Abstract
The antitumor antibiotic mitomycin C is shown to form a covalent complex with calf thymus DNA under anaerobic conditions in the presence of either NADPH cytochrome c reductase/NADPH, xanthine oxidase/NADH, or the chemical reducing system H2/PtO2. Digestion of the complex with DNase I/snake venom diesterase/alkaline phosphatase yields a single mitomycin deoxyguanosine adduct as the major DNA alkylation product, identified as N2-(2'' beta,7''-diaminomitosen-1'' alpha-yl) 2'-deoxyguanosine (Structure 2). Two minor adducts, 2-5% each of the total adduct pool, are isolated and identified as the 1'' beta stereoisomer of 2 (Structure 3), and 10''-decarbamoyl-2 (Structure 7). The same results were obtained with M13 DNA and poly(dG-dC).poly(dG-dC); however, in the latter case, a minor adduct apparently possessing two deoxyguanosine and one mitomycin unit is isolated. Digestion of the covalent mitomycin-calf thymus DNA complex with nuclease P1 yields four dinucleotide adducts, all of which consist of 2 linked at its 3' end to each of the four possible 5' nucleotides (A, T, G, and C). Upon treatment of each dinucleotide adduct with snake venom diesterase/alkaline phosphatase, 2 is released along with the corresponding free nucleoside. In apparent conflict with the present results, previous reports from another laboratory have indicated that modification of calf thymus DNA by mitomycin C under conditions identical to those described here result in the isolation of three mitomycin C mononucleotide adducts possessing linkages of the drug to N2 and O6 of guanine and N6 of adenine. Evidence is shown suggesting that the latter adducts are actually three of the above four dinucleotide derivatives of 2 obtained independently by us and, thus, all of them in fact possess an identical N2-mitosenylguanine adduct moiety. Model-building studies indicate an excellent fit of the guanine N2-linked drug molecule inside the minor groove of B-DNA with no appreciable distortion of the DNA structure.
Full text
PDF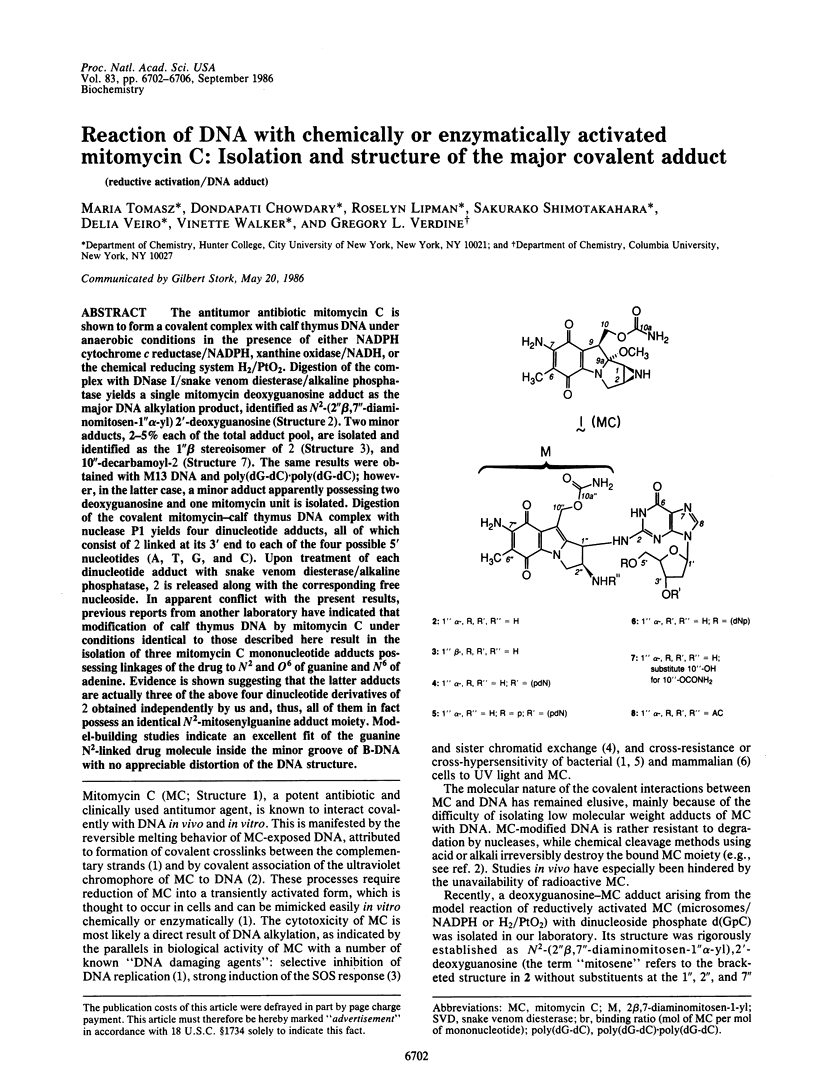


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BOYCE R. P., HOWARD-FLANDERS P. GENETIC CONTROL OF DNA BREAKDOWN AND REPAIR IN E. COLI K-12 TREATED WITH MITOMYCIN C OR ULTRAVIOLET LIGHT. Z Vererbungsl. 1964 Dec 30;95:345–350. doi: 10.1007/BF01268667. [DOI] [PubMed] [Google Scholar]
- Carrano A. V., Thompson L. H., Stetka D. G., Minkler J. L., Mazrimas J. A., Fong S. DNA crosslinking, sister-chromatid exchange and specific-locus mutations. Mutat Res. 1979 Nov;63(1):175–188. doi: 10.1016/0027-5107(79)90114-3. [DOI] [PubMed] [Google Scholar]
- Cohen R. J., Crothers D. M. Preparation and characterization of monodisperse, cross-linked low molecular weight deoxyribonucleic acid. Biochemistry. 1970 Jun 9;9(12):2533–2539. doi: 10.1021/bi00814a021. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Shudo K., Okamoto T. Modification of deoxyribonucleic acid with reductively activated mitomycin C. Structures of modified nucleotides. Chem Pharm Bull (Tokyo) 1983 Mar;31(3):861–869. doi: 10.1248/cpb.31.861. [DOI] [PubMed] [Google Scholar]
- Kaplan D. J., Tomasz M. Altered physiochemical properties of the deoxyribonucleic acid-mitomycin C complex. Evidence for the conformational change in deoxyribonucleic acid. Biochemistry. 1982 Jun 8;21(12):3006–3013. doi: 10.1021/bi00541a031. [DOI] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980 May;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes S. R., Fracasso P. M., Heimbrook D. C., Rockwell S., Sligar S. G., Sartorelli A. C. Role of NADPH:cytochrome c reductase and DT-diaphorase in the biotransformation of mitomycin C1. Cancer Res. 1984 Dec;44(12 Pt 1):5638–5643. [PubMed] [Google Scholar]
- Kinoshita S., Uzu K., Nakano K., Takahashi T. Mitomycin derivatives. 2. Derivatives of decarbamoylmitosane and decarbamoylmitosene. J Med Chem. 1971 Feb;14(2):109–112. doi: 10.1021/jm00284a006. [DOI] [PubMed] [Google Scholar]
- Lipman R., Weaver J., Tomasz M. Electrostatic complexes of mitomycin C with nucleic acids and polyanions. Biochim Biophys Acta. 1978 Dec 21;521(2):779–791. doi: 10.1016/0005-2787(78)90317-9. [DOI] [PubMed] [Google Scholar]
- Mercado C. M., Tomasz M. Circular dichroism of mitomycin-DNA complexes. Evidence for a conformational change in DNA. Biochemistry. 1977 May 3;16(9):2040–2046. doi: 10.1021/bi00628a044. [DOI] [PubMed] [Google Scholar]
- Pan S. S., Andrews P. A., Glover C. J., Bachur N. R. Reductive activation of mitomycin C and mitomycin C metabolites catalyzed by NADPH-cytochrome P-450 reductase and xanthine oxidase. J Biol Chem. 1984 Jan 25;259(2):959–966. [PubMed] [Google Scholar]
- Petrusek R. L., Anderson G. L., Garner T. F., Fannin Q. L., Kaplan D. J., Zimmer S. G., Hurley L. H. Pyrrol[1,4]benzodiazepine antibiotics. Proposed structures and characteristics of the in vitro deoxyribonucleic acid adducts of anthramycin, tomaymycin, sibiromycin, and neothramycins A and B. Biochemistry. 1981 Mar 3;20(5):1111–1119. doi: 10.1021/bi00508a011. [DOI] [PubMed] [Google Scholar]
- Pullman B. Electrostatics of Polymorphic DNA. J Biomol Struct Dyn. 1983 Dec;1(3):773–794. doi: 10.1080/07391102.1983.10507481. [DOI] [PubMed] [Google Scholar]
- Thompson L. H., Rubin J. S., Cleaver J. E., Whitmore G. F., Brookman K. A screening method for isolating DNA repair-deficient mutants of CHO cells. Somatic Cell Genet. 1980 May;6(3):391–405. doi: 10.1007/BF01542791. [DOI] [PubMed] [Google Scholar]
- Tomasz M., Lipman R. Reductive metabolism and alkylating activity of mitomycin C induced by rat liver microsomes. Biochemistry. 1981 Aug 18;20(17):5056–5061. doi: 10.1021/bi00520a036. [DOI] [PubMed] [Google Scholar]
- Tomasz M., Lipman R., Verdine G. L., Nakanishi K. Reassignment of the guanine-binding mode of reduced mitomycin C. Biochemistry. 1986 Jul 29;25(15):4337–4344. doi: 10.1021/bi00363a024. [DOI] [PubMed] [Google Scholar]
- Tomasz M., Mercado C. M., Olson J., Chatterjie N. The mode of interaction of mitomycin C with deoxyribonucleic acid and other polynucleotides in vitro. Biochemistry. 1974 Nov 19;13(24):4878–4887. doi: 10.1021/bi00721a002. [DOI] [PubMed] [Google Scholar]
- Weaver J., Tomasz M. Reactivity of mitomycin C with synthetic polyribonucleotides containing guanine or guanine analogs. Biochim Biophys Acta. 1982 May 31;697(2):252–254. doi: 10.1016/0167-4781(82)90084-7. [DOI] [PubMed] [Google Scholar]