Abstract
Named entity recognition is a crucial component of biomedical natural language processing, enabling information extraction and ultimately reasoning over and knowledge discovery from text. Much progress has been made in the design of rule-based and supervised tools, but they are often genre and task dependent. As such, adapting them to different genres of text or identifying new types of entities requires major effort in re-annotation or rule development. In this paper, we propose an unsupervised approach to extracting named entities from biomedical text. We describe a stepwise solution to tackle the challenges of entity boundary detection and entity type classification without relying on any handcrafted rules, heuristics, or annotated data. A noun phrase chunker followed by a filter based on inverse document frequency extracts candidate entities from free text. Classification of candidate entities into categories of interest is carried out by leveraging principles from distributional semantics. Experiments show that our system, especially the entity classification step, yields competitive results on two popular biomedical datasets of clinical notes and biological literature, and outperforms a baseline dictionary match approach. Detailed error analysis provides a road map for future work.
Keywords: Natural Language Processing, Named entity recognition, Distributional Semantics, UMLS, Chunking
1. Introduction
An overwhelming amount of health and biomedical text is becoming available with the recent adoption of electronic health records, the growing number of biomedical publications, and the exploding prevalence of health information online. At the same time, in the research community, significant efforts have been devoted to creating standard terminologies and knowledge bases hence facilitating extraction of information from and reasoning over raw data. The bottleneck of biomedical information processing thus has shifted from where to collect data and resources to how to make use of the knowledge resources and build scalable models to process large amounts of text. Since much of the data is recorded in narrative and unstructured form, like in clinical notes and biomedical publications, the quality of basic natural language processing (NLP) tools has a critical impact on the performance of higher-level tasks such as information retrieval, information extraction, and knowledge discovery. Biomedical named-entity recognition (BM-NER) 1, sometimes referred to as biomedical concept identification or concept mapping, is a key step in biomedical language processing: terms (either single words or multiple words) of interest are identified and mapped to a pre-defined set of semantic categories. Examples of BM-NER systems include extracting clinical information from radiology reports [1, 2, 3], identifying diseases and drug names in discharge summaries [4, 5, 6], detecting gene and protein mentions in biomedical paper abstracts [7, 8, 9].
In the general domain, named-entity recognition (NER) focuses on identifying names of persons, locations, and organizations in news articles, reports, and even tweets. Thanks to the availability of annotated corpora, supervised learning methods have been widely adopted and prevail unsupervised ones. Such state-of-the-art NER systems have achieved performance as high as human annotators [10, 11]. On their side, BM-NER are getting better with the advant of more annotated corpora to learn from. Recent supervised systems could efficiently find gene names and clinical problems from certain type of texts with above 0.8 F score [12, 6, 13, 14]. Traditional ways of tackling BM-NER range from dictionary matching, heuristic rules, to supervised Hidden Markov Models(HMMs)/Conditional Random Fields(CRFs)-based sequence labeling. The first two approaches do not require training data, but usually involve ad-hoc rules and assumptions that may limit the type of entities and texts to which they could apply. CRF-based labelers have yielded high performance in sequence learning tasks, and are the state of the art for some biological and medical entity recognition tasks. However, the supervised nature of CRF entails a fairly large amount of training data which must be annotated by humans. As a result, it is only applicable in a limited number of settings.
In this paper, we provide a stepwise unsupervised solution to biomedical named-entity recognition. Our approach does not rely on hand-built rules or examples of annotated entities, so it can be adapted to different semantic categories and text genres easily. Instead of supervision, the entity recognition leverages terminologies, shallow syntactic knowledge (noun phrase chunking), and corpus statistics (inverse document frequency and context vectors). Experimental results demonstrate that our method yields competitive results in two popular datasets of different genres, clinical notes and biomedical literature, respectively, and different corresponding entity types.
2. Background
There are two main steps of named entity recognition: detecting boundaries of entity mentions and classifying the mentions into pre-defined semantic categories. The task of entity linking or concept normalization, that is linking a term to a unique concept identifier in a terminology for instance is not typically part of NER, and as such is not the focus of this paper. With sequence labeling models like Hidden Markov Models (HMMs) and Conditional Random Fields (CRFs), the two tasks could be jointly handled taking advantage of the Markov property which models transitions between labels [15, 16]. In an unsupervised framework, however, boundary detection and entity classification are typically conducted separately [17]. In this section we review related work from two perspectives, unsupervised named entity recognition and biomedical named entity recognition, and direct the reader to existing reviews of supervised approaches for NER in the general domain [17].
2.1. Unsupervised Named Entity Recognition
The NLP community has invested a lot of efforts in unsupervised NER. Early work [18, 19] relies on heuristic rules and lexical resources such as WordNet [20]. More recently, Alfonseca and Manandhar formulate named entity classification as a word sense disambiguation task and cluster words based on the words with which they co-occur frequently in online search results [21]. The context word frequency vector, which represents the semantics of words to be classified, is called “signature.” Nadeau et al. present a system of retrieving entity lists by web page wrapper, followed by disambiguation through heuristic rules [22]. Sekine and Nobata give definitions and rule-based taggers for 200 categories of entities, as well as a standard taxonomy of general entities [23]. Shinyama and Sekine observes that named entities often appear synchronously in several news articles, whereas common nouns do not [24]. Exploiting this characteristic, they successfully obtained rare named entities with 90% accuracy just by comparing time series distributions of a word in two newspapers. This technique can be useful in combination with other NER methods.
The second category of methods is relatively new, and is essentially weakly supervised instead of unsupervised. Such methods use a bootstrapping-like technique to strengthen the models, starting from small sets of seed data or rules. The first notable work is done by Collins and Singer, in which a small set of handcrafted rules are predefined as seed rules [25]. The system iteratively labels the dataset based on current rules, and induces more rules with high precisions on found entities. Riloff and Jones introduce mutual bootstrapping that consists of growing a set of entities and a set of contexts in turn [26]. Several improvements and extensions were later proposed following this bootstrapping approach [27, 28, 29]. It is noteworthy that previous works in this category focus only on entity classification, which assume that the named entities have already been correctly extracted from the text.
It is interesting that in many ways, unsupervised named entity recognition systems are enlightened by previous works in word sense disambiguation, especially in classifying extracted entities. On the one hand, the bootstrapping framework in [25] was initially used by [30] for word sense disambiguation; on the other hand, the idea of classifying entities based on their context signatures [21] is also similar with distributional methods in word sense disambiguation [31], in which contexts of mentions are used to determine word senses.
2.2. Biomedical Named Entity Recognition
There are two major reserach directions in BM-NER: finding gene, protein, and related biological or genetic terms, as well as finding disease, drug names, and other medical terms. We use biological NER and medical NER to denote these two research sub-domains respectively. The early NER systems in both fields are typically rule-based or lexicon-based [1, 32, 33, 34, 35, 36, 7], several of which are widely applied. MedLEE is a general natural language processor for clinical texts, encoding and mapping terms to a controlled vocabulary [1]; GENIES is a system extracting molecular pathways from journal articles, which is modified from MedLEE [35]; EDGAR is a natural language processing system that extracts information about drugs and genes relevant to cancer from the biomedical literature [34]; AbGene is one of the most successful NER systems for gene and protein [7]; MetaMap, developed by National Library of Medicine(NLM), is a tool discovering UMLS Metathesaurus concepts referred to in text [36]. Many of these systems highly resort to lexical knowledge resources such as GO [37] and UMLS [38]. More recently cTAKES provides concept identification and normalization to UMLS in clinical texts [39].
Recent years have witnesses the rise of data-driven methods in biomedical named entity recognition with the availability of annotated data sests. In biological NER, the release of the GENIA corpus [40] has pushed forward related research using various supervised learning models, including Support Vector Machines (SVMs) [41, 42, 43], Hidden Markov Models (HMMs) [44], and Conditional Random Fields (CRFs) [8, 45]. The shared task of BioNLP/NLPBA 2004 used GENIA as dataset [46], and 9 teams submitted their NER systems to the event. In the first BioCreAtIvE challenge [47], gene mention identification was the first subtask of task1 [9]. Such shared tasks and workshops continued every year with new challenges, advancing the field with related information extraction tasks such as gene normalization[48] and bio-event extraction[49]. So far, state-of-the-art systems for these datasets are mostly supervised ones based on SVM [41] and CRF [8, 45].
In the medical domain, the first publicly available corpus for NER evaluation was created in the i2b2 challenge 2010 [6]. In this event, 22 supervised and semi-supervised systems were developed for entity extraction, and most of the leading systems used CRF, except for the best performed system[50]. Before the availability of i2b2 corpus, recent research also focus on evaluation on, extension to, and comparison with MetaMap and its predecessor MMTx. Meystre and Haug evaluate MMTx with a automatically retrieved clinical problem list [51]. Abacha and Zweigenbaum make modifications to MetaMap, and compare MetaMap with statistical based methods like CRF and SVMs[12, 52]. Patrick et al. implement a fuzzy matcher which better maps terms to UMLS concepts [53]. Before i2b2 2010, Wang annotates a dataset of clinical progress notes with 11 concept categories, evaluating the performance of CRF on the dataset [54]. They also present a cascading system that combines a CRF, an SVM, and a Maximum Entropy model to reclassify the identified entities in order to reduce misclassification [13]. Most recent advances in clinical entity recognition follow the trend of supervised learning, combined with ensemble system[55] and large scale feature engineering [56, 57].
3. Methods
3.1. Datasets
We evaluate our systems upon two widely accepted datasets: the i2b2 and GENIA corpora. The i2b2 corpus is a set of clinical notes with Problems, Tests, and Treatments annotated as entities, while GENIA corpus is a collection of biomedical literature consisting of biological entities such as DNA, RNA, and protein. i2b2 and GENIA are mainstream datasets for evaluating NER and were leveraged in two major BM-NER shared task events: the i2b2 challenge 20102 and the BioNLP/NLPBA 20043, respectively. Evaluations and other details of these two data sets are given in [6] and [40].
3.1.1. i2b2 Corpus
The i2b2 corpus was created for the i2b2/VA 2010 challenge [6]. The dataset includes discharge summaries from Partners Health Care, Beth Israel Deaconess Medical Center, and University of Pittsburgh Medical Center (denoted in this paper as Partners, Beth, and Pittsburgh for short). Pittsburgh notes were used as test set in i2b2 challenge and the other two sources as training set. All records in the dataset have been fully de-identified and manually annotated for concept, assertion, and relation information. In this paper, only concept annotations are used with three categories of entity annotations: Problem, Treatment and Test.
3.1.2. GENIA Corpus
The GENIA corpus4 is the primary collection of biomedical literature compiled and annotated within the scope of the GENIA project. The corpus was created to support the development and evaluation of information extraction and text mining systems for the domain of molecular biology. The corpus contains Medline abstracts, selected using a PubMed query for the three MeSH terms “human,” “blood cells,” and “transcription factors.” The corpus has been annotated with various levels of linguistic and semantic information. The original GENIA corpus contains 36 classes of entities. A more widely used version of GENIA corpus is the one simplified for the BioNLP/NLPBA shared task, in which entities are grouped into only 5 major classes: protein, DNA, RNA, cell line, cell type. We use these five categories in this paper.
3.2. Methods in a Nutshell
Our methods are partly inspired by [25] through the use of “seed knowledge,” and by [21] through classification based on “signature” similarity. Our approach differs, however, in the following ways: first, besides classifying entities, our method also identifies entities from raw text; second, it leverages existing terminology in lieu of task-specific user defined rules or online information retrieval; second, signature vector computation is refined through the use of TF-IDF weights and adding internal words (words that are inside a term, instead of being part of the context). To our best knowledge, our method is the first general and complete unsupervised NER solution for biomedical text with both entity detection and classification. Furthermore, it is the first time such system is applied to both biological and clinical entities. There are three main steps in our unsupervised NER approach: seed term collection, boundary detection, and entity classification. In the first step, for each target entity class, seed terms are extracted from the UMLS metathesaurus automatically based on mappings from the target class to either UMLS semantic groups, UMLS semantic types, or individual UMLS concepts. In the second step, boundary detection, noun phrase chunking is leveraged under the hypothesis that most entities are strongly correlated with noun phrases (NPs), followed by a filter to get rid of non-entity NPs. In the last step, all the candidate entities identified in the previous step are fed into a classifier to predict their semantic category. The entire workflow is illustrated in Figure 1. No handcrafted rules or training data is needed in our framework, and only the mapping in the first step needs to be adjusted (easily) to generalize the method to other applications.
Figure 1.
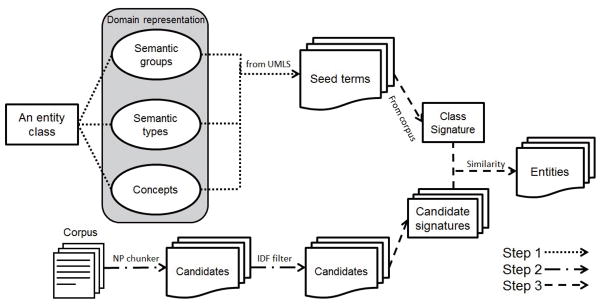
Overall approach to unsupervised biomedical named entity recognition.
3.3. Step 1: Seed Term Collection
The first step in our approach is to collect seed terms for entity classes, upon which signature vectors of the classes will be generated in the third step. The seed term sets are gathered from external terminologies, not the input corpora. In order to make the method general and portable, classes of entities are defined by users by choosing the corresponding UMLS semantic types, semantic groups [58], or specific concepts which best represent the semantic domains of the classes. We call the set of chosen semantic types, semantic groups and concepts the domain representations of classes. Semantic groups and semantic types, which form a hierarchy of categorization, are preferred since they by themselves describe sets of concepts with similar meanings; however, it is not always feasible to represent an entity class with them. In that case we would need specific UMLS concepts for the domain representations. The three entity classes Problem, Treatment, Test in the i2b2 dataset are represented by following semantic types or semantic groups:
Problem: Disorders (Semantic group)
Treatment: Therapeutic or Preventive Procedure (Semantic type) + Clinical Drug (Semantic type)
Test: Laboratory Procedure (Semantic type) + Laboratory or Test Result (Semantic type) + Diagnostic Procedure (Semantic type)
For GENIA, following domain representations are assigned to entity classes:
protein: Amino Acid, Peptide, or Protein (Semantic type)
DNA: C0012854 (UMLS Concept)
RNA: C0035668 (UMLS Concept)
cell type: C0007600 (UMLS Concept)
cell line: C0449475 (UMLS Concept)
Notice that the choices of domain representations might not be accurate (actually, for some entity types like Problem, there is no clear UMLS semantic type). However, as our method allows noises in the seed term set, it is acceptable to pick the most likely representation based on one’s expertise. Once the domain representation is determined for a class, all the UMLS concepts (and their lexical variants) which belong to the representative semantic types or groups are extracted from the UMLS metathesaurus as part of the seed term set for that target entity class. If the domain representation of a class is defined by individual UMLS concepts, then all is-a descendants of those concepts will be included into the seed term set. For example, there is no proper semantic type or semantic group that could be mapped to the entity type “cell type” in the GENIA corpus. Instead, the individual UMLS concept “C0449475: cell type” is a good choice for the representation; thus, we collect all the is-a descendants of C0449475 (including all its lexical variants), as seed terms for “cell type.” A mixed representation of semantic types/groups and UMLS concepts is also allowed for an entity class.
At the end of this step, we will have a dictionary for each target entity class, which we assume to be a set of known terms for that class.
3.4. Step 2: Boundary Detection
The second step is to detect boundaries of entities, collecting candidates for entity classification. In our solution, we hypothesize that entities should be noun phrases (NPs), and use an NP chunker to approximate the set of NPs. Although full parsing is needed to find all NPs in a sentence, chunking is more time efficient and its coverage is quite acceptable in most applications. However, it is clear that not all noun phrases in the text can be entities. In order to remove those noun phrases that are clearly not entities of interest, we employ an inverse document frequency (IDF) based technique to filter candidates generated by the NP chunker. The intuition behind this filter is that noun phrases that are most common in the texts, such as “the patient” and “date of birth,” are very unlikely to be entities. IDF is a measure of whether a term is common or rare across all documents [59]. Given a corpus D of documents (sentences in our case) d and a specific term t, IDF is defined as:
| (1) |
We calculate IDF value for every word in the dataset, and obtain the IDF value for a noun phrase by averaging the IDFs of the words it contains. Then we filter all the candidate NPs whose IDF value is lower than a predetermined threshold (set to 4 in our experiments). The reason of using such averaged IDF for a noun phrase instead of calculating the IDF value of its own directly is to handle the inherent sparsity of the copora: there are much more possible noun phrases than words in a given dataset.
3.5. Step 3: Entity Classification
The intuition of our classification approach is that entities of same class tend to have similar vocabulary and context. For example, in clinical text, the word “pain” is highly likely to be inside an entity of class “Problem” (abdominal pain, incisional pain, back pain, etc.), but not “Treatment” or “Test”; “mg”, as a unit usually used in medication orderings, is likely to be after entities of class “Treatment” (Furosemide 20 mg, Amiodarone 200 mg, etc, in which Furosemide and Amiodarone are entities of treatment). The similarity-based method is primarily used in word sense disambiguation (WSD), assuming that the meaning of a word is closely related to the distribution of words around it [31]. Such distributional semantics have also been applied to several problems in biomedical informatics [60, 61]. In our method, three improvements are made over the original signature-based approaches [21]. First, internal words of the named entity are included in the vector in addition to the context words surrounding the entity. In WSD, occurences of same word are the target for clustering, thus the internal words will always be the same for different mentions; but in entity classification, candidate to be clustered are terms that have different internal words. Second, we do not use any external resources such as web search results used in [21] to generate signature vectors, which means our system can be used independently and is favored in unsupervised BM-NER tasks that no resources could be resorted to. Instead, we leverage the test corpus itself to generate the signatures, since an unannotated test corpus could usually be available when the tool is used. Finally, we use TF-IDF, instead of raw frequency, as weight for a word in their context vectors. The motivation is that TF-IDF is a better measurement of how important a word or term is to a document than raw frequency. As such, words that are more important and decisive will have larger weights in signature vectors.
Signature generation
We use “signature” to denote the vector of internal and context words for a certain object. Such object could be a term (single word or multi-word) or an entity class. Assume the vocabulary (all possible unigrams) contains V unigrams v1, v2, …, vV. For a term t in the text, its signature st is a vector of 2V dimensions:
| (2) |
Values in the vector are calculated as follows:
| (3) |
| (4) |
In above equations, TF function f(t, d) is defined as the raw frequency of term t in d, contextt is defined as the previous two words and following two words of t, wi, wo represent the weights for internal and context words respectively, and D is the set of all sentences in the test data. Figure 2 shows an example of how to build the signature vector for the seed term “abdominal pain”. We suppose this term occur 2 times in the data sets, and all IDF values of words are already calculated.
Figure 2.
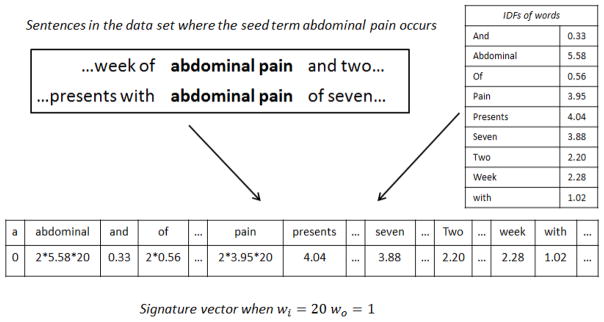
Building a signature vector for the seed term “abdominal pain” from IDF table and corpus, considering previous and following two words as well as internal words, assuming w0 = 1, wi = 20
Then we define the signature of an entity class as the average vector of the signatures of all the seed terms belonging to this category, i.e. each target entity class c, is represented by a single signature vector sc
| (5) |
in which |c| is the number of seed terms belonging to this class.
If a seed term occurs more than once in the corpus, its signature will be obtained simply by averaging signatures of all the mentions. However, if a seed term does not have a mention in the corpus, it will simply be ignored in the computation of the class signature.
Similarity calculation
Once each target entity class has a signature vector computed, and candidate named entities are generated at step 2, similarity between the candidate signature and each class signatures can be computed. The candidate is assigned the class with which it has the highest similarity, as long as the similarity is high enough as determined by a threshold set experimentally. If the candidate’s similarity to all classes is under the threshold, it is removed from the set of recognized named entities.
Similarity is computed by the cosine metric between two signature vectors. Given two vectors v and w, the similarity is defined as:
| (6) |
3.6. Experimental Setup
In our experiments, seed terms are extracted from UMLS version 2012AB. For the boundary detection step, noun phrases are identified through the OpenNLP chunker5, a fast implementation based on maximum entropy model, which is also shown to be a state-of-the-art chunker for biomedical literature [62]. The threshold for the IDF filter is experimentally set to 4 for all evaluated datasets, a reasonable guess of how rare an entity should at least be. For the entity classification step, 20 and 1 are chosen as values of wi and wo, following the intuition that internal words are more informative than context words. The threshold for signature similarity is experimentally set as 0.002, which means if a candidate has similarities with all classes lower than 0.002, it will not be regarded as an entity. This threshold could control the bias between high precision and high recall. In addition, stopwords are removed from all signatures.
All the settings remain the same for both i2b2 and GENIA in order to test the portability of our approach.
Following standard BM-NER evaluation, precision, recall, and F score (both exact and inexact) are reported to measure performance of recognizing a single class of entities. Then micro-F score is calculated to evaluate the overall performance across all entity classes. In exact evaluation, a true positive is an entity recognized with both correct boundary and correct class. In inexact evaluation, which is defined in i2b2 challenge [6], a true positive is an entity with correct class and is at least overlapped with gold standard entity. Therefore, inexact evaluation lowers the requirement for boundary detection and accepts partial matches as correct answers. For an entity class, precision, recall, and F score is defined as follows.
| (7) |
| (8) |
| (9) |
In order to evaluate the overall performance of a recognizer, Micro F is calculated as follows.
| (10) |
| (11) |
| (12) |
We compare the overall performance of our system with a baseline dictionary match system and a supervised system. The dictionary match approach MetaMap 6 is not designed specifically for the datasets we use, but it is proper to be a benchmark since it is also a portable unsupervised system. In our experiments we use the release version 2011v2 with all default settings. The output of MetaMap is processed by choosing entities that are mapped by MetaMap to UMLS concepts or semantic types which are in the domain representation of target semantic classes. Finally, in order to get a sense of how unsupervised approach performs against supervised ones, we also compare our system with the best corresponding supervised systems [6, 41] in the i2b2 2010 challenge and the BioNLP 2004 shared task, which reported performances on Pittsburgh and GENIA respectively.
4. Results
4.1. Datasets
There are 3 and 5 types of entities in i2b2 and GENIA, respectively. Numbers of documents, sentences, and entities are given in Table 1.
Table 1.
Numbers of documents, sentences, and entities in the i2b2 and GENIA corpora.
| Corpus | # Documents | # Sentences | # Entities |
|---|---|---|---|
| i2b2-Pittsburgh | 477 | 27,627 | Problem: 12,586 |
| Treatment: 9,343 | |||
| Test: 9,225 | |||
|
| |||
| i2b2-Beth | 73 | 8,798 | Problem: 4,187 |
| Treatment: 3,072 | |||
| Test: 3,036 | |||
|
| |||
| i2b2-Partners | 97 | 7,517 | Problem: 2,885 |
| Treatment: 1,768 | |||
| Test: 1,570 | |||
|
| |||
| GENIA | 2,000 | 18,546 | protein: 24,966 |
| DNA: 8,557 | |||
| RNA: 719 | |||
| cell type: 6,221 | |||
| cell line: 3,663 | |||
4.2. Step 1: Seed Term Collection
Domain representations and number of seed terms collected according to the representations for entity classes are described in Table 2. For GENIA, “RNA” and “cell type” have relatively small amount of seed terms because the UMLS concepts they map to have limited number of is-a descendants. The class “cell line” has a significantly larger but noisier set of seed terms than other classes in GENIA, which is caused by some incorrect is-a links from lower level concepts to very high level concepts in UMLS. These links bring several high level concepts into the seed term sets, which further introduce more incorrect descendants.
Table 2.
Domain representations for entity classes in the i2b2 and GENIA corpora (ST: semantic type; SG: semantic group; C: concept).
| Dataset | Class | Domain representation | # Seed terms |
|---|---|---|---|
| i2b2 | Problem | Disorders (SG) | 398,725 |
| Treatment | Therapeutic or Preventive Procedure (ST) + Clinical Drug (ST) | 153,084 | |
| Test | Laboratory Procedure (ST) + Laboratory or Test Result (ST) + Diagnostic Procedure (ST) | 66,015 | |
|
| |||
| GENIA | protein | Amino Acid, Peptide, or Protein (ST) | 35,351 |
| DNA | C0012854 (C) | 45,671 | |
| RNA | C0035668 (C) | 1,029 | |
| cell type | C0007600 (C) | 423 | |
| cell line | C0449475 (C) | 264,729 | |
4.3. Step 2: Boundary Detection
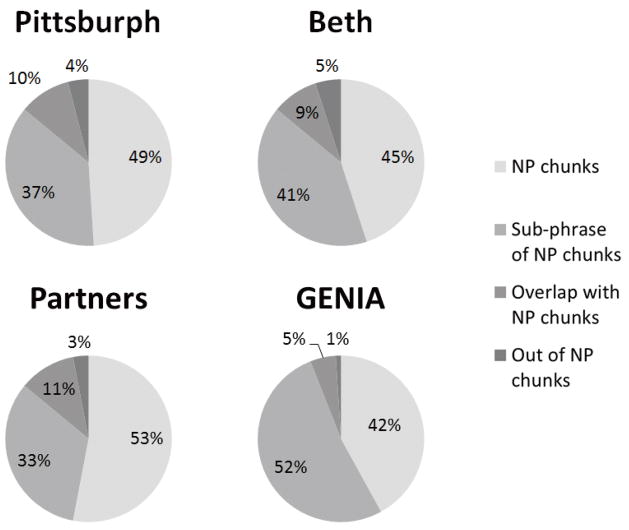
In order to verify the hypothesis that entities are NPs, we report the coverage of noun phrase chunks on entities (Figure 3). In all the three corpora of i2b2 as well as GENIA, around 45% of the entities are NP chunks, and nearly 30% of the entities are part of (but not) NP chunks. Only less than 5% of them are completely out of NP chunks without any overlapping words with them. Thus, if we use the collection of NP chunks as an approximation of entity candidate set, around half of entities will be covered. If we allow fuzzy match (i.e., we do not expect the boundaries to be exactly matched with ground truth), only a very small portion of the entities will be missing.
Figure 3.
Proportions of entities in the corpora that are noun phrases (NPs), sub-phrases of an NP, overlap with an NP, and out of any NP.
To evaluate the effectiveness of the IDF filter followed by the NP chunking, we look into the candidate sets before and after IDF filtering for Pittsburgh dataset. Before IDF filtering, the NP chunker finds 72,768 noun phrases from the text, 15,254 of which are entities in gold standard and 57,514 of which are not. The IDF filter removes 17,058 (30%) incorrect candidates successfully, at the expense of only wrongly removing 967 (6%) NPs that should be entities. This supports our hypothesis that phrases that are too common tend not to be entities, and demonstrates the effectiveness of using averaged IDF value to filter candidates.
4.4. Step 3: Entity Classification
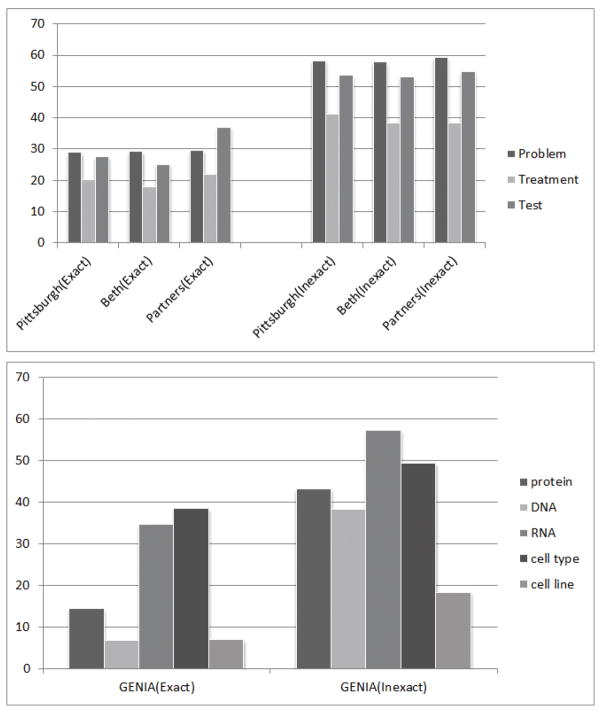
In order to evaluate the entity classification step on its own, an experiment is conducted with gold standard entity boundaries for all the entities in the corpus. In this experiment we assume all entities have already been extracted successfully from text, and our task is only to classify them into categories using signature similarity. Table 3 shows the classification results on Pittsburgh and GENIA. Similar results to Pittsburg are obtained for Beth and Partners, but are not shown for simplicity of presentation. The performance of the target class “cell line” is very low, which is a result of a very noisy seed term set. As discussed before, the UMLS metathesaurus contains a lot of incorrect relationships, which lead to an abnormally large (and probably un-representative) seed term set for the class “cell line.” Since it is a 5-class classification task, the mistakes made on “cell line” also affects the accuracy for the other classes. However, all other GENIA categories reach F scores, as well as overall accuracy, higher than 50%.
Table 3.
Entity classification accuracy on the Pittsburgh and GENIA corpora.
| Dataset | Accuracy | Class | Precision | Recall | F |
|---|---|---|---|---|---|
| Pittsburgh | 69.5 | Problem | 63.9 | 882 | 741 |
| Treatment | 75.0 | 41.6 | 53.7 | ||
| Test | 77.0 | 72.3 | 74.6 | ||
|
| |||||
| GENIA | 53.8 | protein | 87.7 | 54.5 | 67.2 |
| DNA | 52.3 | 59.4 | 55.6 | ||
| RNA | 44.4 | 74.4 | 55.6 | ||
| cell type | 54.8 | 47.6 | 50.9 | ||
| cell line | 12.9 | 43.2 | 19.9 | ||
Overall, the classification of entities shows very good results for all entity classes provided in the datasets, considering that only 34% and 19% of the entities in i2b2 and GENIA respectively could be found in UMLS as entries, which means that the distributional semantics contribute significantly to the coverage of the algorithm.
4.5. Overall System Performance
We compare the overall performance of our system with a baseline unsupervised system and supervised ones in Table 4 (Only Pittsburgh and GENIA are shown, since results on Beth and Partners show exactly the same pattern as that on Pittsburgh). Detailed performance on all the datasets are given in Table 5 and F scores are illustrated in Figure 4. Our system outperforms MetaMap significantly on both clinical and biomedical datasets. As expected, since our system has very weak supervision, it is not as competitive as supervised systems based on SVM or CRF equipped with deep knowledge resources. However, we would emphasize that our method has stable performance on all the datasets, spanning different types of entities and different types of texts.
Table 4.
Overall performance of our system, MetaMap, and the best supervised systems for the i2b2 and BioNLP2004 challenges.
| Dataset | System | Exact Micro F | Inexact Micro F |
|---|---|---|---|
| Pittsburgh | Ours | 26.5 | 53.1 |
| MetaMap | 11.3 | 27.9 | |
| Supervised | 85.2 | 92.4 | |
|
| |||
| GENIA | Ours | 15.2 | 39.5 |
| MetaMap | 7.7 | 19.2 | |
| Supervised | 72.6 | N/A | |
Table 5.
Detailed system performance on the Pittsburgh, Beth, Partners, and GENIA corpora.
| Dataset | Class | Exact | Inexact | ||||
|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | ||
| Pittsburgh | Overall (Micro) | 29.4 | 24.1 | 24.1 | 49.6 | 57.2 | 53.1 |
| Problem | 26.7 | 31.7 | 29.1 | 49.2 | 71.5 | 58.3 | |
| Treatment | 28.6 | 15.9 | 20.4 | 45.4 | 37.9 | 41.3 | |
| Test | 36.9 | 22.1 | 27.7 | 54.6 | 52.6 | 53.6 | |
|
| |||||||
| Beth | Overall (Micro) | 28.8 | 22.6 | 25.3 | 50.5 | 54.1 | 52.2 |
| Problem | 28.1 | 30.5 | 29.3 | 51.5 | 66.1 | 57.9 | |
| Treatment | 27.4 | 13.4 | 18.0 | 45.9 | 33.2 | 38.5 | |
| Test | 31.3 | 21.1 | 25.2 | 51.8 | 54.4 | 53.1 | |
|
| |||||||
| Partners | Overall (Micro) | 30.0 | 29.4 | 29.7 | 48.6 | 60.4 | 53.9 |
| Problem | 26.5 | 33.5 | 29.6 | 50.0 | 72.8 | 59.3 | |
| Treatment | 30.0 | 17.4 | 22.1 | 43.1 | 34.6 | 38.4 | |
| Test | 38.7 | 35.3 | 36.9 | 49.2 | 62.2 | 54.9 | |
|
| |||||||
| GENIA | Overall (Micro) | 15.4 | 15.0 | 15.2 | 37.0 | 42.3 | 39.5 |
| protein | 203 | 113 | 14.5 | 52.8 | 36.7 | 43.3 | |
| DNA | 5.6 | 9.1 | 6.9 | 30.0 | 53.2 | 38.4 | |
| RNA | 29.9 | 41.3 | 34.7 | 48.6 | 69.8 | 57.3 | |
| cell type | 40.7 | 36.7 | 38.6 | 50.4 | 48.7 | 49.5 | |
| cell line | 5.0 | 11.8 | 7.1 | 128 | 33.1 | 18.5 | |
Figure 4.
F-scores on the Pittsburgh, Beth, Partners, and GENIA corpora.
5. Discussion
The strategy to tackle boundary detection and entity classification using a stepwise solution shows much promise, especially considering that our system is unsupervised and highly portable. Our experimental results indicate that seed terms extracted automatically from UMLS act as a good proxy for training data, which equips the model with the expertise necessary to recognize specific entities. For boundary detection, NP chunking, although not perfect, is still a good approximation, followed by an IDF filter which effectively removes unrelated candidates. Finally, it should be highlighted that entity classification based on distributional vector similarity of both internal and external words could yield very competitive and stable performance, even if it does not rely on any training data or heuristics. When all the steps are combined, the overall system outperforms existing unsupervised dictionary match system significantly in all classes of entities in the two datasets.
Our system, while performing worse than supervised ones that rely on training data, has the large advantage that no annotation is required (this is true for the candidate named entities, but even so for the seed terms, which are not manually selected). As such, the level of supervision is very low in our approach: only domain representations that map entity classes to UMLS concepts, semantic types, or semantic groups need to be defined manually.
Furthermore, our method shows great portability and stability since the performance remains good when the target dataset changes from clinical notes to biological papers. It should be highlighted that the workflow and settings (except for the UMLS terminology and domain representations that are chosen in the initial step) do not change when applying our methods to different genres of text and different target semantic classes. Thus, our solution is capable of being applied directly in other unsupervised BM-NER tasks in which parameter tuning is not tolerated.
5.1. Impact of Seed Term Set
Signatures are computed based on mentions of seed terms in the data; thus the quality of seed terms influences if a class signature is truly representative. For example, the class “cell line” has a large seed term set of more than 260,000 terms. However, it is extremely noisy, containing terms like “human chromosomes” that is incorrect itself and misleading in terms of introducing more terms. This is caused by the imperfection of the relationship network of UMLS metathesaurus. On the contrary, “RNA” and “cell type” have small but accurate seed term set, which lead to much better performance in both precision and recall. Note that when the domain representation of an entity class contains only semantic types and semantic groups, performance is always satisfactory and stable, which may indicate that semantic type annotations of UMLS concepts is a more reliable resource than the UMLS relationships for this task.
In order to verify the hypothesis that a more reliable seed term set is beneficial, we replace UMLS with Cell Line Ontology [63] and Cell Ontology[64] targeting cell types and cell lines, and report performance on the two corresponding classes in GENIA. For the class “cell line”, seed term set using all entries in Cell Line Ontology yields inexact precision, recall, and F of 53.8, 59.6, and 56.5 respectively, which are significantly higher than those yielded by extracting seed terms from UMLS and hence supports our hypothesis. Cell Ontology, on the other hand, brings no significant increase over UMLS. Precision, recall, F of recognizing cell types are 51.5, 49.9, and 50.7. However, this may indicate that UMLS is sufficiently reliable as a terminology for cell types.
5.2. Should Entities Be Noun Phrases?
Our experimental results suggest that performance of such a combined system is largely determined by the part of boundary detection, which is the bottleneck of unsupervised named entity recognition. Rule based and distributional semantics based methods have limited potential handling boundary detection with satisfactory accuracy. Syntax based method is usually preferred practically. In this paper, we attack boundary detection by assuming that all entities are noun phrases, which is a reasonable but imperfect assumption. Results show that around 40% entities are inside(but not) noun phrases, which means in exact evaluation our system will automatically miss nearly half of the correct entities. By observing output of the system, we summarize that about 47% and 36% errors (errors = false positives + false negatives) in exact evaluations on Pittsburgh and GENIA respectively are more or less caused by the imperfection of this assumption. Typical errors of this type (we call them chunking related errors) are given as follows.
The first category of chunking related errors is caused by the fact that chunkers are not capable of finding all noun phrases from text. Chunking, by its definition [65], is a shallow parsing step generating non-overlapping phrases. This means that nested NPs will not be found by a chunker. For instance, in the sentence “Sinus node dysfunction s/p pacemaker,” our recognizer labels the whole sentence as an entity of type “Problem,” because the sentence as a whole is a noun phrase identified by the chunker. However, in the gold standard, “sinus node dysfunction” is annotated as an entity of type “problem.” It is clear that “sinus node dysfunction” is a nested NP which could not be found by a chunker. In our error analysis we found that 31% in Pittsburgh and 36% in GENIA of chunking related errors are of this type. In the future work, this type of error could be eliminated by doing full parsing instead of chunking, followed by choosing all NPs in the parse tree as candidates.
The second major category is inconsistency with annotation. One of the most noticeable questions is whether to include determiners in the entities. Determiners are usually annotated inside the entities in i2b2 corpus, but are excluded from entities in GENIA. For instance, in GENIA, “IL-6 gene,” instead of “the IL-6 gene” is annotated, which is inconsistent with output of chunker. A similar type of error is about negations. For example, “no hemodynamically significant lesions bilaterally” is recognized as a noun phrase and an entity, instead of “hemodynamically significant lesions bilaterally” in the ground truth. Negations are excluded from entities in both i2b2 and GENIA, leading to 7% and 9% chunking related errors respectively. From the perspective of information extraction application, errors caused by determiners are insignificant, but negations should be taken care of, possibly by adding a negation detection component to the system.
In these two situations, it is unfair to blame the NP chunker since errors are caused by the limitation of our assumption that all entities are NP chunks. However, chunking errors also contribute to part of the failures, especially in i2b2. According to [62], OpenNLP could reach 89.7% F score of NP chunking on GENIA, which is a quite satisfactory performance. However, since clinical notes are usually more noisy and ambiguous than scientific literature, chunking on i2b2 is much more challenging than on GENIA. Thus, nearly half of the chunking related errors on Pittsburgh are exactly chunking errors.
In summary, the assumption that entities are noun phrases is reasonable and acceptable in a named entity recognition system. However, it could be further improved by considering all NPs instead of only NP chunks, adding negation detections, as well as a more effective chunker (parser) for clinical notes in future work.
5.3. Impact of IDF
IDF is leveraged in two ways in our framework. One is the IDF filter which removes common noun phrases with very low IDF; the other is the TF-IDF weights in signatures. The effectiveness of the two usages could be evaluated by comparing to systems without them. For example, on Pittsburgh, introducing IDF filter and TF-IDF weights independently could make the Inexact F score raise from 45.6 to 49.1 and 50.2 respectively, and a joint usage of them could bring performance increase of 7.5 to 53.1. Both IDF filter and TF-IDF bring visible improvements on the baseline system that does not rely on IDF filter and use only term frequency as signature weights. Similar improvements are obtained for all other data sets. The impact of IDF filter indicates that named entities are unlikely to be the most common noun phrases. This is especially true when the dataset contains multiple documents on different topics that usually contain different keywords. Phrases occurring frequently across all the documents are always general ones like “father”, “date of birth”, “the genome”, etc.
Figure 5 shows the performance on Pittsburgh data set using different IDF filter cutoffs. The peak of performance on GENIA, which is not shown here for the sake of brevity, lies around 4.5. Since the IDF filter is just a pre-processing step to remove spurious candidates with low IDF, the choice of threshold is favored towards lower values so as not to miss too many true candidates. In all experiments, we thus chose a threshold of 4 for the IDF filter.
Figure 5.
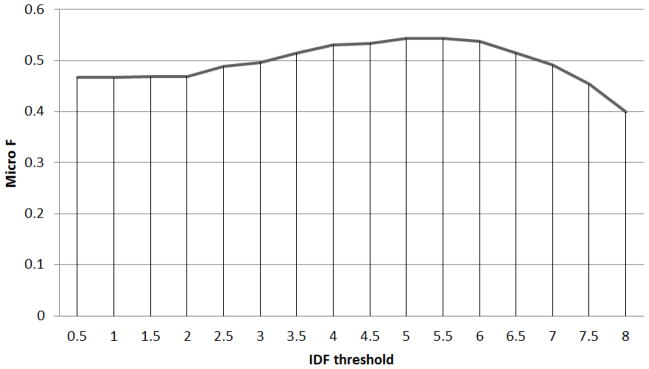
IDF threshold - F score curve on Pittsburgh.
Nevertheless, the exploitation of IDF filter in this paper could be further improved. In our current system, the IDF value of a noun phrase is obtained by averaging IDF values of all the words in the phrase. The reason is to reduce dimensions, especially when the dataset is a small one with limited number of noun phrases, which leads to the situation that most noun phrases appear only once or twice, thus, have similar IDF values. However, such approach is sometimes not so reasonable when a very informative (entity) word is in a long phrase and all the other words are common ones. It is also possible that a long phrase is an entity, but all of the words inside are common ones. For example, “No known drug allergies” is an entity of type Problem in Pittsburgh dataset, but all the words inside the phrase are among the most common ones in the dataset.
5.4. Impact of Internal and Context Words
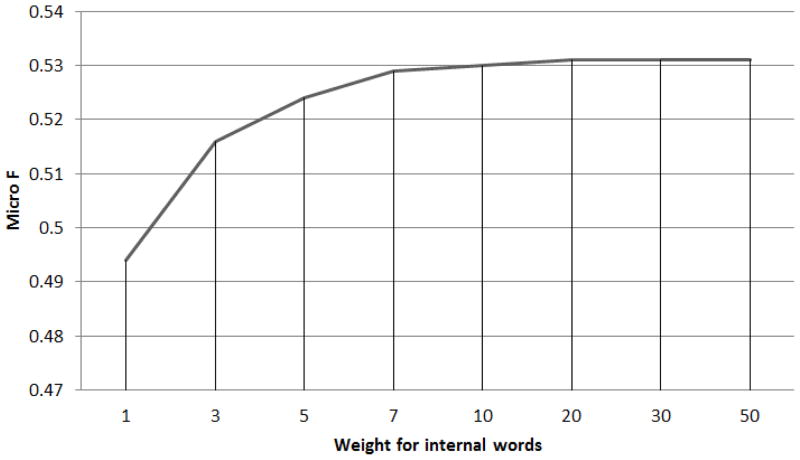
Several previous systems classify term semantics based on context words, which is a typical approach in natural language processing. In our method, we not only resort to context words, but leverage internal ones as well. Moreover, we found that at least in our experimental settings, internal words are more informative than context words. Removing internal words from signature will make the performance (Inexact F) on Pittsburgh drop significantly from 53.1 to 32.9. It is interesting that the system using only internal words(F 44.1) outperforms the system using only context words(F 32.9), indicating that internal words are somehow more helpful in deciding entity types. This is because judging type of an entity by its internal words is essentially doing a fuzzy dictionary match between seed terms and candidates. If an entity contains a word, say “pain”, that occurs frequently inside seed terms of a certain entity class (Problem), it is highly likely that the entity belong to that class (Problem). Combining internal and context words is more effective than either relying only on internal words, which ignores context information, or only on context words, which does not make fully use of the seed terms as a dictionary. The use of internal words is a vital change to distributional semantics, which traditionally only focus on contextual information of objects.
Figure 6 indicates that overall performance increases as the weights for internal words get larger, until it plateaus around 20. The same phenomenon is observed on the GENIA corpus, with the plateau starting around 20 as well. Thus, 20 was chosen experimentally as the weight for internal words when building signature vectors in all our experiments.
Figure 6.
Internal word weight - F score curve on Pittsburgh.
5.5. Impact of Data for Signature Generation
A major limitation of our method is the need of a fairly large test dataset for signature generation, which hinders the method to be employed in online settings in which users input single or multiple sentence at a time instead of a corpus. This problem could be solved by using a backup corpus to generate signatures. However, such a backup corpus should be of the same type of text as the target input, since distributions of terms and their context words in distinct types of text have dramatic differences. In order to validate the efficacy of such backup corpus, we tested on Pittsburgh using Beth, Partners, and GENIA respectively for signature generation, and got Inexact F scores of 52.7, 53.3, and 21.2. Comparing with the original system using signatures generated from Pittsburgh itself (F = 53.1), using corpora of the same type (discharge summaries) from i2b2 corpus (Beth and Partners) does not change, even increase in the case of Partners, the performance on Pittsburgh, but using GENIA will decrease the F score dramatically. It should be emphasized that such backup corpus does not need any annotation as well, and such a raw text set is often easier to collect.
A possible alternative is relying only on internal words when signatures are generated, which could be extracted from seed terms directly. Results have showed that such compromise does not harm the results so significantly as discarding internal words.
Although our system is fully unsupervised and as such its comparison to supervised approach might be unfair, we wanted to have confidence that the approach does not overfit the input corpus, and the signature knowledge gained from one data set is applicable to other similar corpora as well. 10-fold cross validation was conducted on Pittsburgh and GENIA data sets. On Pittsburgh, average performance over the 10 folds yielded 23.9 exact micro F (stdev=0.12) and 53.0 inexact micro F (stdev=0.10). On GENIA, our system yielded average exact micro F of 15.1 (stdev=0.08) and inexact micro F of 39.0 (stdev=0.11). The results have no significant difference from the performance reported in section 4.5, which indicates that generating signatures from other sources than the target data is acceptable as long as they are of the same genre of text. In addition, the experiments on Pittsburgh with signatures generated from Beth and Partners act similarly as cross validation, and also indicate that the performance of our system is not a result of over-fitting on the source dataset, and can cross from one corpus to another within the same domain and genre. While the signatures are domain dependent, they are not data set dependent.
6. Conclusion
Biomedical named entity recognition (BM-NER) is a challenging task in biomedical natural language processing. In this paper, we design a framework which provides a stepwise solution to BM-NER, including a seed term extractor, an NP chunker, an IDF filter, and a classifier based on distributional semantics. In our framework, shallow syntactic analysis and lexical semantics are properly exploited in different phases. Our method does not rely on any rules, heuristics, or training data, which makes it easy to be applied in different settings and applications. Experimental results on two mainstream biomedical datasets demonstrate the effectiveness and generalizability of our methods. For individual steps, we show that quality of seed term sets is an important factor of a successful system, and the usage of NPs as entity candidates is a reasonable approximation to boundary detection. After filtering candidates with IDF filter, our distributional similarity based classifier shows competitive performance on entity classification, taking advantage of both internal and context information. Finally, this paper envisions possible improvements on the approach, including nested NPs as candidates, better chunker for medical text, better domain representations, and improved IDF values of phrases.
Acknowledgments
This work is supported by an NSF award #1027886 (NE). Any opinions, findings, or conclusions are those of the authors and do not necessarily reflect the views of the funding organizations.
Footnotes
In this paper, without further explanation, “biomedical entity”, “entity”, and “named entity” are all referring to biomedical entities.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman C, Alderson P, Austin J, Cimino J, Johnson S. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hripcsak G, Friedman C, Alderson P, DuMouchel W, Johnson S, Clayton P, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Annals of Internal Medicine. 1995;122(9):681. doi: 10.7326/0003-4819-122-9-199505010-00007. [DOI] [PubMed] [Google Scholar]
- 3.Fiszman M, Chapman W, Aronsky D, Evans R, Haug P. Automatic detection of acute bacterial pneumonia from chest x-ray reports. Journal of the American Medical Informatics Association. 2000;7(6):593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of biomedical informatics. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 5.Melton G, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. Journal of the American Medical Informatics Association. 2005;12(4):448–457. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzuner O, South B, Shen S, DuVall S. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanabe L, Wilbur W. Tagging gene and protein names in biomedical text. Bioinformatics. 2002;18(8):1124–1132. doi: 10.1093/bioinformatics/18.8.1124. [DOI] [PubMed] [Google Scholar]
- 8.Settles B. Biomedical named entity recognition using conditional random fields and rich feature sets. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications; Association for Computational Linguistics; 2004. pp. 104–107. [Google Scholar]
- 9.Yeh A, Morgan A, Colosimo M, Hirschman L. Biocreative task 1a: gene mention finding evaluation. BMC bioinformatics. 2005;6(Suppl 1):S2. doi: 10.1186/1471-2105-6-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkel J, Grenager T, Manning C. Incorporating non-local information into information extraction systems by gibbs sampling. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics; Association for Computational Linguistics; 2005. pp. 363–370. [Google Scholar]
- 11.Ratinov L, Roth D. Design challenges and misconceptions in named entity recognition. Proceedings of the Thirteenth Conference on Computational Natural Language Learning; Association for Computational Linguistics; 2009. pp. 147–155. [Google Scholar]
- 12.Abacha A, Zweigenbaum P. Medical entity recognition: a comparison of semantic and statistical methods. Proceedings of BioNLP 2011 Workshop; Association for Computational Linguistics; 2011. pp. 56–64. [Google Scholar]
- 13.Wang Y, Patrick J. Cascading classifiers for named entity recognition in clinical notes. Proceedings of the Workshop on Biomedical Information Extraction; Association for Computational Linguistics; 2009. pp. 42–49. [Google Scholar]
- 14.Morgan A, Lu Z, Wang X, Cohen A, Fluck J, Ruch P, Divoli A, Fundel K, Leaman R, Hakenberg J, et al. Overview of biocreative ii gene normalization. Genome biology. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G, Su J. Named entity recognition using an hmm-based chunk tagger. Proceedings of the 40th Annual Meeting on Association for Computational Linguistics; Association for Computational Linguistics; 2002. pp. 473–480. [Google Scholar]
- 16.McCallum A, Li W. Early results for named entity recognition with conditional random fields, feature induction and web-enhanced lexicons. Proceedings of the seventh conference on Natural language learning at HLT-NAACL; 2003; Association for Computational Linguistics; 2003. pp. 188–191. [Google Scholar]
- 17.Nadeau D, Sekine S. A survey of named entity recognition and classification. Lingvisticae Investigationes. 2007;30(1):3–26. [Google Scholar]
- 18.Rau L. Extracting company names from text. Artificial Intelligence Applications, 1991; Proceedings., Seventh IEEE Conference on; IEEE; 1991. pp. 29–32. [Google Scholar]
- 19.Coates-Stephens S. The analysis and acquisition of proper names for the understanding of free text. Computers and the Humanities. 1992;26(5):441–456. [Google Scholar]
- 20.Fellbaum C. Wordnet, Theory and Applications of Ontology: Computer Applications. 2010. pp. 231–243. [Google Scholar]
- 21.Alfonseca E, Manandhar S. An unsupervised method for general named entity recognition and automated concept discovery. Proceedings of the 1st International Conference on General WordNet; Mysore, India. 2002. pp. 34–43. [Google Scholar]
- 22.Nadeau D, Turney P, Matwin S. Unsupervised named-entity recognition: Generating gazetteers and resolving ambiguity. Advances in Artificial Intelligence. 2006:266–277. [Google Scholar]
- 23.Sekine S, Nobata C. Definition, dictionaries and tagger for extended named entity hierarchy. Proceedings of the Language Resources and Evaluation Conference (LREC); 2004. pp. 1977–1980. [Google Scholar]
- 24.Shinyama Y, Sekine S. Named entity discovery using comparable news articles. Proceedings of the 20th international conference on Computational Linguistics; Association for Computational Linguistics; 2004. p. 848. [Google Scholar]
- 25.Collins M, Singer Y. Unsupervised models for named entity classification. Proceedings of the Joint SIGDAT Conference on Empirical Methods in Natural Language Processing and Very Large Corpora; 1999. pp. 100–110. [Google Scholar]
- 26.Riloff E, Jones R, et al. Learning dictionaries for information extraction by multi-level bootstrapping. Proceedings of the National Conference on Artificial Intelligence; JOHN WILEY & SONS LTD; 1999. pp. 474–479. [Google Scholar]
- 27.Cucchiarelli A, Velardi P. Unsupervised named entity recognition using syntactic and semantic contextual evidence. Computational Linguistics. 2001;27(1):123–131. [Google Scholar]
- 28.Etzioni O, Cafarella M, Downey D, Popescu A, Shaked T, Soderland S, Weld D, Yates A. Unsupervised named-entity extraction from the web: An experimental study. Artificial Intelligence. 2005;165(1):91–134. [Google Scholar]
- 29.Elsner M, Charniak E, Johnson M. Structured generative models for unsupervised named-entity clustering. Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics; Association for Computational Linguistics; 2009. pp. 164–172. [Google Scholar]
- 30.Yarowsky D. Unsupervised word sense disambiguation rivaling supervised methods. Proceedings of the 33rd annual meeting on Association for Computational Linguistics; Association for Computational Linguistics; 1995. pp. 189–196. [Google Scholar]
- 31.Jurafsky D, Martin J, Kehler A. Speech and language processing: an introduction to natural language processing, computational linguistics and speech recognition. Vol. 2. MIT Press; 2002. [Google Scholar]
- 32.Fukuda K, Tsunoda T, Tamura A, Takagi T, et al. Toward information extraction: identifying protein names from biological papers. Pac Symp Biocomput. 1998;707:707–718. [PubMed] [Google Scholar]
- 33.Proux D, Rechenmann F, Julliard L, Pillet V, Jacq B, et al. Detecting gene symbols and names in biological texts: a first step toward pertinent information extraction. GENOME INFORMATICS SERIES. 1998:72–80. [PubMed] [Google Scholar]
- 34.Rindflesch T, Tanabe L, Weinstein J, Hunter L. Edgar: extraction of drugs, genes and relations from the biomedical literature. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing, NIH Public Access; 2000. p. 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. Genies: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17(suppl 1):S74–S82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- 36.Aronson A. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. Proceedings of the AMIA Symposium; American Medical Informatics Association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, Davis A, Dolinski K, Dwight S, Eppig J, et al. Gene ontology: tool for the unification of biology. Nature genetics. 2000;25(1):25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.SGK, MJJ, OPV ZJ, SS, K-SKC, CCG Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Ohta T, Tateisi Y, Tsujii J. Genia corpusa semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(suppl 1):i180–i182. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 41.GuoDong Z, Jian S. Exploring deep knowledge resources in biomedical name recognition. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications; Association for Computational Linguistics; 2004. pp. 96–99. [Google Scholar]
- 42.Kazama J, Makino T, Ohta Y, Tsujii J. Tuning support vector machines for biomedical named entity recognition. Proceedings of the ACL-02 workshop on Natural language processing in the biomedical domain; Association for Computational Linguistics; 2002. pp. 1–8. [Google Scholar]
- 43.Mitsumori T, Fation S, Murata M, Doi K, Doi H. Gene/protein name recognition based on support vector machine using dictionary as features. BMC bioinformatics. 2005;6(Suppl 1):S8. doi: 10.1186/1471-2105-6-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao S. Named entity recognition in biomedical texts using an hmm model. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications; Association for Computational Linguistics; 2004. pp. 84–87. [Google Scholar]
- 45.McDonald R, Pereira F. Identifying gene and protein mentions in text using conditional random fields. BMC bioinformatics. 2005;6(Suppl 1):S6. doi: 10.1186/1471-2105-6-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, Ohta T, Tsuruoka Y, Tateisi Y, Collier N. Introduction to the bio-entity recognition task at jnlpba. Proceedings of the international joint workshop on natural language processing in biomedicine and its applications; 2004. pp. 70–75. [Google Scholar]
- 47.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of biocreative: critical assessment of information extraction for biology. BMC bioinformatics. 2005;6(Suppl 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solt I, Gerner M, Thomas P, Nenadic G, Bergman CM, Leser U, Hakenberg J. Gene mention normalization in full texts using gnat and linnaeus. Proceedings of the BioCreative III Workshop; Bethesda, USA. 2010. pp. 134–139. [Google Scholar]
- 49.Kim J-D, Ohta T, Pyysalo S, Kano Y, Tsujii J. Overview of bionlp’09 shared task on event extraction. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing: Shared Task; Association for Computational Linguistics; 2009. pp. 1–9. [Google Scholar]
- 50.Sea K, deBruijn B, Cherry C. Nrc at i2b2: one challenge, three practical tasks, nine statistical systems, hundreds of clinical records, millions of useful features. Proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data; 2010. [Google Scholar]
- 51.Meystre S, Haug P. Comparing natural language processing tools to extract medical problems from narrative text. AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2005. p. 525. [PMC free article] [PubMed] [Google Scholar]
- 52.Abacha A, Zweigenbaum P. Automatic extraction of semantic relations between medical entities: a rule based approach. Journal of Biomedical Semantics. 2011;2(Suppl 5):S4. doi: 10.1186/2041-1480-2-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patrick J, Wang Y, Budd P. Automatic mapping clinical notes to medical terminologies. Australasian Language Technology Workshop; 2006. p. 75. [Google Scholar]
- 54.Wang Y. Annotating and recognising named entities in clinical notes. Proceedings of the ACL-IJCNLP 2009 Student Research Workshop; Association for Computational Linguistics; 2009. pp. 18–26. [Google Scholar]
- 55.Kang N, Afzal Z, Singh B, Van Mulligen EM, Kors JA. Using an ensemble system to improve concept extraction from clinical records. Journal of Biomedical Informatics. 2012;45(3):423–428. doi: 10.1016/j.jbi.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 56.de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. Journal of the American Medical Informatics Association. 2011;18(5):557–562. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang B, Cao H, Wu Y, Jiang M, Xu H. Clinical entity recognition using structural support vector machines with rich features. Proceedings of the ACM sixth international workshop on Data and text mining in biomedical informatics; ACM; 2012. pp. 13–20. [Google Scholar]
- 58.McCray A, Burgun A, Bodenreider O, et al. Aggregating umls semantic types for reducing conceptual complexity. Studies in health technology and informatics. 2001;(1):216–220. [PMC free article] [PubMed] [Google Scholar]
- 59.Manning C, Raghavan P, Schütze H. Introduction to information retrieval. Vol. 1. Cambridge University Press; Cambridge: 2008. [Google Scholar]
- 60.Cohen T, Widdows D. Empirical distributional semantics: Methods and biomedical applications. Journal of biomedical informatics. 2009;42(2):390. doi: 10.1016/j.jbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pivovarov R, Elhadad N. A hybrid knowledge-based and data-driven approach to identifying semantically similar concepts. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang N, van Mulligen E, Kors J. Comparing and combining chunkers of biomedical text. Journal of biomedical informatics. 2011;44(2):354–360. doi: 10.1016/j.jbi.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Sarntivijai S, Xiang Z, Meehan TF, Diehl AD, Vempati U, Schürer SC, Pang C, Malone J, Parkinson HE, Athey BD, et al. Cell line ontology: Redesigning the cell line knowledgebase to aid integrative translational informatics. ICBO; p. 833. [Google Scholar]
- 64.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome biology. 2005;6(2):R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tjong Kim Sang E, Buchholz S. Introduction to the conll-2000 shared task: Chunking. Proceedings of the 2nd workshop on Learning language in logic and the 4th conference on Computational natural language learning; Association for Computational Linguistics; 2000. pp. 127–132. [Google Scholar]