Abstract
Objective
African Americans, East Asians, and Hispanics with systemic lupus erythematosus (SLE) are more likely to develop renal disease than SLE patients of European descent. We investigated whether European genetic ancestry protects against the development of lupus nephritis and explored genetic and socioeconomic factors that might explain this effect.
Methods
This was a cross-sectional study of 1906 adults with SLE. Participants were genotyped for 126 single nucleotide polymorphisms (SNPs) informative for ancestry. A subset of participants was also genotyped for 80 SNPs in 14 candidate genes for renal disease in SLE. We used logistic regression to test the association between European ancestry and renal disease. Analyses adjusted for continental ancestries, socioeconomic status, and candidate genes.
Results
Participants (n=1906) had on average 62.4% European, 15.8% African, 11.5% East Asian, 6.5% Amerindian, and 3.8% South Asian ancestry. Among participants, 34% (n=656) had renal disease. A 10% increase in European ancestry was associated with a 15% reduction in the odds of having renal disease after adjustment for disease duration and sex (OR 0.85, 95% CI 0.82-0.87, p=1.9 × 10−30). Adjusting for other genetic ancestries, measures of socioeconomic status, or SNPs in genes most associated with renal disease (IRF5 (rs4728142), BLK (rs2736340), STAT4 (rs3024912), ITGAM (rs9937837) and HLA-DRB1*0301 and DRB1*1501, p<0.05) did not substantively alter this relationship.
Conclusion
European ancestry is protective against the development of renal disease in SLE, an effect independent of other genetic ancestries, common risk alleles, and socioeconomic status.
Introduction
African Americans, Hispanics, and East Asians with systemic lupus erythematosus (SLE) are significantly more likely to develop renal disease than SLE patients of European descent (1-3). Though there are striking differences in the risk of lupus nephritis across ethnic groups, the reasons for such differences remain less clear. Poverty and limited access to medical care are associated with the development of renal disease in SLE (5, 6). Genetic factors may also predispose to lupus nephritis, though parsing genetic and environmental factors using traditional epidemiologic techniques can be complex (4, 7).
Genetic ancestry, a quantitative description of the founding population groups from which an individual is descended, provides a tool for understanding how population genetics may influence the risk of renal disease in SLE. In a prior study of SLE in an ethnically diverse population, genetic ancestry appeared to explain some of the differential risk in lupus nephritis across ethnic groups, though that study used a small number of genetic markers to define genetic ancestry and provided a limited evaluation of the role of genetic ancestry (8). Even within relatively homogenous groups, though, genetic differences may be associated with varying disease characteristics in SLE. For example, among European Americans with SLE in the United States, southern European ancestry, compared to northern European ancestry, was associated with a higher risk of renal disease (9). Together, these studies suggest that ancestral genetics are important in the development of specific disease features in SLE and may contribute to varying risk of lupus nephritis across ethnicities.
In addition to ancestral genetics, polymorphisms in individual genes have been associated with lupus nephritis. Recent studies have identified variants in STAT4, TNFAIP3, ITGAM, and specific HLA alleles not only as risk factors for SLE, but also for renal disease in SLE (4, 10-12). While variation in these genes may contribute to the development of lupus nephritis, the relationship between the risk conferred by these genes and risk associated with ancestral genetics has not been fully elucidated.
In this study, we investigated whether the genetic ancestry of participants in a multiethnic SLE case collection is associated with risk of renal disease in SLE. We hypothesized that European ancestry would be associated with a lower risk of renal disease. We further hypothesized that this association would remain after adjusting for measures of socioeconomic status, but would be partially explained by variation in candidate genes for lupus nephritis.
Patients and Methods
Participants
All participants met the revised American College of Rheumatology (ACR) classification criteria for SLE and SLE diagnosis was verified by medical records review. These SLE patients were previously enrolled in one of two case collections developed to study the genetics of SLE: the University of California, San Francisco (UCSF) Lupus Genetics Project Case Collection (n=1836), and the Autoimmune Biomarkers Collaborative Network (ABCoN) case collection (n=139). A subset of UCSF participants was also enrolled in a longitudinal study of SLE known as the Lupus Outcomes Study, or LOS (n=1028). Participants were recruited from a variety of settings including academic medical centers (UCSF and Johns Hopkins University), community hospitals in the San Francisco Bay Area, and lupus support groups. The Institutional Review Boards at the University of California, San Francisco and Johns Hopkins University approved this study and all participants provided informed consent prior to enrollment.
Measurements
Outcome Variables
Our primary outcome variable was the presence of renal disease. “Renal disease” was defined according to the ACR classification criteria for SLE as >0.5 grams of proteinuria per day or 3+ protein on urine dipstick analysis documented in the medical record (13). We also considered those participants with biopsy-proven lupus nephritis to have renal disease.
Covariates
Covariates included disease duration, sex, and socioeconomic status. Disease duration was established by review of medical records and was defined as the time from formal diagnosis until entry into either the UCSF or ABCoN genetic studies. To characterize socioeconomic status, we examined educational attainment, personal income, and neighborhood poverty. Education was self-reported and grouped into categories ranging from less than a high school degree to a professional or doctoral degree. Household income was also self-reported and grouped into six categories ranging from below $20,000/year to > $100,000/year. To capture neighborhood poverty, we used geocoded data from the US decennial census to determine the percentage of individuals living in poverty in a participant’s neighborhood. We dichotomized this variable into participants living in a neighborhood with >30% of residents living below 125% of the federal poverty level and those living with <30% of the neighborhood below this level. This cut point of 30% represents the top decile of neighborhood poverty in the LOS dataset and also corresponds to a federal “poverty area” designation (14). All socioeconomic status (SES) measures described here were available for those participants enrolled in the LOS. Data on education and income, but not neighborhood poverty, were available for ABCoN participants.
Genotyping and Quality Control (QC)
DNA was collected from blood or from saliva using Oragene DNA sample collection kits (DNAGenotek) when blood was not available. Participants were genotyped in one of three groups. Participants of self-identified non-European ancestry (n=936) were genotyped for a set of 384 SNPs using a custom BeadXpress assay (Illumina). A subset of 613 participants of self-identified European ancestry had been previously genotyped for a genome-wide association study of SLE susceptibility using the HumanHap550 BeadChip (Illumina) (15). An additional set of 426 European ancestry participants was genotyped for 12,000 single nucleotide polymorphisms (SNPs) using a custom iSELECT Infinium II array (Illumina) (16).
SNPs were excluded if they were invariant or missing in >10% of the study population. We also excluded any individuals who had >10% of their genotype data missing (n=35), a first-degree familial relationship to another participant (n=15), who had duplicate samples (n=10) or had all phenotype and demographic data missing (n=4).
Ancestry Estimation
We used a set of 104 post-QC SNPs common to all three genotyping platforms to determine the ancestral populations from which participants were descended (17). These SNPs provide information about participants’ genetic ancestry and are known as ancestry informative markers (AIMs). The AIMs used here were unlinked SNPs with an r2 <0.5, and were distributed throughout the autosomal genome.
We estimated continental ancestry using the program STRUCTURE (18, 19). When estimating population membership, we included genotype data from 502 individuals of established European, West African, East Asian, South Asian, and Amerindian ancestry to improve ancestry differentiation and interpretation. These reference population genotypes were derived from either HapMap genotypes or from selected genotyping using TaqMan assays. The reference population genotypes included 116 European American (30 CEU HapMap and 86 from 4 grandparent defined European Americans), 102 sub-Saharan African (60 Yoruba HapMap, 19 Bini West African, and 23 Kanuri West African), 105 Amerindian (50 Mayan, 26 Quechuan, and 29 Nahua), 115 East Asian (36 Japanese HapMap, 36 Han Chinese HapMap and 43 from other East Asian population groups) and 64 South Asian Indian samples. All SNPs chosen had identical genotyping results using TaqMan assays and Illumina genotyping (610K platform) for a subset of 40 subjects.
We ran STRUCTURE first assuming six major continental populations. We then excluded outliers with >15% membership in the sixth population (Oceanic, n=5) and re-estimated ancestry assuming five populations. We also applied principal components analysis (PCA) to the AIMs genotype data to define ancestry for the entire study population. For this analysis, the group of 503 individuals of known ancestry were again included. We performed PCA using the EIGENSOFT software program under default parameters (20).
Candidate Gene Selection
We examined a set of 64 post-QC SNPs in 14 genes common to the 384 SNP platform and the 550K platform as candidate genes for renal disease in SLE (see Supplementary Table 1 for full list of genes and SNPs). Participants genotyped on the 12K platform were excluded from candidate gene analyses as there was insufficient overlap between SNPs on that platform and the 384 SNP platform. The 14 genes examined here were all established SLE susceptibility genes and we hypothesized that established risk alleles for SLE may also be associated with renal disease in SLE.
A subset of participants had been previously genotyped for HLA-DRB1 alleles. Data on HLA type were available only for participants of self-reported European American or African American ethnicities. We chose to examine two HLA-DRB1 alleles, *0301 and *1501, which have been associated with the development of SLE. Like other SLE risk alleles, these HLA types may not only confer risk for the development of SLE, but also renal disease, and thus were chosen as candidate genes (21).
Data Analysis
We used descriptive statistics including medians and proportions to characterize the demographic and clinical characteristics of the study population. Differences in demographic and clinical characteristics among self-identified ethnic groups were assessed using the chi-squared test and the Kruskal-Wallis test. Differences in percent ancestry among those with and without renal disease were assessed using the Mann-Whitney U test.
We tested for associations between candidate gene SNPs and renal disease using logistic regression in PLINK, adjusting for ancestry, represented by the first three principal components (22). Logistic regression assumed an additive genetic model. We also tested associations between the HLA-DRB1 alleles *0301 and *1501 and renal disease using logistic regression adjusting for the first three principal components.
We focused subsequent analyses on European ancestry, since it was the most common ancestry type, was present among most participants, and associated with the largest effect size when ancestries were modeled individually (see results) or when European ancestry was jointly modeled with each other ancestry (data not shown). We assessed the association between European ancestry, derived from our STRUCTURE estimates, and renal disease using logistic regression, adjusting for disease duration and sex.
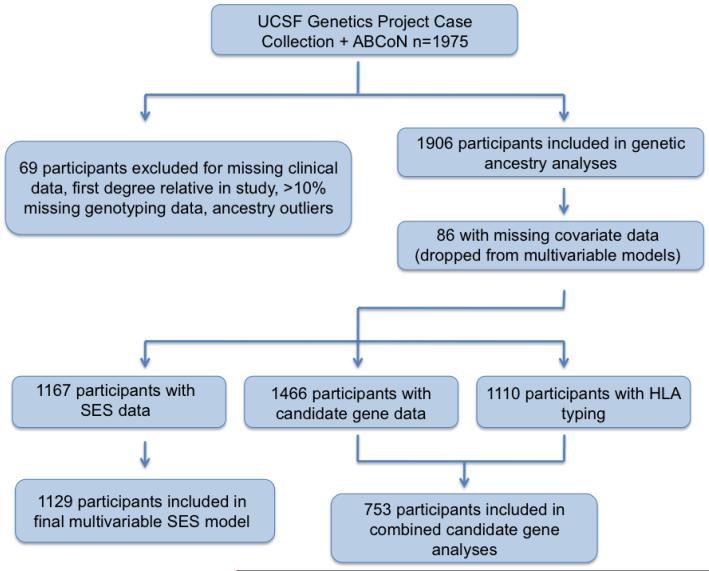
Using this basic logistic regression model, we then tested whether adjusting for a series of covariates could explain any apparent relationship between European ancestry and renal disease. In successive models, we adjusted for other ancestries, represented by principal components 2 and 3, socioeconomic status using education, income, and neighborhood poverty, and specific candidate genes (Figure 1).
Figure 1.
Study Design. This figure illustrates the groups of participants included in this study as well as the analytic design.
When examining the role of candidate genes, we selected the top SNP in each gene associated with renal disease in bivariate analysis using a p value of <0.05. We deliberately used this liberal p value as we were interested in generating a more inclusive list of SNPs that might mediate the relationship between genetic ancestry and renal disease. We then included all selected SNPs in a multivariable model of renal disease and European ancestry, adjusting for disease duration, sex, and other genetic ancestries for this model. SNPs were retained in the final model only if the logistic term p-value was <0.05. Finally, the HLA-DRB1*0301 and HLA-DRB1*1501 alleles were added to the model for the subset of subjects having HLA-DRB1 typing (Figure 1).
For multivariable models we tested logistic model fit using the Hosmer Lemeshow goodness-of-fit test, the link test, and graphically, using lowess smoothers. To improve fit, we performed a power transformation of disease duration using the Box-Tidwell method and utilized continuous and positive-negative terms for principal components. Analyses used Stata/MP version 9.2 (College Station, TX). To compare models after adding or removing variables, we used the Bayesian information criterion (BIC), with a decrease of 10 units between models indicating that a variable strongly improves the model.
Lastly, we used a k-means cluster analysis as an alternate approach to examining the association between ancestry and renal disease. We first grouped participants into ancestrally similar clusters using a k-means cluster analysis of the first three PC values. Participants were grouped into four clusters with this technique. We removed outliers from each cluster prior to any analyses (total 29 subjects). We then calculated the odds ratio for renal disease associated with a 10% increase in European ancestry within each cluster. We combined odds ratios, adjusted for disease duration and gender, across clusters via random effects meta-analysis.
Results
This study included 1975 SLE cases, 139 of which were from the ABCoN case collection and 1836 of which were from the UCSF Genetics Project Case Collection. After removal of subjects not meeting quality control criteria and ancestry outliers, and after removing subjects for whom data were missing, 1820 participants were used in our main multivariable analyses.
The majority of participants were female (n=1744, 92%). Participants of European descent comprised 53% of the sample (n=1009), African Americans 17% (n=331), Hispanics 13% (n=247), East Asians 11% (n=214), and unknown or other ethnicities 6% (n=105). The median disease duration was 6 years (IQR 2-13). Among participants, 34% (n=656) had a history of renal disease. Self-identified East Asian participants had the highest prevalence of renal disease (59%) while those of European descent had the lowest (23%) (Table 1).
Table 1. Demographic and Clinical Characteristics of 1906 SLE Cases.
| Characteristic | All (n=1906) | European American (n=1009) |
African American (n=331) |
Hispanic (n=247) |
East Asian (n=214) |
Other (n=105) | p* |
|---|---|---|---|---|---|---|---|
| Female, n (%) | 1744 (92) | 921 (91) | 310 (94) | 226 (92) | 194 (91) | 96 (91) | 0.73 |
| Median disease duration, years (IQR) | 6 (2-13) | 7 (2-14) | 8 (4-15) | 5 (2-10) | 6 (3-13) | 5 (1-11) | 0.0001 |
| Renal Disease, n (%) | 656 (34) | 231 (23) | 156 (47) | 100 (41) | 127 (59) | 42 (40) | <0.0005 |
| Education,** n (%) | |||||||
| Less than HS | 56 (5) | 14 (2) | 27 (11) | 8 (7) | 3 (3) | 4 (6) | <0.0005 |
| HS | 214(18) | 102 (16) | 55 (23) | 28 (26) | 19 (17) | 10 (16) | |
| Some college | 473 (40) | 255 (39) | 101 (43) | 53 (50) | 33 (29) | 31 (49) | |
| College | 264 (23) | 169 (26) | 27 (12) | 11 (10) | 45 (40) | 12 (19) | |
| Professional | 160 (14) | 108 (17) | 26 (11) | 7 (7) | 13 (11) | 6 (10) | |
| Household income, n (%) | |||||||
| < $20,000 | 210 (20) | 80 (14) | 90 (41) | 14 (15) | 17 (17) | 9 (16) | <0.0005 |
| $20,000-40,000 | 229 (22) | 126 (21) | 45 (21) | 26 (28) | 13 (13) | 19 (33) | |
| $40,000-60,000 | 195 (18) | 113 (19) | 35 (16) | 21 (23) | 15 (15) | 11 (19) | |
| $60,000-80,000 | 144 (14) | 90 (15) | 20 (9) | 10 (11) | 17 (17) | 7 (12) | |
| $80,000-100,000 | 103 (10) | 66 (11) | 11 (5) | 12 (13) | 11 (11) | 3 (5) | |
| >$100,00 | 178 (17) | 117 (20) | 16 (7) | 9 (10) | 27 (27) | 9 (16) | |
| Lives in a neighborhood with >30% residents in poverty, n (%) |
79 (9) | 32 (6) | 22 (30) | 13 (17) | 4 (5) | 8 (17) | <0.0005 |
Group differences evaluated using chi square test or Kruskal Wallis test
HS=high school. “Some college” includes an associate’s degree, technical degree, or an attempted but not completed 4 year degree.
Among participants for whom socioeconomic data were available (n=1167), 23% had a high school education or less, 40% had some college education or a technical degree, and 37% had a college diploma or professional degree. Overall, 9% of participants lived in a neighborhood where >30% of residents earned less than 125% of the income designated as the federal poverty level. Neighborhood poverty was highly correlated with self-reported ethnicity with 30% of African Americans and 17% of Hispanics living in poor neighborhoods (Table 1).
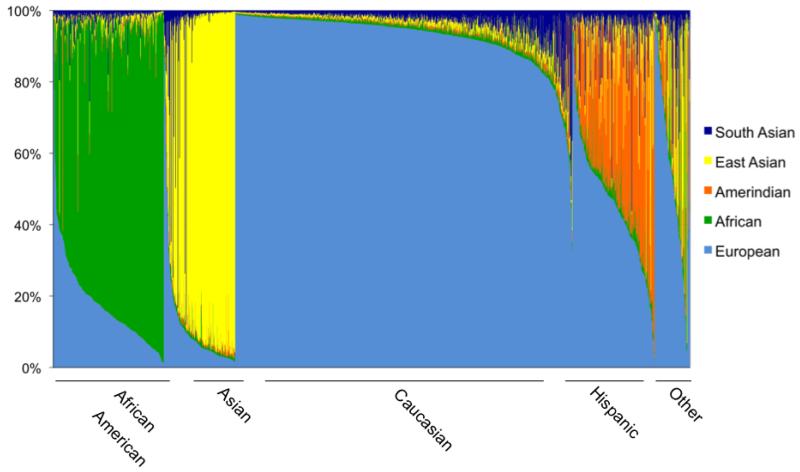
Ancestry estimation using STRUCTURE revealed that the population had on average 62.4% European, 15.8% African, 11.5% East Asian, 6.5% Amerindian, and 3.8% South Asian ancestry (Figure 2). Using PCA, we observed that the first three PCs occurred above the Eigenvalue plateau and explained 85% of the variance explained by the first ten PCs. We found a high degree of correlation between the first PC and percent European ancestry, with a Pearson correlation coefficient of 0.96 (p<0.00005). Using self-identified ethnicity, we determined that PC2 distinguished African ancestry from East Asian and Amerindian ancestries. PC3 separated East Asian and Amerindian ancestries (Supplementary Figure 1).
Figure 2.
Genetic Ancestry Estimation for 1906 Study Participants. Each vertical bar along the x-axis represents an individual participant; bars are shaded according participants’ genetic ancestry. Individuals are grouped along the x-axis according to self-identified ethnicity. SAS=South Asian, EAS=East Asian, AMI=Amerindian, AFR=African, EURA=European.
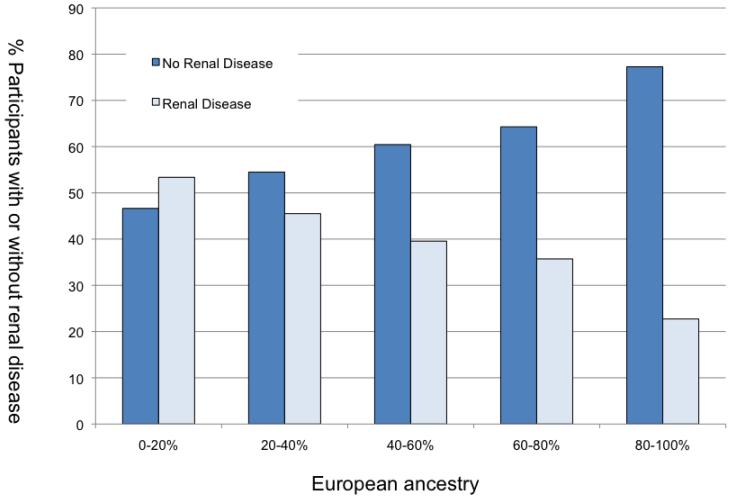
In bivariate analysis, all ancestries except South Asian ancestry showed evidence of positive or negative association with renal disease (p for Mann-Whitney U Test <0.00005). The odds ratio associated with a change from 0 to 100% European ancestry was 0.19 (95% CI 0.15-0.25). The OR associated with a 0 to 100% change in African ancestry was 2.14 (95% CI 1.55-2.94), East Asian ancestry 4.63 (95% CI 3.20-6.71) and Amerindian ancestry 3.86 (95% CI 2.00-7.47). Figure 3 demonstrates that with increasing European ancestry, the percent of participants with renal disease correspondingly decreased.
Figure 3.
Renal Disease Status by Percent European Ancestry. Participants are grouped along the x-axis according to percent European ancestry; the height of the bar represents the proportion of participants with or without renal disease.
Multivariable logistic regression analyses are shown in Table 2. We found that a 10% increase in European ancestry was associated with a 15% reduction in the odds of having renal disease after adjustment for disease duration and sex (OR 0.85, 95% CI 0.82-0.87, p=1.9 × 10−30). This model implies that subjects with 100% European ancestry have a 5-fold lower odds of developing renal disease than those with no European ancestry (OR 0.19, 95% CI 0.15-0.25). A 25% increase in European ancestry, corresponding to an additional grandparent of European origin, would result in a 34% reduction in the odds of developing renal disease (OR 0.66, 95% CI 0.62-071). A 50% increase in European ancestry would result in a 56% reduction in the odds of developing renal disease (OR 0.44, 95% CI 0.38-0.50). We also adjusted for other ancestries, represented by PC2 and PC3, and found that adding other ancestries to our model did not substantively change the relationship between European ancestry and renal disease (OR for 10% increase in European ancestry 0.84, 95% CI 0.78-0.90, p=3.1 × 10−13, table 2).
Table 2. Association between European Ancestry and Renal Disease among SLE Cases.
| Model | N | OR for 10% ↑ in Euro Ancestry |
95% CI | p-value |
|---|---|---|---|---|
| European ancestry + disease duration, sex (Model 1) |
1820* | 0.85 | 0.82-0.87 | 1.9×10−30 |
| Model 1 + other genetic ancestries | 1820 | 0.82 | 0.77-0.87 | 3.1×10−13 |
| Model 1 + educational attainment | 1129 | 0.83 | 0.80-0.86 | 5.5×10−24 |
| Model 1 + IRF5 (rs4728142), BLK (rs2736340), STAT4 (rs3024912) + other genetic ancestries |
1413 | 0.84 | 0.78-0.90 | 5.1×10−7 |
| Model 1 + IRF5 (rs4728142), BLK (rs2736340), STAT4 (rs3024912), HLA DRB1*0301, *1501, other genetic ancestries† |
753 | 0.82 | 0.69-0.98 | 0.032 |
Though the initial study population included 1906 participants, 86 were dropped from multivariable analyses (85 due to missing disease duration data, one due to missing sex data).
Only SNPs with p<0.05 in multivariable models were retained in the final model.
We compared our main model to a similar model using self-reported ethnicity rather than genetic ancestry as the primary predictor. Though self-reported European ethnicity was highly associated with a lower risk of renal disease, this model had a higher Bayesian information criterion (BIC) than the model using genetic ancestry, suggesting that genetic ancestry data provides a better model fit (data not shown).
We tested multiple measures of socioeconomic status as factors that may mediate the relationship between ancestry and renal disease. In bivariate analysis, neither income, education, nor neighborhood poverty was associated with renal disease. In multivariable models, adjusting for educational attainment did not alter the relationship between European ancestry and renal disease (OR for 10% increase in European ancestry 0.83, 95% CI 0. 0.80-0.86, p=5.5 × 10−24 Table 2), though educational attainment approached statistical significance in the model with a p-value of 0.04 (education variable not shown in Table 2). We compared the BIC from our initial model and the model that included education and found that adding data on education did not improve the model, as the BIC increased by 3. Leaving in education and omitting European ancestry strongly weakened the model with a BIC that increased by 40. Similarly, adding neighborhood poverty and individual income to our multivariable model did not substantively alter the association between European ancestry and lupus nephritis or contribute to model fit (results not shown).
Next we examined whether variation in genes associated with renal disease or specific HLA-DRB1 alleles could explain the relationship between European ancestry and renal disease. Among the 64 candidate gene SNPs examined, SNPs in IRF5, BLK, STAT4, ITGAM and HLA-DRB1*0301 and DRB1*1501 showed some evidence of association with SLE (unadjusted p<0.05, Supplementary Table 1). Though these SNPs were not definitively associated with lupus nephritis, we included them as possible explanatory covariates in our multivariate model. We thus adjusted our multivariable model for the top SNPs in these genes, retaining only those SNPs that were significant at p<0.05 in the final model. We found that the odds ratio for renal disease associated with a 10% increase in European ancestry was similar to that seen in other analyses (OR for 10% increase in European ancestry 0.84, 95% CI 0.78-0.90, p-5.1×10−7). The BIC increased by 6 when these SNPs were added to the model. A model with only these SNPs but no European ancestry information was clearly inferior, with a BIC that increased by 18. Adding HLA alleles to the model significantly reduced our sample size and widened the confidence intervals, though the point estimate for the odds ratio associated with European ancestry did not change substantially (OR=0.82, 95% CI 0.69-0.98, p=0.032, table 2). Adding HLA data to our model did lower the BIC by 2 suggesting a somewhat improved fit.
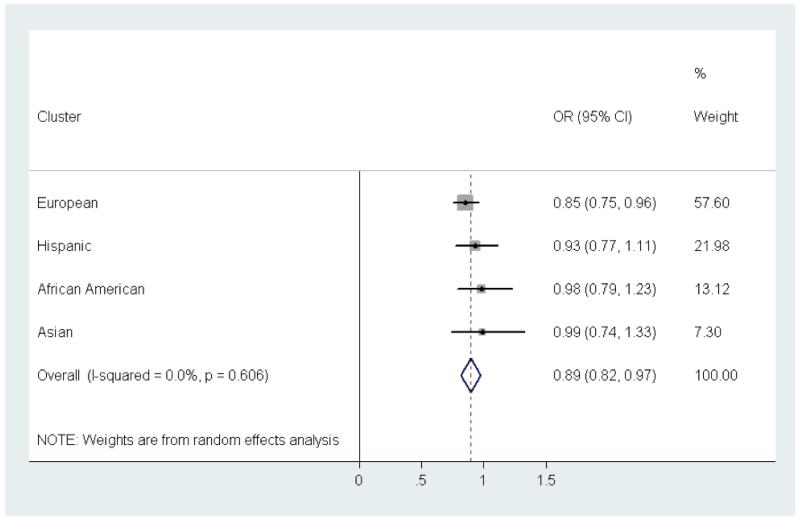
Lastly, we examined the association between European ancestry and renal disease within clusters of ancestrally similar participants defined by principal components. The cluster analysis revealed four groups, which corresponded to self-reported European American, Hispanic, African American, and East Asian ethnicities. Within each cluster, we examined the association between European ancestry and renal disease using logistic regression. Within the European American cluster, a 10% increase in European ancestry was associated with a 15% reduction in the odds of developing renal disease, similar to what we had seen in previous analyses. Though not statistically significant, the point estimates for the odds ratios within the Hispanic, African American, and East Asian clusters were all less than one. This result suggests that even within these ethnic groups, as the proportion of European ancestry increases, the risk of lupus nephritis may decrease. The test for heterogeneity of ORs across these groups was not significant (p=0.61) and supported combining these results via meta-analysis. The combined OR per 10% European ancestry was 0.89 (95% CI 0.82-0.97), p=2.5 × 10−107 (Figure 4).
Figure 4.
Meta Analysis of PC-Based Clusters. Using a K means cluster analysis of ancestry data, participants were grouped into four clusters that reflected similar ancestral genetics. The forest plot here depicts the odds ratios (ORs) for renal disease per 10% increase in European ancestry within each cluster. The overall OR was derived by combining cluster-specific ORs using a random-effects model.
Discussion
Epidemiologic studies of SLE performed as early as the 1960s have noted that SLE is not only more common, but also more severe among minority populations in the United States (23). Subsequent studies have confirmed this finding and generated vigorous debate over whether environmental or genetic factors might account for these differences. Using traditional epidemiologic methods to disentangle genetics and environment, though, has often proven difficult (24-26). Here we have demonstrated that European ancestry is an important predictor of renal disease risk in SLE, independent of socioeconomic status and other ancestries. Our approach does not assume that ethnic groups are homogeneous but rather specifically capitalizes on genetic heterogeneity within groups as well as genetic commonality among groups to more effectively dissect the relationship between ethnicity, genetics, and disease risk. Using this approach, we have demonstrated the importance of genetic ancestry in the development of lupus nephritis in a diverse cohort of SLE patients.
While European ancestry may be protective against the development of lupus nephritis, the mechanisms by which this effect operates remain more difficult to elucidate. Adjusting for candidate genes for lupus nephritis did not markedly attenuate the association between European ancestry and renal disease. There are several possible explanations for this observation. First, we examined a limited set of candidate genes. Other genes not included here may account for the observed association between ancestry and renal disease. Indeed, some recent studies have shown that variants of MYH9/APOL1 appear to be risk factors for glomerulosclerosis in African Americans and Hispanics (27, 28). A recent study examining the association between MYH9 and lupus nephritis found that particular SNPs in MYH9 were in fact associated with lupus nephritis in European populations as well as among the Gullah, a genetically isolated African American population. This finding, though, was not replicated among other African Americans, Asians, or Hispanics (29). Subsequent studies have failed to confirm that MYH9 plays a critical role in ethnic differences in the development of lupus nephritis (30). Given this uncertainty about the role of MYH9 in lupus nephritis across ethnic populations, it seems unlikely that the addition of MYH9 alone to our models would explain the effect of European ancestry.
It is also possible that genetic ancestry does not reflect single gene differences but rather captures the more subtle genetic effects of many genes with small effect sizes that vary across populations. Similarly, European ancestry may capture gene-ancestry or gene-environment interactions, genetic phenomena that could alter disease characteristics across populations but are more difficult to quantify. Ancestry thus may function as a “summary measure” of these small effects, which, though important, are more challenging to detect and describe.
Lastly, the association between European ancestry and lupus nephritis may not reflect genetic effects at all but rather socioeconomic or social factors. We have adjusted for other ancestries, and thus the cultural and environmental factors that may be associated with these ancestries, and for several measures of socioeconomic status in our analyses. Still, our results may still be confounded by non-genetic factors. The social aspects of ethnicity are complex and may not be completely captured by relatively simple measures of socioeconomic status (31). Unmeasured environmental factors could account for our results. Furthermore, for historical reasons, even among admixed populations, socioeconomic status and ancestry remain intertwined and residual confounding may persist (32, 33). Though we cannot fully exclude the possibility that unmeasured environmental factors account for our observed results, it is nonetheless interesting that SES was not associated with lupus nephritis in bivariate analysis and adjusting for multiple measures of SES had little effect on our results. These findings may suggest that SES, as measured by education, income, and neighborhood poverty, plays a much smaller role in the development of renal disease in SLE compared to ancestral genetics.
This study has some important limitations. First, we examined a small number of SNPs, which, as noted, may not adequately capture genes that do account for differences in risk of renal disease associated with ancestry. Second, we did not have candidate gene data available for all participants. We excluded the subset of participants genotyped on the 12K platform, thereby reducing our sample size. Similarly, we had HLA-DRB1 data available for only self-identified European Americans and African Americans. The limited sample size makes detecting smaller effects difficult. Our participants belonged to two distinct cohorts and most of the participants from the ABCoN cohort were African American. This demographic imbalance could have introduced a selection bias if participants were classified or treated differently in each cohort. We performed a sensitivity analysis excluding ABCoN participants and found that our main results did not change, making significant selection bias unlikely. Lastly, as noted, though we adjusted for several measures of socioeconomic status, unmeasured environmental covariates or residual confounding may account for our results.
In summary, we have identified European ancestry as a factor important in the development of renal disease in SLE. These results suggest that genetics plays a critical role in determining susceptibility to lupus nephritis. Further studies aimed at parsing ancestry-specific risk may help identify important risk alleles and ultimately may improve our understanding of both the factors leading to the development of renal disease in SLE and the patients most at risk for developing this complication.
Supplementary Material
Acknowledgments
Support: Support for this project was provided by the following NIH sponsored grants: P60 AR053308, K24 AR0217, M01 RR0079, R01 AR44804, RR 024130, AR 43727 and by a UCSF Dean’s Research Grant, a Kirkland Scholar Award, and an American College of Rheumatology Research and Education Foundation Physician Scientist Development Award.
Footnotes
Financial Disclosures: Robert Graham and Timothy Behrens are full-time employees of Genentech, Inc. No other authors have financial disclosures.
References
- 1.Alarcon GS, McGwin G, Jr., Petri M, Reveille JD, Ramsey-Goldman R, Kimberly RP. Baseline characteristics of a multiethnic lupus cohort: PROFILE. Lupus. 2002;11(2):95–101. doi: 10.1191/0961203302lu155oa. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon GS, Friedman AW, Straaton KV, Moulds JM, Lisse J, Bastian HM, et al. Systemic lupus erythematosus in three ethnic groups: III. A comparison of characteristics early in the natural history of the LUMINA cohort. LUpus in MInority populations: NAture vs. Nurture. Lupus. 1999;8(3):197–209. doi: 10.1191/096120399678847704. [DOI] [PubMed] [Google Scholar]
- 3.Seligman VA, Lum RF, Olson JL, Li H, Criswell LA. Demographic differences in the development of lupus nephritis: a retrospective analysis. Am J Med. 2002;112(9):726–9. doi: 10.1016/s0002-9343(02)01118-x. [DOI] [PubMed] [Google Scholar]
- 4.Morel L. Genetics of human lupus nephritis. Semin Nephrol. 2007;27(1):2–11. doi: 10.1016/j.semnephrol.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Ward MM. Access to care and the incidence of endstage renal disease due to systemic lupus erythematosus. J Rheumatol. 2010;37(6):1158–63. doi: 10.3899/jrheum.091199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastian HM, Roseman JM, McGwin G, Jr., Alarcon GS, Friedman AW, Fessler BJ, et al. Systemic lupus erythematosus in three ethnic groups. XII. Risk factors for lupus nephritis after diagnosis. Lupus. 2002;11(3):152–60. doi: 10.1191/0961203302lu158oa. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez M, Alarcon GS, Calvo-Alen J, Andrade R, McGwin G, Jr., Vila LM, et al. A multiethnic, multicenter cohort of patients with systemic lupus erythematosus (SLE) as a model for the study of ethnic disparities in SLE. Arthritis Rheum. 2007;57(4):576–84. doi: 10.1002/art.22672. [DOI] [PubMed] [Google Scholar]
- 8.Alarcon GS, Bastian HM, Beasley TM, Roseman JM, Tan FK, Fessler BJ, et al. Systemic lupus erythematosus in a multi-ethnic cohort (LUMINA) XXXII: [corrected] contributions of admixture and socioeconomic status to renal involvement. Lupus. 2006;15(1):26–31. doi: 10.1191/0961203306lu2260oa. [DOI] [PubMed] [Google Scholar]
- 9.Richman IB, Chung SA, Taylor KE, Kosoy R, Tian C, Ortmann WA, et al. European population substructure correlates with systemic lupus erythematosus endophenotypes in North Americans of European descent. Genes Immun. 2010;11(6):515–21. doi: 10.1038/gene.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet. 2008;4(5):e1000084. doi: 10.1371/journal.pgen.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bates JS, Lessard CJ, Leon JM, Nguyen T, Battiest LJ, Rodgers J, et al. Meta-analysis and imputation identifies a 109 kb risk haplotype spanning TNFAIP3 associated with lupus nephritis and hematologic manifestations. Genes Immun. 2009;10(5):470–7. doi: 10.1038/gene.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim-Howard X, Maiti AK, Anaya JM, Bruner GR, Brown E, Merrill JT, et al. ITGAM coding variant (rs1143679) influences the risk of renal disease, discoid rash and immunological manifestations in patients with systemic lupus erythematosus with European ancestry. Ann Rheum Dis. 2010;69(7):1329–32. doi: 10.1136/ard.2009.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 14.Trupin L, Tonner MC, Yazdany J, Julian LJ, Criswell LA, Katz PP, et al. The role of neighborhood and individual socioeconomic status in outcomes of systemic lupus erythematosus. J Rheumatol. 2008;35(9):1782–8. [PMC free article] [PubMed] [Google Scholar]
- 15.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358(9):900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 16.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 21.Barcellos LF, May SL, Ramsay PP, Quach HL, Lane JA, Nititham J, et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 2009;5(10):e1000696. doi: 10.1371/journal.pgen.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegel M, Reilly EB, Lee SL, Fuerst HT, Seelenfreund M. Epidemiology of Systemic Lupus Erythematosus: Time Trend and Racial Differences. Am J Public Health Nations Health. 1964;54:33–43. doi: 10.2105/ajph.54.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uribe AG, Alarcon GS. Ethnic disparities in patients with systemic lupus erythematosus. Curr Rheumatol Rep. 2003;5(5):364–9. doi: 10.1007/s11926-003-0022-8. [DOI] [PubMed] [Google Scholar]
- 25.Ward MM. Examining health disparities in systemic lupus erythematosus. Arthritis Rheum. 2001;44(12):2711–4. doi: 10.1002/1529-0131(200112)44:12<2711::aid-art457>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 26.Demas KL, Costenbader KH. Disparities in lupus care and outcomes. Curr Opin Rheumatol. 2009;21(2):102–9. doi: 10.1097/BOR.0b013e328323daad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–84. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behar DM, Rosset S, Tzur S, Selig S, Yudkovsky G, Bercovici S, et al. African ancestry allelic variation at the MYH9 gene contributes to increased susceptibility to non-diabetic end-stage kidney disease in Hispanic Americans. Hum Mol Genet. 2010;19(9):1816–27. doi: 10.1093/hmg/ddq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin CP, Adrianto I, Lessard CJ, Kelly JA, Kaufman KM, Guthridge JM, et al. Role of MYH9 and APOL1 in African and non-African populations with lupus nephritis. Genes Immun. 2011 doi: 10.1038/gene.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freedman BI, Edberg JC, Comeau ME, Murea M, Bowden DW, Divers J, et al. The non-muscle Myosin heavy chain 9 gene (MYH9) is not associated with lupus nephritis in African Americans. Am J Nephrol. 2010;32(1):66–72. doi: 10.1159/000314688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braveman PA, Cubbin C, Egerter S, Chideya S, Marchi KS, Metzler M, et al. Socioeconomic status in health research: one size does not fit all. JAMA. 2005;294(22):2879–88. doi: 10.1001/jama.294.22.2879. [DOI] [PubMed] [Google Scholar]
- 32.Gravlee CC, Non AL, Mulligan CJ. Genetic ancestry, social classification, and racial inequalities in blood pressure in Southeastern Puerto Rico. PLoS One. 2009;4(9):e6821. doi: 10.1371/journal.pone.0006821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Florez JC, Price AL, Campbell D, Riba L, Parra MV, Yu F, et al. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia. 2009;52(8):1528–36. doi: 10.1007/s00125-009-1412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.