient input. Bcd-dependent hb transcription is a highly dynamic process and it becomes turned off quickly (in the order of just a few minutes) after the embryo enters the interphase of nuclear cycle 14 (Liu and Ma, 2013a; Liu and Ma, 2013b). It has been proposed that this dynamic property represents a mechanism that enables the embryo to benefit directly and faithfully from the scaling properties of the Bcd gradient through the formation of a scaled hb boundary near mid embryo (Liu and Ma, 2013a). Our analysis of the eve expression pattern (Fig. 6C-E) shows that the robust hb expression pattern in the embryos under current investigation (Fig. 6A,B) is further propagated downstream of the AP patterning network.
The role of hb in AP patterning is well documented (Hülskamp et al., 1990; Struhl et al., 1992; Yu and Small, 2008). It has been suggested that the only role of Bcd relevant to thoracic formation is to promote an hb expression stripe near mid embryo (Wimmer et al., 2000). The aforementioned special relationship between Bcd and hb also makes hb particularly relevant to scaling because it can funnel the maternally derived information about embryo size into a zygotically operating patterning network. The hypothesis of the existence of ‘relevant’ target genes capable of faithfully and directly interpreting the scaling properties of maternally derived morphogen gradients places a special emphasis on the expression patterns of such genes. Meanwhile, it lessens an absolute requirement of the exact shape of a morphogen gradient unless, as documented (He et al., 2010a; Liu and Ma, 2011), an altered gradient shape can affect the expression properties of a relevant target gene(s). This hypothesis represents a new conceptual framework that goes beyond the textbook version of how the Bcd morphogen gradient controls AP patterning because it also underscores the roles of other inputs and cross-regulatory mechanisms (La Rosée et al., 1997; Schaeffer et al., 2000; Manu et al., 2009; Ochoa-Espinosa et al., 2009; Löhr et al., 2010; Porcher and Dostatni, 2010; Chen et al., 2012; Jaeger et al., 2012). According to this new framework, the primary role of a maternal morphogen gradient would be to ‘inform’ the embryo of its own size through concentration-dependent activation of one (or few) ‘relevant’ target gene(s), as opposed to directly specifying the boundary positions of all (or many) patterning genes as theorized in the classical framework (Wolpert, 1969; Roth and Lynch, 2012). Our ‘relevant-targets’ hypothesis is supported by recent findings of the Drosophila dorsal-ventral patterning system, in which the scaling properties of the Dorsal morphogen gradient are faithfully interpreted only by its target genes twist and snail (Chahda et al., 2013; Garcia et al., 2013). It remains to be seen how generally this ‘relevant-targets’ operational strategy is deployed by developmental systems to achieve the formation of robust and scaled patterns.
Our findings reported here and in a previous study (Cheung et al., 2011) together offer a valuable glimpse into the existence of naturally occurring operational paths towards a developmental goal within a given species. They directly advance our knowledge about how, at a molecular level, developmental canalization can be achieved (Waddington, 1942). The inbred lines used in our studies were derived from random crosses of flies collected from the wild and force-selected for egg size extremes, followed by extensive sister-brother crosses (Miles et al., 2011). The artificial selection and inbreeding process can thus be viewed as a ‘reshuffling’ of the naturally occurring genetic variation to yield a specific trait under selection, in this case, egg size extremes. The fact that the large-egg line has acquired special Bcd gradient properties that had not been directly selected for indicates that these properties play important roles to counterbalance the selected egg size extreme. Our DNA sequencing data (supplementary material Table S4) show that the large-egg line contains variations in genes known to have a role in the anterior localization of bcd mRNA (although their functional relevance remains to be determined). These sequencing results demonstrate that the Drosophila melanogaster populations in the wild must have a rich genetic diversity that underpins the potential of achieving a robust and scaled AP patterning outcome via distinct operational paths.
MATERIALS AND METHODS
Fly strains
Wild-caught Drosophila melanogaster females were used in an artificial selection procedure based on the size of the eggs laid (Miles et al., 2011). Individual inbred lines from selected population cages were established through subsequent brother-sister crosses (Cheung et al., 2011; Miles et al., 2011). The current report focuses specifically on embryos from two inbred lines: Line 2.49.3 and Line 9.31.2, which have large and small eggs, respectively. In a previous study (Cheung et al., 2011), embryos from another pair of inbred lines were used, Line 2.46.4 and Line 9.17.1, which laid large and small eggs, respectively. All embryos used in the current study were collected on grape juice-agar plates at 25°C under standard conditions. The embryos used in bcd mRNA FISH experiments were from “0 to 1-hour” collections and those in Bcd immunostaining and other FISH experiments were from “0 to 4-hour” collections. The collected embryos were dechorinated, fixed, devitellenized and stored using established protocols as previously described (Cheung et al., 2011).
Antibody staining and fluorescence in situ hybridization
The fluorescence immunostaining to detect Bcd in whole-mount embryos was performed as described previously (He et al., 2008; Cheung et al., 2011). Primary and secondary antibodies were rabbit polyclonal anti-Bcd (1:400, Santa Cruz Biotechnology) and goat anti-rabbit AlexaFluor 594 (1:400, Invitrogen). Procedures for FISH analysis detecting bcd or hb mRNA were according to Cheung et al. (Cheung et al., 2011) and Liu and Ma (Liu and Ma, 2011), respectively. For FISH, mouse anti-digoxigenin (Roche) and goat anti-mouse AlexaFluor 555 (Invitrogen) were used as primary and secondary antibodies, respectively. All imaging in these analyses (except for eve FISH) was performed on Zeiss Imager Z1 ApoTome microscope in conjunction with a Zeiss Plan 10× Aprochromat objective and the corresponding software, Axiovision 4.5. Imaging for all embryos in these analyses was focused on the midsagittal plane and no adjustments were made during the experiment. Wherever data were used to make direct comparisons in these analyses, it was ensured that both the experimental and imaging steps were performed side-by-side for the relevant samples.
Measurements of eve expression boundaries in whole-mount embryos were performed as described previously (Miles et al., 2011). Briefly, embryos were collected and fixed as before (Kosman et al., 1998) with the exception of replacing Proteinase K with SDS (Lott et al., 2007; Miles et al., 2011). Embryos were stained by both a fluorescently labeled probe for detecting eve mRNA and Sytox Green (Invitrogen) for nuclear counterstain, mounted in 90% glycerol + n-Propyl galate, and imaged on the Zeiss AxioPlan 2 microscope (Carl Zeiss) with a Vti Infinity 3 confocal (Visitech International). Captured images were then processed and eve expression boundary positions (defined as the trough-to-peak midpoint positions) recorded for individual embryos as previously described (Manu et al., 2013).
Data analysis
Embryos at early nuclear cycle 14 were used to quantify the Bcd gradient profiles. In order to properly measure and subtract background fluorescence signals, embryos from bcdE1 females were mixed with the experimental embryos (He et al., 2008; Cheung et al., 2011). Embryos used for quantitative data analysis were chosen according to a series of criteria as previously described (He et al., 2008; Cheung et al., 2011). The raw fluorescence images were oriented such that the dorsal side faced upwards and the anterior faced to the left. The intensity data were captured through the use of a digital sliding circle, within the nuclear layer, that recorded the level of fluorescence and position along the AP axis of the embryo (He et al., 2008). A similar approach was used to capture the fluorescence intensities used to establish hb mRNA FISH profiles. In this case, a sliding circle along the dorsal side of the embryo within the cytoplasmic layer was used (Liu and Ma, 2011; Liu and Ma, 2013a). Each hb FISH intensity profile was normalized to the average peak values found in the anterior (x/L<0.4) of an embryo. The background level of an embryo was defined as the mean of the lowest intensity values.
To measure the amount of bcd mRNA in embryos from Lines 2.49.3 and 9.31.2, fluorescence images of embryos were captured and selected for analysis as previously described (Cheung et al., 2011). For each embryo, a mask was generated using the DAPI counterstain, the non-specific background signal of which formed an easily discernible outline; only the pixel intensity information bounded by this mask was analyzed in subsequent steps. The contained pixels were segregated into two populations according to Otsu’s method and a contour outline was generated from the resulting threshold (Otsu, 1979; Cheung et al., 2011). To obtain the background intensity within the signal area of an embryo, the signal-specific contour was transposed to the posterior of the embryo (Fig. 4A,B). The reported signal intensity, corresponding to the total amount of bcd mRNA (a.u.), was defined as the aggregate intensity value, within the anterior signal area, subtracted by that of the posterior; subtraction was performed on individual embryos based on the background specific to an embryo. The signal area was defined as the total area contained within the specific signal contour and was used to quantify the distribution of bcd mRNA. All calculations were performed using Matlab (R2011b, MathWorks) or Excel (Microsoft).
DNA sequencing
Genomic DNA purified from adult flies of Line 2.49.3 was amplified by PCR. The PCR products were then sequenced using the ABI PRISM 3730xl DNA Analyzer, a capillary electrophoresis system, at the Genetic Variation and Gene Discovery Core Facility of Cincinnati Children’s Hospital Medical Center. The compiled DNA sequencing data were aligned against the public database (FlyBase) and all deviations detected were confirmed by visual inspections of the sequencing chromatograph profiles at the relevant locations. Supplementary material Table S3 lists primers used in PCR amplifications and DNA sequencing and supplementary material Table S4 lists the sequencing results.
Supplementary Material
Acknowledgments
We thank members of our groups for their insightful discussions, Junbo Liu for technical assistance, and Feng He for analytical assistance. We thank Sascha Dalessi and Sven Bergmann for personal communication and for providing the computer code to generate the data shown in Fig. 5. We thank Sven Bergmann and anonymous reviewers for their constructive suggestions that improved the presentation of this work.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
M.K. and J.M. initiated the study; D.C. and J.M. developed the concepts and approaches; D.C. and C.M. performed the experiments and data analyses; C.M. and M.K. provided the inbred lines; D.C. and J.M. interpreted the data; D.C. generated all figures and contributed to writing; J.M. supervised the work and wrote the paper.
Funding
This work was supported in part by grants from the National Institutes of Health (NIH) [R01GM101373 to J.M.; R01GM078381 to M.K.]; and the National Science Foundation (NSF) [IOS-0843424 to J.M.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.098640/-/DC1
References
- Baena-Lopez L. A., Nojima H., Vincent J. P. (2012). Integration of morphogen signalling within the growth regulatory network. Curr. Opin. Cell Biol. 24, 166–172 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi D., Shilo B. Z., Barkai N. (2011). Scaling of morphogen gradients. Curr.Opin. Genet. Dev. 21, 704–710 [DOI] [PubMed] [Google Scholar]
- Bergmann S., Sandler O., Sberro H., Shnider S., Schejter E., Shilo B. Z., Barkai N. (2007). Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS Biol. 5, e46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M., Nüsslein-Volhard C. (1988). The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7, 1749–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahda J. S., Sousa-Neves R., Mizutani C. M. (2013). Variation in the dorsal gradient distribution is a source for modified scaling of germ layers in Drosophila. Curr. Biol. 23, 710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xu Z., Mei C., Yu D., Small S. (2012). A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149, 618–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung D., Miles C., Kreitman M., Ma J. (2011). Scaling of the Bicoid morphogen gradient by a volume-dependent production rate. Development 138, 2741–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore M. A., Mann R. S. (2008). The control of size in animals: insights from selector genes. Bioessays 30, 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalessi S., Neves A., Bergmann S. (2012). Modeling morphogen gradient formation from arbitrary realistically shaped sources. J. Theor. Biol. 294, 130–138 [DOI] [PubMed] [Google Scholar]
- Day S. J., Lawrence P. A. (2000). Measuring dimensions: the regulation of size and shape. Development 127, 2977–2987 [DOI] [PubMed] [Google Scholar]
- de Lachapelle A. M., Bergmann S. (2010). Precision and scaling in morphogen gradient read-out. Mol. Syst. Biol. 6, 351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Wang W., Lu L. J., Ma J. (2010). A two-dimensional simulation model of the bicoid gradient in Drosophila. PLoS ONE 5, e10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. (1988). A gradient of bicoid protein in Drosophila embryos. Cell 54, 83–93 [DOI] [PubMed] [Google Scholar]
- Drocco J. A., Grimm O., Tank D. W., Wieschaus E. (2011). Measurement and perturbation of morphogen lifetime: effects on gradient shape. Biophys. J. 101, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., St Johnston D. (2004). Seeing is believing: the bicoid morphogen gradient matures. Cell 116, 143–152 [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Elphick L., Nüsslein-Volhard C., St Johnston D. (1994). Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221–1232 [DOI] [PubMed] [Google Scholar]
- Flatt T. (2005). The evolutionary genetics of canalization. Q. Rev. Biol. 80, 287–316 [DOI] [PubMed] [Google Scholar]
- Gamberi C., Peterson D. S., He L., Gottlieb E. (2002). An anterior function for the Drosophila posterior determinant Pumilio. Development 129, 2699–2710 [DOI] [PubMed] [Google Scholar]
- Garcia M., Nahmad M., Reeves G. T., Stathopoulos A. (2013). Size-dependent regulation of dorsal-ventral patterning in the early Drosophila embryo. Dev. Biol. 381, 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis E. R., Lehmann R. (1992). Localization of nanos RNA controls embryonic polarity. Cell 71, 301–313 [DOI] [PubMed] [Google Scholar]
- Gregor T., Bialek W., de Ruyter van Steveninck R. R., Tank D. W., Wieschaus E. F. (2005). Diffusion and scaling during early embryonic pattern formation. Proc. Natl. Acad. Sci. USA 102, 18403–18407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T., Tank D. W., Wieschaus E. F., Bialek W. (2007a). Probing the limits to positional information. Cell 130, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T., Wieschaus E. F., McGregor A. P., Bialek W., Tank D. W. (2007b). Stability and nuclear dynamics of the bicoid morphogen gradient. Cell 130, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T., McGregor A. P., Wieschaus E. F. (2008). Shape and function of the Bicoid morphogen gradient in dipteran species with different sized embryos. Dev. Biol. 316, 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm O., Coppey M., Wieschaus E. (2010). Modelling the Bicoid gradient. Development 137, 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Wen Y., Deng J., Lin X., Lu L. J., Jiao R., Ma J. (2008). Probing intrinsic properties of a robust morphogen gradient in Drosophila. Dev. Cell 15, 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Saunders T. E., Wen Y., Cheung D., Jiao R., ten Wolde P. R., Howard M., Ma J. (2010a). Shaping a morphogen gradient for positional precision. Biophys. J. 99, 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Wen Y., Cheung D., Deng J., Lu L. J., Jiao R., Ma J. (2010b). Distance measurements via the morphogen gradient of Bicoid in Drosophila embryos. BMC Dev. Biol. 10, 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Ren J., Wang W., Ma J. (2011). A multiscale investigation of bicoid-dependent transcriptional events in Drosophila embryos. PLoS ONE 6, e19122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Ren J., Wang W., Ma J. (2012). Evaluating the Drosophila Bicoid morphogen gradient system through dissecting the noise in transcriptional bursts. Bioinformatics 28, 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegdé J., Stephenson E. C. (1993). Distribution of swallow protein in egg chambers and embryos of Drosophila melanogaster. Development 119, 457–470 [DOI] [PubMed] [Google Scholar]
- Hendrikse J. L., Parsons T. E., Hallgrímsson B. (2007). Evolvability as the proper focus of evolutionary developmental biology. Evol. Dev. 9, 393–401 [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B., Wieschaus E., Leibler S. (2002). Establishment of developmental precision and proportions in the early Drosophila embryo. Nature 415, 798–802 [DOI] [PubMed] [Google Scholar]
- Howard M., ten Wolde P. R. (2005). Finding the center reliably: robust patterns of developmental gene expression. Phys. Rev. Lett. 95, 208103 [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Pfeifle C., Tautz D. (1990). A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Krüppel and knirps in the early Drosophila embryo. Nature 346, 577–580 [DOI] [PubMed] [Google Scholar]
- Irion U., St Johnston D. (2007). bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 445, 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J., Surkova S., Blagov M., Janssens H., Kosman D., Kozlov K. N., Manu, Myasnikova E., Vanario-Alonso C. E., Samsonova M., et al. (2004). Dynamic control of positional information in the early Drosophila embryo. Nature 430, 368–371 [DOI] [PubMed] [Google Scholar]
- Jaeger J., Manu, Reinitz J. (2012). Drosophila blastoderm patterning. Curr. Opin. Genet. Dev. 22, 533–541 [DOI] [PubMed] [Google Scholar]
- Johnstone O., Lasko P. (2001). Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35, 365–406 [DOI] [PubMed] [Google Scholar]
- Kosman D., Small S., Reinitz J. (1998). Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 208, 290–294 [DOI] [PubMed] [Google Scholar]
- La Rosée A., Häder T., Taubert H., Rivera-Pomar R., Jäckle H. (1997). Mechanism and Bicoid-dependent control of hairy stripe 7 expression in the posterior region of the Drosophila embryo. EMBO J. 16, 4403–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander A. D. (2007). Morpheus unbound: reimagining the morphogen gradient. Cell 128, 245–256 [DOI] [PubMed] [Google Scholar]
- Lander A. D. (2011). Pattern, growth, and control. Cell 144, 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Davidson E. H. (2005). Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102, 4936–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz H. D. (2009). Follow the mRNA: a new model for Bicoid gradient formation. Nat. Rev. Mol. Cell Biol. 10, 509–512 [DOI] [PubMed] [Google Scholar]
- Little S. C., Tkačik G., Kneeland T. B., Wieschaus E. F., Gregor T. (2011). The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 9, e1000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ma J. (2011). Fates-shifted is an F-box protein that targets Bicoid for degradation and regulates developmental fate determination in Drosophila embryos. Nat. Cell Biol. 13, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ma J. (2013a). Uncovering a dynamic feature of the transcriptional regulatory network for anterior-posterior patterning in the Drosophila embryo. PLoS ONE 8, e62641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ma J. (2013b). Dampened regulates the activating potency of Bicoid and the embryonic patterning outcome in Drosophila. Nat. Commun. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., He F., Ma J. (2011). Morphogen gradient formation and action: insights from studying Bicoid protein degradation. Fly (Austin) 5, 242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löhr U., Chung H. R., Beller M., Jäckle H. (2010). Bicoid—morphogen function revisited. Fly (Austin) 4, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott S. E., Kreitman M., Palsson A., Alekseeva E., Ludwig M. Z. (2007). Canalization of segmentation and its evolution in Drosophila. Proc. Natl. Acad. Sci. USA 104, 10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. (1988). cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature 336, 595–598 [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Luk S. K., Kilpatrick M. (1991). Protein encoded by the exuperantia gene is concentrated at sites of bicoid mRNA accumulation in Drosophila nurse cells but not in oocytes or embryos. Genes Dev. 5 12B, 2455–2466 [DOI] [PubMed] [Google Scholar]
- Manu, Surkova S., Spirov A. V., Gursky V. V., Janssens H., Kim A. R., Radulescu O., Vanario-Alonso C. E., Sharp D. H., Samsonova M., et al. (2009). Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 7, e1000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Ludwig M. Z., Kreitman M. (2013). Sex-specific pattern formation during early Drosophila development. Genetics 194, 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcey D., Watkins W. S., Hazelrigg T. (1991). The temporal and spatial distribution pattern of maternal exuperantia protein: evidence for a role in establishment but not maintenance of bicoid mRNA localization. EMBO J. 10, 4259–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A., Hayward P. (2006). Filtering transcriptional noise during development: concepts and mechanisms. Nat. Rev. Genet. 7, 34–44 [DOI] [PubMed] [Google Scholar]
- Miles C. M., Lott S. E., Hendriks C. L., Ludwig M. Z., Manu, Williams C. L., Kreitman M. (2011). Artificial selection on egg size perturbs early pattern formation in Drosophila melanogaster. Evolution 65, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Frohnhöfer H. G., Lehmann R. (1987). Determination of anteroposterior polarity in Drosophila. Science 238, 1675–1681 [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A., Yu D., Tsirigos A., Struffi P., Small S. (2009). Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc. Natl. Acad. Sci. USA 106, 3823–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. (1979). A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 [Google Scholar]
- Patel N. H., Lall S. (2002). Precision patterning. Nature 415, 748–749 [DOI] [PubMed] [Google Scholar]
- Porcher A., Dostatni N. (2010). The bicoid morphogen system. Curr. Biol. 20, R249–R254 [DOI] [PubMed] [Google Scholar]
- Porcher A., Abu-Arish A., Huart S., Roelens B., Fradin C., Dostatni N. (2010). The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development 137, 2795–2804 [DOI] [PubMed] [Google Scholar]
- Rice S. A. (1985). Diffusion-Limited Reactions, Amsterdam: Elsevier Science Publishers B.V. [Google Scholar]
- Roth S., Lynch J. (2012). Does the Bicoid gradient matter? Cell 149, 511–512 [DOI] [PubMed] [Google Scholar]
- Schaeffer V., Killian D., Desplan C., Wimmer E. A. (2000). High bicoid levels render the terminal system dispensable for Drosophila head development. Development 127, 3993–3999 [DOI] [PubMed] [Google Scholar]
- Simpson-Brose M., Treisman J., Desplan C. (1994). Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell 78, 855–865 [DOI] [PubMed] [Google Scholar]
- Spirov A., Fahmy K., Schneider M., Frei E., Noll M., Baumgartner S. (2009). Formation of the bicoid morphogen gradient: an mRNA gradient dictates the protein gradient. Development 136, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Struhl K., Macdonald P. M. (1989). The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57, 1259–1273 [DOI] [PubMed] [Google Scholar]
- Struhl G., Johnston P., Lawrence P. A. (1992). Control of Drosophila body pattern by the hunchback morphogen gradient. Cell 69, 237–249 [DOI] [PubMed] [Google Scholar]
- Su T. T., O’Farrell P. H. (1998). Size control: cell proliferation does not equal growth. Curr. Biol. 8, R687–R689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. H. (1942). Canalization of development and the inheritance of aquired characters. Nature 150, 563–565 [Google Scholar]
- Wartlick O., Kicheva A., González-Gaitán M. (2009). Morphogen gradient formation. Cold Spring Harb. Perspect. Biol. 1, a001255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartlick O., Mumcu P., Jülicher F., Gonzalez-Gaitan M. (2011). Understanding morphogenetic growth control - lessons from flies. Nat. Rev. Mol. Cell Biol. 12, 594–604 [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Struhl G. (1991). RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67, 955–967 [DOI] [PubMed] [Google Scholar]
- Wimmer E. A., Carleton A., Harjes P., Turner T., Desplan C. (2000). Bicoid-independent formation of thoracic segments in Drosophila. Science 287, 2476–2479 [DOI] [PubMed] [Google Scholar]
- Wolpert L. (1969). Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 [DOI] [PubMed] [Google Scholar]
- Yang X., Xu T. (2011). Molecular mechanism of size control in development and human diseases. Cell Res. 21, 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Small S. (2008). Precise registration of gene expression boundaries by a repressive morphogen in Drosophila. Curr. Biol. 18, 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.